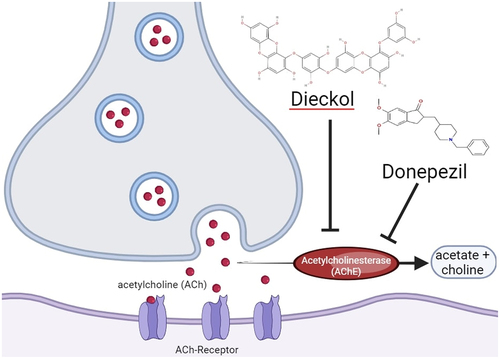
ABSTRACT
Alzheimer’s disease (AD) is a common brain disease associated with cognitive impairment and dementia. donepezil, an acetylcholinesterase (AChE) inhibitor drug as a commercial AD drug represents a non-cost-effective treatment with the toxic effects reported. As the prevalence of AD increases, the development of effective therapeutic treatments is urgently required. Laminaria digitata is a brown seaweed claimed to be able to prevent and treat neurodegenerative diseases. Therefore, this study measured and compared the binding affinity and toxicity of seven common phytoconstituents in Laminaria digitata against acetylcholinesterase (AChE) with those of donepezil using a molecular docking approach. The binding free energy values of donepezil, dieckol, eckol, fucodiphlorethol G, 7-Phloroecol, laminaran, alginic acid, and fucoidan with acetylcholinesterase (AChE) were −12.3, −13.5, −10.5, −8,7, −9.7, −8.0, −10.3, and −7.4 kcal/mol. All ligands constantly interacted with the AChE amino acid residues, namely Tyr124. Dieckol, with the strongest and most stable interaction, is classified as class IV toxicity, with an LD50 value of 866 mg/kg. It has aryl hydrocarbon receptor (AhR) and mitochondrial membrane potential (MMP) toxicity at certain doses. Theoretically, based on Lipinski’s rule, dieckol is likely to have poor absorption and permeation properties; therefore, several considerations during the drug discovery process are needed.
Introduction
Alzheimer’s disease is a common brain disease, with an estimated 60–80% of cases. It mostly affects older age groups with symptoms of comprehensive dementia, memory loss, cognitive impairment, executive dysfunction, personality, and behavioral disorders. Alzheimer’s is becoming a major public health issue (Alzheimer’s [Citation1]. Donepezil is one of the drugs used to treat Alzheimer’s symptoms. Donepezil works as an acetylcholinesterase inhibitor, which is responsible for converting acetylcholine (ACh) into an inactive form, namely acetate and choline. In Alzheimer’s patients, acetylcholine levels tend to be low; therefore, inhibiting this enzyme can increase acetylcholine levels in the brain and reduce cognitive symptoms [Citation2]. However, donepezil is a non-cost-effective treatment, and its toxic effects have been reported, including confusion, diaphoresis, and bradycardia [Citation3]. Therefore, the development of drugs from natural ingredients based on the inhibition of inhibiting acetylcholinesterase (AChE) is feasible [Citation4].
Based on the results of previous research, several plants that can interact with enzymes and receptors have potential to prevent various neurological, cardiovascular diseases, and improve memory, learning, cognitive skills due to its antioxidant properties by review and animal test [Citation5,Citation6]. In addition to targeting AChE, targeting the insulin pathway and mitochondrial function have been considered as an intervention strategy for Alzheimer’s, for example metformin as an antidiabetic drug can protect against cognitive decline and improve cognitive function [Citation7,Citation8].
Laminaria digitata is a brown seaweed claimed to be able to prevent and treat neurodegenerative diseases; therefore, it is related to Alzheimer’s disease [Citation9]. This activity is supported by phytoconstituents that are commonly present, such as eckol; fucodiphlorethol G, 7-phloroeckol, dieckol; laminaran; alginic acid; and fucoidan () [Citation10–12]. Based on previous research, several compounds that have been identified may be involved in AChE inhibition. Therefore, this study will ensure the previous research review by measuring and comparing the binding affinity of seven phytoconstituents in Laminaria digitata against acetylcholinesterase (AChE) with that of donepezil using a molecular docking approach. Phytoconstituents with the greatest affinity and most stable bond were identified. The selected phytoconstituents were subjected to in silico toxicity testing against 17 targets to detect additional possible harmful effects beyond those found in previous studies.
Table 1. The 2D and 3D structures of ligands.
Molecular docking plays a major role in structure-based molecular biology and computer-based drug design by demonstrating the ability of small molecules to interact with proteins or receptors at the nanoscale level. Molecular docking technology enables the study of protein binding sites in biochemical processes [Citation13]. This method can be used as a preliminary test for drug discovery in further research.
