Abstract
Determination of stock structure is an important component of fisheries management; incorporation of molecular genetic data is an effective method for assessing differentiation among putative populations. We examined genetic variation in Southern Flounder Paralichthys lethostigma within and between the U.S. South Atlantic and Gulf of Mexico basins to improve our understanding of the scale of population structure in this wide-ranging species. Analysis of amplified fragment length polymorphism (AFLP) fingerprints and analysis of mitochondrial DNA (mtDNA) control region sequences found clear divergence between ocean basins. Based on mtDNA sequences, no genetic differentiation was detected within the U.S. South Atlantic at spatial scales that were broad (among states: North Carolina, South Carolina, Georgia, and Florida) or fine (among estuarine regions within North Carolina). Increased genetic resolution was observed with AFLP fingerprint data, and we found significant subdivision between nearly all Southern Flounder geographic populations, suggesting the presence of finer-scale genetic population structure within the U.S. South Atlantic. However, AFLP genetic cluster analysis also revealed evidence for a high degree of mixing within the Atlantic basin; patterns of variation, which included genetic similarity between South Carolina and Gulf of Mexico samples, were not aligned closely with geography. We examined the partitioning of genetic variation among groups by using analyses of molecular variance and found no evidence that North Carolina Southern Flounder, which are managed on the state level as a unit stock, are differentiated from the remainder of U.S. South Atlantic Southern Flounder. Our findings indicate only weak structure and the potential for basinwide mixing among Atlantic Southern Flounder, suggesting that cooperation among U.S. South Atlantic states will be essential for the effective assessment of stock dynamics and future management plans.
Accurate identification of stock structure is essential for effective fisheries management but remains a challenge for both managers and scientists (Begg et al. Citation1999; NCDMF Citation2005, Citation2013). Examination of genetic variation across a species' range is a commonly used approach for defining interbreeding populations, and recent advances in molecular techniques have contributed to an expanded interest in the field of fisheries genetics (Ward Citation2000; Hauser and Carvalho Citation2008; Waples et al. Citation2008). Marine species were once assumed to be panmictic, with long-distance dispersal capability and a lack of obvious barriers to gene flow in the ocean resulting in low genetic differentiation among putative populations (Ward et al. Citation1994; Waples Citation1998). However, highly polymorphic molecular markers have revealed significant genetic structure in marine fish at multiple geographic scales (Ruzzante et al. Citation1999; Knutsen et al. Citation2003; O'Reilly et al. Citation2004; McCairns and Bernatchez Citation2008), and Knutsen et al. Citation(2011) suggested that even low levels of population subdivision in marine organisms can be biologically meaningful. Maintenance of subtle genetic stock differentiation may be important in conserving local diversity and adaptive variation. Consequently, exploitation of fish populations can lead to reductions in genetic diversity, which may impact the ability of depleted stocks to recover (Hutchinson et al. Citation2003). Incorporating population genetic analyses into fisheries management and considering both historical and contemporary patterns of molecular diversity can help to promote sustainability and preserve evolutionary processes in the face of increasing anthropogenic impacts (Avise Citation1992; Crandall et al. Citation2000; Conover et al. Citation2006; Hauser and Carvalho Citation2008).
The Southern Flounder Paralichthys lethostigma is an economically important flatfish that occurs throughout estuarine and coastal ocean waters in the northern Gulf of Mexico (hereafter, Gulf) from northern Mexico to Florida (FL) and in the U.S. South Atlantic (hereafter, Atlantic) from FL to North Carolina (NC). The species supports a thriving recreational fishery across its range and is also a valuable commercial finfish resource in some areas, particularly in NC, where commercial landings averaged 1.5 million kg (3.3 million lb) annually between 1991 and 2007 (NCDMF Citation2013). In fact, the NC stock has experienced elevated commercial harvest rates since at least 1991; although a fishery management plan (NCDMF Citation2005, Citation2013) was first established in 2005, the most recent stock assessment concluded that NC Southern Flounder remain overfished and are still undergoing overfishing (Takade-Heumacher and Batsavage Citation2009).
Mature Southern Flounder inhabiting estuarine waters migrate offshore annually to spawn during the late-fall and winter months, with recruitment of larvae into estuarine nursery grounds occurring in late winter and early spring (Wenner et al. Citation1990). Adults typically return to estuarine and nearshore coastal waters after spawning (Monaghan Citation1996), but recent studies have found that Southern Flounder migration patterns are more variable, as some adults remain offshore through the late-spring and summer months (Watterson and Alexander Citation2004; Taylor et al. Citation2008). In addition, previous tagging studies indicate that most individuals display limited movement when occupying estuarine habitats and remain in close proximity to their tagging sites (Wenner et al. Citation1990; Monaghan Citation1996; Craig and Rice 2008). Based mainly on the estuarine tagging results, the current fisheries management plan considers Southern Flounder in NC to be a unit stock (NCDMF Citation2013). However, there is also evidence of extensive southward movement, as some individuals tagged in NC estuaries were found in other Atlantic states (Monaghan Citation1996; Craig and Rice 2008). Prior to estuarine recruitment, Southern Flounder remain in a pelagic larval stage for a moderately long duration (∼45 d), which increases the potential for long-range dispersal. The unknown level of adult mixing in the offshore environment combined with the long duration of the larval stage implies that the genetic population structure of Southern Flounder could be more homogeneous than currently assumed. The use of molecular methods should provide increased knowledge of Southern Flounder population structure and connectivity, thereby contributing to effective management decisions for the long-term sustainability of the species.
Previous population genetics studies of Southern Flounder have been focused on the Gulf, but there has been no comprehensive study spanning the species' range within the Atlantic. When examining allozyme variation in Southern Flounder, Blandon et al. Citation(2001) found that Gulf populations were significantly differentiated from Atlantic populations, and a break in allele frequencies on the coast of Texas between Galveston Bay and Matagorda Bay indicated the presence of population structure within the Gulf. Updated studies using mitochondrial DNA (mtDNA) and microsatellites again revealed high divergence between Gulf and Atlantic populations of Southern Flounder, but in contrast to the allozyme survey, little differentiation was detected within the Gulf (Anderson and Karel Citation2012; Anderson et al. Citation2012). Given the low levels of within-basin variability detected in previous studies, we applied a powerful genetic tool in an attempt to better characterize diversity and stock structure of Southern Flounder in the Atlantic basin.
We examined Southern Flounder populations by using the amplified fragment length polymorphism (AFLP) technique (Vos et al. Citation1995). This method rapidly generates a genetic fingerprint consisting of hundreds of highly reproducible DNA fragments that are scattered across the genome and that can be compared among individuals and populations without the need for prior characterization of an organism's genome (Bensch and Åkesson Citation2005). The large number of AFLPs produced allows for assessment of genome-wide variation when analyzing population structure, which can be more informative than targeting mtDNA markers (which are inherited as a single haploid locus). It has been claimed that the information content of an AFLP study is comparable to that obtained from a small number of highly informative microsatellite loci, which are commonly used but require substantial development time and cost (Campbell et al. Citation2003; Meudt and Clarke Citation2007; Sønstebø et al. Citation2007). Although previously underutilized in studies of animals (reviewed by Bensch and Åkesson Citation2005), AFLPs have become increasingly common in studies of aquatic populations, including commercially important fish and invertebrate species both in aquaculture (e.g., Channel Catfish Ictalurus punctatus: Mickett et al. Citation2003; Olive Flounder Paralichthys olivaceus: Liu et al. Citation2005) and in natural populations (e.g., common shrimp Crangon crangon: Weetman et al. Citation2007; Striped Mullet Mugil cephalus: Liu et al. Citation2009). For comparison with a more established method, we also sequenced a fragment of the mtDNA control region, which is a commonly used molecular marker. The control region has been identified as highly variable in teleosts (Lee et al. Citation1995) and was previously used by Anderson et al. Citation(2012) to examine Southern Flounder population structure.