Material and methods
Materials
This research is an in silico molecular docking method using AutoDock Tools 1.5.7, AutoDock Vina version 1.2.0, Pymol version 2.5.4, and Ligplot version 2.2.
Crystal structure preparation and method validation
The type of receptors used was Recombinant Human Acetylcholinesterase Donepezil (PDB 4EY7) obtained from the Protein Data Bank website https://www.rcsb.org/. Protein was prepared using AutoDock Tools by separating part A of the protein. This is because Part A is the receptor-active site. Part A, which has been separated, is cleaned of unnecessary molecules such as water, donepezil, and other metabolites so that only amino acids remain; then, it is saved as PDB and PDBQT files. Donepezil was isolated from the receptor and saved as PDB and PDBQT files. The next step was redocking between donepezil and the receptor to validate the method using Autodock Tools with grid box values at center x, y, and z respectively −13.987, −43.906, and 27.108 with a spacing of 1 Å. The original ligand was docked ten times with an accuracy of 16. Subsequently, an RMSD test was carried out using Pymol software.
Ligands preparation
The ligands used in this study were seven phytoconstituents from Laminaria digitata, including Eckol; Fucodiphlorethol G; 7-Phloroecol; dieckol, Laminaran; Alginic Acid; and Fucoidan. Each structure was downloaded from the PubChem database https://pubchem.ncbi.nlm.nih.gov/ in 3D SDF form and converted to PDB form using the Chimera software. Then, each ligand was prepared (water or unnecessary compounds were removed and hydrogen atoms were added) using AutoDock Tools and saved in the PDBQT file.
Molecular docking
The seven ligands and receptor proteins that have been prepared are converted into PDBQT form, then input into the software AutoDock Vina, and each was docked according to the validation method that has been established.
Molecular docking visualization
The ligand-protein complex was formed using PyMOL software, and the results were downloaded as PDB files. The results were visualized using the Ligplot software.
Toxicity test
Dieckol is the best candidate ligand that can bind to the receptor because it has the lowest affinity among all ligands tested. Dieckol toxicity was evaluated using the Protox Web Server by accessing the Protox Web Server database at http://tox.charite.de. Then, select ‘TOX PREDICTION,’ enter ‘Dieckol’ in the column, and begin searching. Select the desired toxicity test and click on Start Toxin Prediction. Wait a few moments until the toxicity prediction results page appears, such as the LD50, average similarity, and toxicity prediction.
Results and discussion
In contemporary drug development through computational methods, Molecular Docking has emerged as a pivotal component. This methodology centers on forecasting the atomic-level interaction between a minute molecule and a protein. Such an approach facilitates the examination of the actions of diminutive compounds, including nutraceuticals, within the specific binding location of a designated protein, thus elucidating the intrinsic biochemical mechanisms governing this interaction. Initially, the conformations of the ligands are sampled based on the active site of the protein. Subsequently, these conformations undergo a ranking process guided by a scoring function. Theoretically, sampling algorithms aim to replicate experimental binding modes, with the obtained conformations then assessed and ranked according to a predetermined scoring function. The computational electrostatic properties of the ligand-receptor complex can be evaluated, scrutinized, and forecasted via the docking investigation [Citation14].
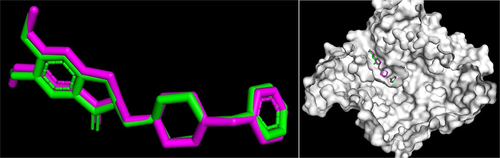
Recombinant Human Acetylcholinesterase Donepezil (PDB 4EY7) on chain A was used as the active site of the receptor. This protein was chosen because of its good resolution (2.35 Å and is related to human receptors [Citation15]. The validation process was carried out using the redocking method for donepezil and AChE protein. The validation success parameter can be determined from the RMSD value or the deviation value of the protein-ligand interactions in the crystal structure before and after docking. The results of the validation method were considered good and valid if the RMSD value was ≤ 2 Å [Citation16]. The validation result in this study was 0.331 Å (). These results showed that the structure and position of the atoms in the docking ligand were not significantly different, and the method was valid.
Figure 1. Position of the redocked ligand (Purple) with the crystallographic ligand (Green) of the donepezil crystallographic ligand.