In this study, our main objective was to genotype Southern Flounder individuals from across the species' range by using both AFLPs and mtDNA to address whether distinct genetic populations are present within the Atlantic, particularly whether Southern Flounder in NC represent a unique stock. As evidenced by previous studies, we expected divergence between Gulf and Atlantic populations, but we attempted to determine whether finer structure exists within ocean basins and whether this structure alters our assumptions regarding the level of mixing and warrants a change in the spatial scale of management. In addition, we assessed the viability of the AFLP method in comparison with more traditional mtDNA sequencing for determining stock structure in a widely distributed marine species.
METHODS
Southern Flounder tissue samples were collected from multiple estuarine sites across four states (NC, South Carolina [SC], Georgia [GA], and FL) spanning the current range of the species in the Atlantic (). North Carolina was subdivided into three regions (north [NCN], central [NCC], and south [NCS]) to examine genetic population structure within state waters. Samples were obtained from both fishery-independent and fishery-dependent sources in partnership with state fishery management agencies and commercial fishers within each state and were also obtained from licensed seafood dealers. Sampling was conducted from 2010 to 2013, and sampled fish ranged from age 0 to age 3 (96% were age 0–1), with an average TL of 360 mm (range = 166–622 mm). Additional tissue samples, provided by J. D. Anderson (Texas Parks and Wildlife Department), were collected from two sites () in the Gulf (Apalachicola, FL [APFL]; and San Antonio Bay, Texas [SBTX]). Sampling methods for Gulf collections are described by Anderson et al. Citation(2012). Fin clips and gill tissue were excised and preserved in either a salt-saturated dimethyl sulfoxide solution (Atlantic samples) or a 70% solution of ethanol (Gulf samples) and were stored at 4°C. Genomic DNA was extracted from fin clips by using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, California) in accordance with the manufacturer's protocols. Gill tissue was extracted only occasionally, when fin clips were damaged or not readily available.
FIGURE 1. Map of Southern Flounder sampling locations in the U.S. South Atlantic and the Gulf of Mexico. Atlantic locations are represented here on a state scale, although multiple sites were sampled within each state (NC = North Carolina; SC = South Carolina; GA = Georgia; FL = Florida). North Carolina was subdivided into three regions (separated by dashed lines; NCN = NC north [Albemarle Sound]; NCC = NC central [Neuse River–Pamlico River estuary]; NCS = NC south [New River and Cape Fear River estuaries]). Gulf of Mexico samples originated from two specific locations (black circles; APFL = Apalachicola, Florida; SBTX = San Antonio Bay, Texas). Sample size (n) for amplified fragment length polymorphisms is shown in bold; n for mitochondrial DNA control region sequences is shown in italics.
![FIGURE 1. Map of Southern Flounder sampling locations in the U.S. South Atlantic and the Gulf of Mexico. Atlantic locations are represented here on a state scale, although multiple sites were sampled within each state (NC = North Carolina; SC = South Carolina; GA = Georgia; FL = Florida). North Carolina was subdivided into three regions (separated by dashed lines; NCN = NC north [Albemarle Sound]; NCC = NC central [Neuse River–Pamlico River estuary]; NCS = NC south [New River and Cape Fear River estuaries]). Gulf of Mexico samples originated from two specific locations (black circles; APFL = Apalachicola, Florida; SBTX = San Antonio Bay, Texas). Sample size (n) for amplified fragment length polymorphisms is shown in bold; n for mitochondrial DNA control region sequences is shown in italics.](/cms/asset/fa8d337f-9794-4236-8568-d78427931087/umcf_a_1037473_f0001_b.gif)
The AFLPs were generated by using the AFLP Plant Mapping Protocol (Applied Biosystems, Inc. [ABI], Foster City, California) for regular plant genomes. We produced DNA fingerprints for 12 selective primer pairs (chosen from an earlier screen of 24 primer pairs): EcoRI-ACA and MseI-CAA; EcoRI-ACC and MseI-CAC; EcoRI-ACC and MseI-CAG; EcoRI-ACC and MseI-CAT; EcoRI-ACT and MseI-CAA; EcoRI-ACT and MseI-CAC; EcoRI-ACT and MseI-CAT; EcoRI-ACT and MseI-CTC; EcoRI-AGC and MseI-CAA; EcoRI-AGC and MseI-CTT; EcoRI-AGG and MseI-CTA; and EcoRI-AGG and MseI-CTG. Fragments were sized along with GeneScan 500 ROX size standard (Life Technologies, Carlsbad, California) on an ABI 3130XL Genetic Analyzer at the DNA Analysis Core Facility (Center for Marine Science, University of North Carolina, Wilmington). To test for reproducibility of AFLP fingerprints, one individual from each sampling location was subjected to repeat analysis across all 12 selective primer pairs beginning from genomic DNA. Fragments between 75–500 bp were scored automatically for presence/absence with a default peak calling threshold of 50 relative fluorescence units (rfu) by using ABI GeneMapper version 4.0. Peak heights were normalized across all samples for each primer pair, with only polymorphic loci considered. To further reduce noise and genotyping error, the individual loci in the AFLP profiles were filtered to a locus selection threshold of 200 rfu and a phenotype calling threshold of 20% following Whitlock et al. Citation(2008). In this AFLP profile optimization procedure, loci with an average peak height below 200 rfu are eliminated from analysis, and presence/absence scoring is based on a threshold of 20% of the average peak height for each locus. The complete AFLP profiles (i.e., prior to optimization) were also considered in this analysis but provided similar results and are not reported here.
A fragment of the mtDNA control region was amplified by using the primers L-PROF (5′-AACTCTCACCCCTAGCTCCCAAAG-3′; Meyer et al. Citation1994) and PL526H (5′-AAAAGAGAACCCCTTACCCG-3′), the latter of which is a Southern Flounder-specific primer designed by one of us (M. A. McCartney, unpublished). The PCR was conducted as follows: an initial denaturation step of 94°C for 1 min, followed by 35 amplification cycles (94°C for 1 min; 58°C for 30 s; and 72°C for 2 min) and a final extension of 72°C for 5 min. Each 25-µL reaction volume contained 1.25 units of MyTaq DNA Polymerase (Bioline, London, UK), 1 × MyTaq Red Reaction Buffer (Bioline), 0.5 µM of each primer, and 1 µL of genomic DNA (5–900 ng). The PCR products were purified by using a StratPrep PCR Purification Kit (Agilent Technologies, Santa Clara, California) and were cycle sequenced in the forward direction (L-PROF) with ABI BigDye Terminator version 3.1. Sequencing was conducted on the same equipment as described for AFLPs. Sequences were edited, trimmed, and aligned manually in Sequencher version 4.10.1 (Gene Codes Corporation, Ann Arbor, Michigan). Control region sequences were deposited in GenBank under accession numbers KP331548–KP331677.
STRUCTURE version 2.3.4 (Pritchard et al. Citation2000) was used to assign individual Southern Flounder to an inferred number of genetically distinct clusters (K) present in the AFLP data set. Because AFLPs are dominant markers that are scored as the presence or absence of peaks, there is no distinction between homozygous and heterozygous states. To account for this ambiguity, we implemented the model by Falush et al. Citation(2007) for analyzing dominant markers in STRUCTURE. A recessive AFLP profile (consisting of all peak absences) was input into the data set, and the model evaluated all possible genotypes (i.e., for a peak presence phenotype, the genotypes presence/presence and presence/absence are both considered). Based on previous studies that demonstrated a lack of structure among Southern Flounder populations within ocean basins (Anderson and Karel Citation2012; Anderson et al. Citation2012), we assumed that populations were admixed and that allele frequencies across populations were correlated. We did not provide the geographic origin of samples a priori. Five replicates for each K-value from 1 to 8 were tested, with 10,000 burn-in iterations followed by 50,000 Markov-chain Monte Carlo iterations. STRUCTURE HARVESTER (Earl and vonHoldt Citation2012) was used to implement the Evanno et al. Citation(2005) method for identifying the most likely number of clusters (i.e., K) in a data set. All replicates from each K-value identified by this method were then combined using the “FullSearch” algorithm in CLUMPP version 1.1.2 (Jakobsson and Rosenberg Citation2007). DISTRUCT version 1.1 (Rosenberg Citation2004) was used to convert CLUMPP outputs into bar plots that showed each individual's probability of assignment to the genetic clusters.