Laminaria digitata was chosen because of its benefits in neurodegenerative diseases related to Alzheimer’s disease [Citation9]. It contains for about 5–12% polyphenolic compounds called phlorotannins [Citation12], while other studies report about 4,5% of the dry mass of Laminaria digitata [Citation17]. It also contains for about 38–61% polysaccharides [Citation17]. Compounds with similar structures can show similar binding activities, and when interacting at the same location on a protein or receptor, they are likely to have the same biological mechanism [Citation18]. From the total of 7 tested ligands, 4 of them such as Eckol, Fucodiphlorethol G, 7-Phloroecol, and Dieckol is a phlorotannin group [Citation12], while 3 of them such as Laminaran, Alginic Acid, and Fucoidan is a polysaccharide group [Citation19]. All of the tested ligands and donepezil can inhibit the same protein, namely acetylcholinesterase. The strength of the interaction between molecules can be estimated from the affinity or binding free energy values. The lower the affinity value, the stronger the interaction between the molecules. In addition, the more negative the binding free energy value, the more stable the bond between the ligand and receptor. This indicates that the binding process occurs more easily and spontaneously [Citation20].
In this study, the binding free energy values of Eckol, Fucodiphlorethol G, 7-Phloroecol, Dieckol, Laminaran, Alginic Acid, and Fucoidan against Acetylcholinesterase (AChE) were −10.5, −8,7, −9.7, −13.5, −8.0, −10.3, and −7.4 kcal/mol. The binding free energy value of Donepezil was −12.3 kcal/mol. Based on these results, dieckol has the lowest binding free energy value compared to the other tested ligands and donepezil; therefore, its binding strength and stability with the receptor are better than those of donepezil and other tested ligands. Meanwhile, Fucoidan had the highest binding free energy value, so that the strength and stability of its binding to the receptor were worse than those of the other tested ligands (). This is caused by the chemical structure of fucoidan, which has only one aromatic ring, compared to the chemical structure of dieckol, which has the most aromatic rings (). Aromatic compounds are very rigid and stable owing to the resonance or delocalization of pi electrons. This stability arises from the even distribution of electron density throughout the ring, resulting in a lower overall energy state [Citation21]. Generally, the more aromatic the rings in a compound, the more complex the interactions that occur in the molecule [Citation22].
Table 2. Binding free energy and amino acid residues of ligands.
Based on these results, dieckol was shown to bind AChE more stable than donepezil. This supports the results of previous studies on the possible involvement of dieckol acetylcholinesterase inhibition in improving memory [Citation23]. Acetylcholine (ACh) is an important neurotransmitter involved in learning, memory, and other higher-order behaviors. ACh is released by neurons as a form of communication between nerve cells, and AChE converts ACh into acetate and choline after signal transmission, to stop excessive stimulation. In neurodegenerative diseases, such as Alzheimer’s, there is a buildup of beta-amyloid plaques and neurofibrillary tangles, thereby reducing the amount of acetylcholine released by neurons, inhibiting communication between nerve cells, disrupting synapse function, and damaging nerve cells. However, AChE continues to function as usual in converting ACh after nerve signal transmission, so the ACh levels are lower. It causes inflammation, oxidative stress, and cognitive and autonomic disorders [Citation24,Citation25. Donepezil binds to acetylcholinesterase (AChE) and inhibits the conversion of ACh to increase the binding of ACh and receptors [Citation2]. Dieckol, which is able to interact more stable and stronger with AChE, also has the potential to be developed with the same mechanism of action as donepezil ().
Nevertheless, this potential inhibitory activity against AChE is supported by IC50 data from several previous research. The reference drug donepezil showed significant inhibition against AChE with IC50 0.021 µM [Citation26]. Dieckol exhibited potent AChE inhibitory activity with IC50 values of 5.69 µM, while Echol with IC50 10.03 µM [Citation27]. β-glucan/laminaran also exhibited inhibition of AChE by Ellman’s method with IC50 0.68 μg/µl [Citation28]. The IC50 value of Alginic acid against AChE was 19.33 μg/mL [Citation28]. Fucoidan had potential for inhibitory activity against AChE with IC50 values of 75.2 µM [Citation29]. Fucodiphloroethol G exhibited a significantly inhibitory activity with IC50 range of 27.80–55.12 µM [Citation30]. There is no IC50 data for 7-phloroeckol against AChE, but there is IC50 data for 7-phloroeckol against α-glucosidase 49.5 μM and α-amylase 50 µM [Citation31]. Inhibition of the α-glucosidase enzyme has the potential to reduce glucose levels in the brain, inhibit nerve cell damage, and reduce the formation of beta-amyloid plaques, which is one of the main characteristics of Alzheimer’s [Citation32].
The most significant feature of AChE is its narrow and deep structure. Interestingly, the active site of AChE is lined with conserved aromatic residues. The AChE active site is divided into several sub-sites: catalytic sites (CAS) such as Ser203, His447, and Glu202; Anionic sites (AS) such as Trp86, Tyr133, Tyr337, and Phe338; Acyl binding pockets (ABP) such as Phe295, Phe297, and Trp236; Oxyanion holes (OH) such as Gly121, Gly122, and Ala204; Peripheral anionic sites (PAS) such as Tyr72, Asp74, Tyr124, Trp286, and Tyr34 [Citation33]. The strength of the interactions between molecules can also be predicted from the number of interactions between the amino acid residues and ligands. The greater the number of interacting amino acid residues, the stronger the ligand interaction with the target protein [Citation34].
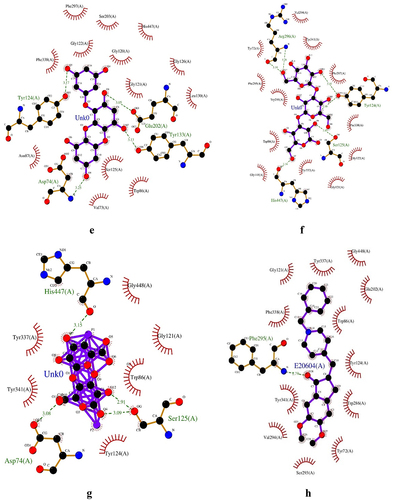
Donepezil interacted with 13 amino acid residues, including Gly121, Tyr337, Gly448, Glu202, Phe338, Trp86, Phe295, Tyr341, Val294, Ser293, Tyr72, Trp286, and Tyr124, which contain a hydrogen bond in the amino acid residue Phe295 (). This is the same as in previous studies [Citation35]. Dieckol interacted with 13 amino acid residues, including Gln291, Ser293, Ser203, Glu292, Leu289, Trp286, Tyr341, Phe,338, Tyr124, Phe295, Gly122, Phe297, and His447, which contain a hydrogen bond in the amino acid residues Gln291, Ser293, and Ser203 (). Previous studies have reported a binding free energy of Dieckol and AChE of −9.5 kcal/mol with hydrogen bonds at Asn233, Thr238, Arg296, and His405 [Citation27]. Eckol interacted with 17 amino acid residues, including Ser203, His447, Phe297, Gly120, Gly122, Phe338, Gly121, Leu130, Gly126, Tyr124, Asn87, Asp74, Ser125, Val173, Trp86, Glu202, and Tyr133, which have hydrogen bonds in the amino acid residues Tyr124, Glu202, Asp74, and Tyr133 (). Previous studies reported a binding free energy of Eckol and AChE of −8.8 kcal/mol with hydrogen bonds at Thr83, Trp86, Tyr124, and Ser125 [Citation27]. Fucoidan interacted with 10 amino acid residues, Gly121, Glu202, Ser203, Tyr124, Tyr341, His447, Tyr337, Trp86, Gly120, and Phe338, which contain a hydrogen bond in the amino acid residues Ser203, Gly121, Tyr124, and Glu202 (). Previous studies reported a binding free energy of Fucoidan and AChE of −8.22 kcal/mol with hydrogen bonds at Tyr337, Asp74, His447, and Tyr124 [Citation29].
Figure 3. Fucodiphlorethol G (a); 7-Phloroecol (b); Dieckol (c); Fucoidan (d); Eckol (e); Laminaran (f); Alginic Acid (g); Donepezil (h).
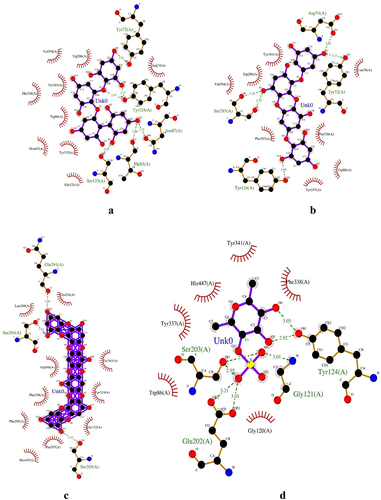
7-Phloroecol interacted with 11 amino acid residues, Asp74, Tyr341, Trp286, Val294, Ser293, Phe297, Tyr124, Tyr337, Trp86, Leu76, and Tyr72, which contain hydrogen bonds in the amino acid residues Tyr124, Tyr72, Ser293, and Asp74 (). Alginic Acid interacted with 9 amino acid residues, His447, Gly448, Gly121, Trp86, Tyr337, Tyr341, Ser125, Tyr124, and Asp74, which contain hydrogen bonds in the amino acid residues His447, Ser125, and Asp74 (). Fucodiphlorethol G interacted with 14 amino acid residues, including Tyr72, Val294, Trp286, Tyr341, Phe338, Trp86, His447, Tyr337, Gly121, Ser125, Thr83, Asn87, Tyr124, and Asp74, which contain hydrogen bonds in the amino acid residues Tyr124, Tyr72, Thr83, Asn87, and Ser125 (). Laminaran interacted with 15 amino acid residues, including Tyr72, Phe295, Trp286, Trp86, Gly448, His447, Tyr337, Gly121, Ser125, Phe338, Tyr124, Phe297, Tyr341, Val294, and Arg296, which contain hydrogen bonds in the amino acid residues Arg296, His447, Tyr124, and Ser125 (). There is no previous study on acetylcholinesterase (AChE) binding interaction with 7-Phloroecol, Alginic Acid, Fucodiphlorethol G, and Laminaran using molecular docking; therefore, this research becomes a preliminary study for further research.