We used Arlequin version 3.5.1.2 (Excoffier and Lischer Citation2010) to compute the genetic differentiation index FST between all pairs of geographic populations for both AFLP and mtDNA data sets. We applied a Bonferroni adjustment to correct for multiple FST comparisons by using software from Lesack and Naugler Citation(2011). Arlequin was also used to conduct analyses of molecular variance (AMOVAs; Excoffier et al. Citation1992). An AMOVA examines the partitioning of genetic differentiation within and among geographical subpopulations. We used this framework to test two different regional groupings for both AFLPs and mtDNA: (1) ocean-basin-scale structure (Gulf versus Atlantic) and (2) structure within the Atlantic (NCN, NCC, and NCS versus SC, GA, and FL). To further investigate the patterns observed from our STRUCTURE results, we also considered a third grouping for AMOVA: subdivision within NC (NCN and NCC versus NCS, SC, GA, and FL). For all analyses conducted in Arlequin, AFLPs were coded as haplotypic data. Arlequin does not account for dominant marker systems, so the calculated fixation indices for AFLP data are based on peak phenotype (presence/absence) frequencies rather than allele frequencies. Despite this, Arlequin is often used for analysis of AFLPs (Svensson et al. Citation2004; Bensch and Åkesson Citation2005; Timmermans et al. Citation2005; Parchman et al. Citation2006; Toews and Irwin Citation2008); the degree of differentiation and their significance values are meaningful, but direct comparisons with FST values from co-dominant data sets should be avoided.
Mitochondrial DNA control region phylogeny in Southern Flounder was inferred with NETWORK version 4.6.1.2 (Fluxus Engineering; www.fluxus-engineering.com) by using the median joining method (Bandelt et al. Citation1999) to generate a haplotype network. Network branching patterns were postprocessed with maximum parsimony (Polzin and Daneshmand Citation2003). The mtDNA control region nucleotide composition and pairwise differences were calculated using MEGA version 6 (Tamura et al. Citation2013). Pairwise differences for the AFLP data set were also determined with MEGA. DnaSP version 5.10 (Librado and Rozas Citation2009) was used to assign mtDNA control region haplotypes and to calculate molecular diversity indices and Tajima's D-statistic (Tajima Citation1989). Sites with gaps were not considered for diversity calculations but were included in all other analyses.
RESULTS
When scored for all 201 samples across the 12 primer pair combinations, 671 polymorphic AFLP loci were identified. In the eight individuals for which the AFLP procedure was repeated, the AFLP profiles were 98.8% reproducible across all loci. Using the method of Evanno et al. Citation(2005) to interpret the AFLP results from STRUCTURE, we identified a strong peak in ΔK at a K-value of 2 and a smaller but distinct secondary peak in ΔK at a K-value of 5. At a K of 2, we observed that each geographic population showed strong membership to one of two distinct genetic clusters representing either the Gulf or the Atlantic (). In addition, the Atlantic samples from NCS and southward, particularly SC, displayed partial assignment to the genetic cluster dominating the Gulf locales. At a K of 5, the assignments remained similar to those produced at a K-value of 2, but SC ancestry was instead shared specifically with the western Gulf, and NCS samples displayed some shared assignment with the major genetic cluster found in the Gulf.