Based on the summary of the results above (), all the ligands interacted with the amino acid residues Tyr124. These PAS interactions involve π–π stacking interactions and π cations, thereby increasing the strength, stability, and affinity of the enzyme substrate [Citation36]. Dieckol is a candidate compound with the best interaction with AChE and is present in approximately 23.4% of dry mass plants [Citation12]. We observed that dieckol has a molecular weight of 742.55 Da, 11 hydrogen bond donors, 30 hydrogen bond acceptors, and an octanol/water partition coefficient (logP) of 7,62. This result completes the information obtained from previous research [Citation37].
Several safety and toxicity studies of Dieckol have been discussed in several previous studies by in silico, in vitro and in vivo. In silico study of dieckol against SARS-CoV-2 main protease has binding free energy −11.4 kcal/mol with acute oral toxicity 0.544 kg/mol without carcinogenicity and AMES toxicity [Citation38]. Toxicity studies of 50 μM dieckol in zebrafish embryos showed no conspicuous adverse effects and did not generate any heartbeat rate disturbances [Citation39]. Another study that administering oral dieckol 750 mg/day to Beagle dogs for 15 days showed soft stools on days 3 and 13, but no deaths or abnormal clinical signs were observed [Citation40]. A recent randomized controlled trial of 500 mg/day of dieckol supplements administered to 24 participants for 1 week showed no serious adverse effects [Citation41].
The toxicity of chemicals can be predicted by in silico studies using a prototype web. Advancement of computational research offers significant benefits to regulatory requirements and risk assessments as well as to the pharmaceutical industry for assessing the safety profile of a drug candidate. This web server inputs two-dimensional chemical structures and reports a chemical’s probable toxicity profile with a confidence score and an overall toxicity radar chart along with the most similar compounds with known toxicity [Citation42]. Dieckol, which is found to have the best interaction with AChE, can be tested by in vitro first. This toxicity data can complement previous dieckol toxicity data.
In this study, dieckol toxicity was tested using 17 targets, including hepatotoxicity, mutagenicity, carcinogenicity, immunotoxicity, aryl hydrocarbon receptor (AhR), Androgen Receptor (AR), Androgen Receptor Ligand Binding Domain (AR-LBD), Aromatase, Estrogen Receptor Alpha (ER), Estrogen Receptor Ligand Binding Domain (ER-LBD), Peroxisome Proliferator Activated Receptor Gamma (PPAR-gamma), nuclear factor (erythroid-derived 2)-like 2/antioxidant responsive element (nrf2/ARE), heat shock factor response element (HSE), Mitochondrial Membrane Potential (MMP), phosphoprotein (tumor suppressor) p53, and ATPase family AAA domain-containing protein 5 (ATAD5). This study aims to complement dieckol toxicity data from previous studies that only used certain toxicity targets, which is useful for detecting possible additional harmful effects beyond those found in previous studies.
However, according to the results of this in silico study, dieckol did not show toxic activity against any of the tested targets, except for AhR and MMP, as shown in . Dieckol is classified as having class IV toxicity with an LD50 value of 866 mg/kg. Category IV toxicity is nontoxic and not an irritant [Citation43]. The dose-dependent toxicity of a compound can be correlated with the logarithm of its partition coefficient between octanol and water (log P). Compounds with high logP or lipophilicity tend to more easily penetrate cell membranes, including AhR and MMP membranes, so the accumulation of these compounds in the lipid bilayer membranes will disrupt the physiological function of the receptor at certain doses [Citation44].
Table 3. Safety and toxicity results of Dieckol.
Based on Lipinski’s rule of five (RO5), pharmacokinetic drugability can be predicted based on the criteria of the number of hydrogen bond donors (HBDs), hydrogen bond acceptors (HBAs), molecular weight (MW), polar surface area (PSA), and logP value. This rule states that good in vivo drug absorption and permeation is more probable when the chemical structure has two or more criteria such as MW not greater than 500, HBDs not greater than 5, HBAs not greater than 10, PSA not greater than 140 Å, and log p value not greater than 5 [Citation45]. Theoretically, compared with Lipinski’s rule, dieckol is likely to have poor absorption and permeation properties; therefore, several considerations during the drug discovery process, including parenteral drug administration, absorption enhancer addition, or nanoparticle-delivered dosage forms, are needed [Citation46].
Molecular docking is faster and more efficient for virtual screening to predict the interactions between molecules. However, further research is needed regarding conformational changes and structural fluctuations that affect the structural stability of a molecule, including the potential energy, kinetic energy, and thermodynamic parameters, to understand conformational transitions [Citation47].
Conclusion
Phytoconstituents in Laminaria digitata such as Eckol, Fucodiphlorethol G, 7-Phloroecol, Dieckol, Laminaran, Alginic Acid, and Fucoidan, were shown to bind to the active site of acetylcholinesterase (AChE) with binding free energy values of −10.5, −8,7, −9.7, −13.5, −8.0, −10.3, and −7.4 kcal/mol. The dieckol’s interaction is stronger and more stable than the other tested ligands including donepezil as a commercial Alzheimer’s drug which has a binding free energy value of −12.3 kcal/mol. Interactions between ligands and amino acid residues are also diverse. Donepezil has 13 amino acid residue interactions, dieckol has 13 amino acid residue interactions, eckol has 17 amino acid residue interactions, fucoidan has 10 amino acid residue interactions, 7-phloroecol has 11 amino acid residue interactions, alginic acid has 9 amino acid residue interactions, fucodiphlorethol G has 14 amino acid residues, and laminaran has 15 amino acid residue interactions. All ligands constantly interact with the Tyr124 amino acid residues. In addition, a dieckol toxicity test on 17 targets showed that the toxicity results were classified as class IV, with an LD50 value of 866 mg/kg. It has Aryl hydrocarbon receptor (AhR) and mitochondrial membrane potential (MMP) toxicity at certain doses. According to Lipinski’s rule, dieckol is likely to have poor absorption and permeation properties. Therefore, several considerations during the drug discovery process are needed.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data generated during the study is available at https://doi.org/10.6084/m9.figshare.25765011.
Additional information
Funding
References
- Association A. Alzheimer’s disease facts and figures. Alzheimer’s Dementia. 2023;19(4):1598–11. doi: 10.1002/alz.13016
- Chen ZR, Huang JB, Yang SL, et al. Role of cholinergic signaling in Alzheimer’s disease. Molecules. 2022;27(6):1–23. doi: 10.3390/molecules27061816
- Cacabelos R. Donepezil in Alzheimer’s disease: From conventional trials to pharmacogenetics. Neuropsychiatr Dis Treat. 2007;3(3):303–333. doi: 10.1038/nm1067
- Son M, Park C, Rampogu S, et al. Discovery of novel acetylcholinesterase inhibitors as potential candidates for the treatment of Alzheimer’s disease. Int J Mol Sci. 2019;20:1000. doi: 10.3390/ijms20041000
- Moreira J, Machado M, Dias-Teixeira M, et al. The neuroprotective effect of traditional Chinese medicinal plants—A critical review. Acta Pharm Sin B. 2023;13(8):3208–3237. doi: 10.1016/j.apsb.2023.06.009
- Wang F, Wang F, Chen T. Secondary metabolites of Galactomyces geotrichum from Laminaria japonica ameliorate cognitive deficits and brain oxidative stress in D-galactose induced Alzheimer’s disease mouse model. Nat Prod Res. 2021;35(23):5323–5328. doi: 10.1080/14786419.2020.1753738