FIGURE 2. Population structure for Southern Flounder as estimated with amplified fragment length polymorphisms (AFLPs). Each individual is represented as a vertical line along the x-axis, and individuals are grouped into geographic populations. Each color represents a different inferred genetic cluster, and the probability of assignment to each of the K genetic clusters is indicated by the proportion of color along the y-axis: (A) K-value of 2 and (B) K-value of 5. Results for the AFLP data set after removing the primer pair EcoRI-AGC–MseI-CTT are also presented: (C) K-value of 2 and (D) K-value of 4. Location codes are defined in .
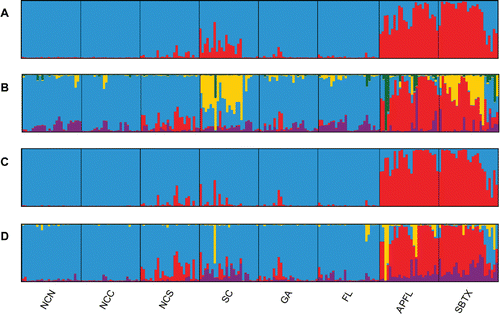
Because Southern Flounder from SC unexpectedly showed common ancestry with those sampled in the western Gulf (SBTX), we examined population pairwise distances for individual AFLP primer pairs in an attempt to identify the source of these similarities. For each primer pair, we compared (1) the average pairwise distance among geographic population pairs that included either SC or SBTX to (2) the average pairwise distance among all other geographic population pairs (i.e., those without SC or SBTX). The percent difference between these two metrics was used to assess the unusual levels of genetic differentiation in SC and the western Gulf. The average percent difference was 6.09% (SD = 5.93) across 11 of the 12 primer pairs. The remaining primer pair (EcoRI-AGC–MseI-CTT) displayed higher levels of differentiation, with a percent difference of 56.77%, and was likely responsible for the similarities observed between SC and SBTX samples. Subsequently, we conducted a second STRUCTURE analysis of the AFLP data set by using the same methods as previously described, except that we excluded the 38 loci from the primer pair EcoRI-AGC–MseI-CTT. A strong peak in ΔK was identified at a K of 2, and a smaller but distinct peak was observed at a K of 4. At the K-value of 2, the pronounced similarity between SC and SBTX was no longer present, but at the K of 4, a shared partial assignment among SC samples and Gulf samples was again detected (). Overall patterns of genetic structure were similar to those observed prior to removal of the EcoRI-AGC–MseI-CTT primer pair from the AFLP data set.
Comparisons of FST for Southern Flounder AFLPs showed significant differentiation between all but four pairs of geographic populations (FST = 0.020–0.133, P < 0.01; ). The four population pairs that were not significantly differentiated after Bonferroni adjustment were all within-Atlantic comparisons (NCN versus GA; NCN versus FL; NCC versus FL; and GA versus FL). In general, FST values for Atlantic versus Gulf population pairs were larger than within-basin comparisons, although FST values for SC versus NCC and for SC versus NCS were comparable to between-basin values. When comparing Southern Flounder from the Gulf and Atlantic basins, AMOVA identified significant differences at all levels of hierarchical population structure (). However, within-population variation accounted for 89.9% of the differences (FST = 0.101, P < 0.0001). When partitioning differentiation in the Atlantic alone (NCN, NCC, and NCS versus SC, GA, and FL), 96.6% of the variation was due to within-population differences (FST = 0.034, P < 0.0001; ). There was no differentiation between these two Atlantic regions. There was also no differentiation between regions when considering NCN and NCC as a separate group relative to NCS, SC, GA, and FL.
TABLE 1. Population pairwise FST comparisons based on amplified fragment length polymorphism profiles (above diagonal) and mitochondrial DNA control region haplotypes (below diagonal) from Southern Flounder sampled in the U.S. South Atlantic (NCN = North Carolina north; NCC = North Carolina central; NCS = North Carolina south; SC = South Carolina; GA = Georgia; FL = Florida) and in the Gulf of Mexico (APFL = Apalachicola, Florida; SBTX = San Antonio Bay, Texas). Values without superscript letters were not significant.