- Austad SN, Ballinger S, Buford TW, et al. Targeting whole body metabolism and mitochondrial bioenergetics in the drug development for Alzheimer’s disease. Acta Pharm Sin B. 2022;12(2):511–531. doi: 10.1016/j.apsb.2021.06.014
- Trushina E, Trushin S, Hasan MF. Mitochondrial complex I as a therapeutic target for Alzheimer’s disease. Acta Pharm Sin B. 2022;12(2):483–495. doi: 10.1016/j.apsb.2021.11.003
- Hannan MA, Dash R, Haque MN, et al. Neuroprotective potentials of marine algae and their bioactive metabolites: Pharmacological insights and therapeutic advances. Mar Drugs. 2020;18(7):347. doi: 10.3390/md18070347
- El-Beltagi HS, Mohamed AA, Mohamed HI, et al. Phytochemical and potential properties of seaweeds and their recent applications: A review. Mar Drugs. 2022;20(6):342. doi: 10.3390/md20060342
- Pakidi CS, Suwoyo HS. Potensi dan Pemanfaatan Bahan Aktif Alga Cokelat Sargassum Sp. Octopus. 2016;5(2):488–498. doi: 10.101/p.algae.2023.05.013
- Venkatesan J, Keekan K, Anil S, et al. Phlorotannins. Elsevier. 2019:3. doi: 10.1016/B978-0-08-100596-5.22360-3
- Torres PHM, Sodero ACR, Jofily P, et al. Key topics in molecular docking for drug design. Int J Mol Sci. 2019;20(18):1–29. doi: 10.3390/ijms20184574
- Agu PC, Afiukwa CA, Orji OU, et al. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci Rep. 2023;13(1):1–18. doi: 10.1038/s41598-023-40160-2
- Singh V, Mujwar S, Singh M, et al. Computational studies to understand the neuroprotective mechanism of action basil compounds. Molecules. 2023;28(20):7005. doi: 10.3390/molecules28207005
- Khasanah NU, Wardani GA, Mardianingrum R. Synthesis and computational study of Bis-(1-(3-Chlorobenzoyl)- 3-Phenylthiourea) cobalt (III) as anticancer candidate. J Kim Sains Apl. 2023;26(7):238–248. doi: 10.14710/jksa.26.7.238-248
- Vissers AM, Caligiani A, Sforza S, et al. Phlorotannin composition of Laminaria digitata. Phytochemical Anal. 2017;28(6):487–495. doi: 10.1002/pca.2697
- Park M, Baek SJ, Park SM, et al. Comparative study of the mechanism of natural compounds with similar structures using docking and transcriptome data for improving in silico herbal medicine experimentations. Brief Bioinform. 2023;24(6). doi: 10.1093/bib/bbad344
- Zayed A, Al-Saedi DA, Mensah EO, et al. Fucoidan ’ s molecular targets: A comprehensive review of its unique and multiple targets accounting for promising bioactivities supported by in silico studies. Mar Drugs. 2024;22(1):29. doi: 10.3390/md22010029
- Mohanty M, Mohanty PS. Molecular docking in organic, inorganic, and hybrid systems: a tutorial review. Springer. 2023;154(7):683–707. doi: 10.1007/s00706-023-03076-1
- Yasuda Y. Organic chemistry aromatic compounds: understanding the fragrant world of organic chemistry. 2023;6:43–45. doi: 10.37532/jmoc.2023.6(3).43-45
- Lanzarotti E, Defelipe LA, Marti MA, et al. Aromatic clusters in protein-protein and protein-drug complexes. J Cheminform. 2020 May 8;12(1):30. doi: 10.1186/s13321-020-00437-4
- Myung CS, Shin HC, Hai YB, et al. Improvement of memory by dieckol and phlorofucofuroeckol in ethanol-treated mice: Possible involvement of the inhibition of acetylcholinesterase. Arch Pharm Res. 2005;28(6):691–698. doi: 10.1007/BF02969360
- Metaxas A, Kempf SJ. Neurofibrillary tangles in Alzheimer′s disease: elucidation of the molecular mechanism by immunohistochemistry and tau protein phospho-proteomics. Neural Regen Res. 2016;11(10):1579–1581. doi: 10.4103/1673-5374.193234
- Marucci G, Buccioni M, Ben DD, et al. Efficacy of acetylcholinesterase inhibitors in Alzheimer’s disease. Neuropharmacology. 2021;190:108352. doi: 10.1016/j.neuropharm.2020.108352
- Saǧlık BN, Osmaniye D, Çevik UA, et al. Design, synthesis, and structure–activity relationships of thiazole analogs as anticholinesterase agents for alzheimer’s disease. Molecules. 2020;25(18):4312. doi: 10.3390/molecules25184312
- Lee J, Jun M. Dual BACE1 and cholinesterase inhibitory effects of phlorotannins from ecklonia cava-an in vitro and in silico study. Mar Drugs. 2019;17(2):1–15. doi: 10.3390/md17020091
- Haider A, Inam W, Khan SA, et al. Β-glucan attenuated scopolamine induced cognitive impairment via hippocampal acetylcholinesterase inhibition in rats. Brain Res. 2016;1644:141–148. doi: 10.1016/j.brainres.2016.05.017
- Subaraja M, Krishnan DA, Hillary VE, et al. Fucoidan serves a neuroprotective effect in an Alzheimer’s disease model. Front Biosci - Elite. 2020;12(1):1–34. doi: 10.2741/E855
- Sang VT, Hung ND, Se-Kwon K. Pharmaceutical properties of marine polyphenols: An overview. Acta Pharm Sci. 2019;57(2):217–242. doi: 10.23893/1307-2080.APS.05714
- Mateos R, Pérez-Correa JR, Domínguez H. Bioactive properties of marine phenolics. Mar Drugs. 2020;18(10):1–65. doi: 10.3390/md18100501
- Chen WN, Tang KS, Yeong KY. Potential roles of α-amylase in Alzheimer’s disease: Biomarker and drug target. Curr Neuropharmacol. 2021;20(8):1554–1563. doi: 10.2174/1570159x20666211223124715
- Moghadam B, Ashouri M, Roohi H, et al. Computational evidence of new putative allosteric sites in the acetylcholinesterase receptor. J Mol Graph Model. 2021;107:107981. doi: 10.1016/j.jmgm.2021.107981
- Dhakal A, McKay C, Tanner JJ, et al. Artificial intelligence in the prediction of protein-ligand interactions: recent advances and future directions. Brief Bioinform. 2022;23(1):1–23. doi: 10.1093/bib/bbab476
- Ham Sembiring M, Nursanti O, Aisyah Rahmania T. Molecular docking and toxicity studies of nerve agents against acetylcholinesterase (AChE). J Recept Signal Transduction Res. 2023;43(5):115–122. doi: 10.1080/10799893.2023.2298899
- Girgin M, Isik S, Kantarci-Carsibasi N, et al. Proposing novel natural compounds against Alzheimer’s disease targeting acetylcholinesterase. PLOS ONE. 2023;18(4):1–23. doi: 10.1371/journal.pone.0284994
- Aatif M, Muteeb G, Alsultan A, et al. Dieckol and its derivatives as potential inhibitors of SARS-CoV-2 spike protein (UK strain: VUI 202012/01): a computational study. Mar Drugs. 2021;19(5):242. doi: 10.3390/md19050242
- Rahman MM, Islam MR, Akash S, et al. In silico investigation and potential therapeutic approaches of natural products for COVID-19: Computer-aided drug design perspective. Front Cell Infect Microbiol. 2022;12(August):1–28. doi: 10.3389/fcimb.2022.929430
- Kang MC, Cha SH, Wijesinghe WA, et al. Protective effect of marine algae phlorotannins against AAPH-induced oxidative stress in zebrafish embryo. Food Chem. 2013;138(2–3):950–955. doi: 10.1016/j.foodchem.2012.11.005
- Yang H, Yun M-S, Kim J-Y, et al. Acute oral toxicity of Phlorotannins in beagle dogs. Korean Journal of Fisheries and Aquatic Sciences. 2014;47(4):356–362. doi: 10.5657/KFAS.2014.0356
- Um MY, Kim JY, Han JK, et al. Phlorotannin supplement decreases wake after sleep onset in adults with self-reported sleep disturbance: a randomized, controlled, double-blind clinical and polysomnographic study. Phytother Res. 2018;32(4):698–704. doi:https://doi.org/10.1002/ptr.6019
- Banerjee P, Eckert AO, Schrey AK, et al. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46(W1):W257–W263. doi: 10.1093/nar/gky318
- Gadaleta D, Vuković K, Toma C, et al. SAR and QSAR modeling of a large collection of LD50 rat acute oral toxicity data. J Cheminform. 2019;11(1):1–16. doi: 10.1186/s13321-019-0383-2
- Heipieper HJ, Martínez PM. Toxicity of Hydrocarbons to Microorganisms. In: Krell T, editor. Cellular Ecophysiology of Microbe. Handbook of Hydrocarbon and Lipid Microbiology. Cham: Springer; 2016. doi: 10.1007/978-3-319-20796-4_45-1
- Benet LZ, Hosey CM, Ursu O, et al. BDDCS, the rule of 5 and drugability. Adv Drug Deliv Rev. 2016;101:89–98. doi: 10.1016/j.addr.2016.05.007
- Ezike TC, Okpala US, Onoja UL, et al. Advances in drug delivery systems, challenges and future directions. Heliyon. 2023;9(6):e17488. doi: 10.1016/j.heliyon.2023.e17488
- Manandhar S, Sankhe R, Priya K, et al. Molecular dynamics and structure-based virtual screening and identification of natural compounds as wnt signaling modulators: possible therapeutics for Alzheimer’s disease. Mol Divers. 2022;26(5):2793–2811. doi: 10.1007/s11030-022-10395-8