TABLE 2. Results from analyses of molecular variance based on amplified fragment length polymorphisms in Southern Flounder. Populations are grouped at two different geographic scales: (1) ocean basin (Gulf of Mexico versus U.S. South Atlantic) and (2) U.S. South Atlantic (North Carolina versus South Carolina, Georgia, and Florida; *P < 0.05; **P < 0.0001).
Across 260 samples, the final mtDNA control region alignment for Southern Flounder was 480 bp, with 88 segregating sites and a total of 130 unique haplotypes. This included a single-base-pair insertion/deletion that was present in two haplotypes, each composed of one individual from different sites. Excluding sites with gaps, overall haplotype diversity was 0.977 (SD = 0.0036) and nucleotide diversity was 0.011 (SD = 0.0003), with an average of 5.06 nucleotide changes between individuals (). Tajima's D-statistic was estimated to test for neutrality of nucleotide substitutions; the values were significantly negative for both ocean basins (Atlantic: D = −2.08, P < 0.05; Gulf: D = −1.97, P < 0.05), which may be indicative of Southern Flounder population expansion (Fu Citation1997).
TABLE 3. Molecular diversity measures for populations of Southern Flounder sequenced at the mitochondrial DNA control region (excluding sites with gaps). Number of samples (n), number of polymorphic sites excluding gaps (Np), number of haplotypes (Nh), haplotype diversity (Hd), and nucleotide diversity (π) are given for each site. Location codes are defined in
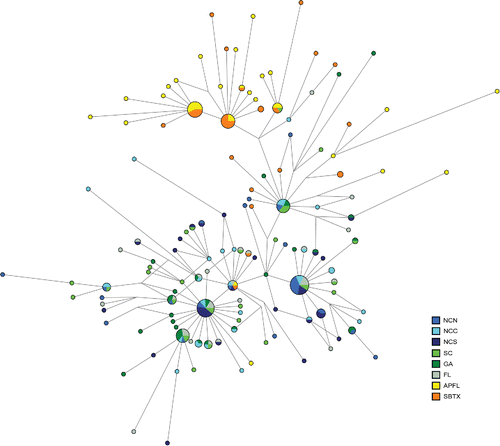
Twenty-nine mtDNA control region haplotypes were common among individual Southern Flounder, and three of the haplotypes were shared between the Gulf and Atlantic. The majority of the haplotypes were singletons, most of which differed from common haplotypes by only one or two nucleotide substitutions. The control region haplotype network showed a pattern of divergence between Southern Flounder in the two ocean basins, although separation was not complete (). The majority of Gulf haplotypes were separated from Atlantic haplotypes by at least three nucleotide changes, but there were Atlantic haplotypes present in the Gulf cluster and vice versa. There was no clear pattern of finer-scale structure within the Atlantic cluster based on the haplotype network.
FIGURE 3. Median joining network of mitochondrial DNA control region haplotypes identified in Southern Flounder. Circles represent haplotypes; colored wedges indicate geographic populations. Circles and wedges are proportional to the sample size, and the length of the connection is proportional to the number of nucleotide changes between haplotypes. Location codes are defined in .
Pairwise population comparisons using the mtDNA control region showed strong differentiation (FST = 0.37–0.45, P < 0.0001) between all pairs of populations compared across basins (). Three pairs of populations within the Atlantic (GA and NCS; GA and NCN; and SC and NCN) were also significantly differentiated, although FST values were much smaller (FST = 0.037–0.052, P < 0.05). None of these within-Atlantic comparisons remained significant after a Bonferroni correction was applied, and no other pairs of populations within the Atlantic or Gulf were found to be significantly differentiated with any test. When comparing the Gulf and Atlantic, the AMOVA for the mtDNA control region showed that the majority of variation was explained by differences within populations (59.3%; FST = 0.407, P < 0.0001), although a large portion of the differentiation was due to differences between ocean basins (40.2%; FCT [genetic differences among groups] = 0.402, P < 0.05; ). Within the Atlantic, almost all of the variation was explained by differences within populations (98.2%; FST = 0.018, P > 0.05), and only a small portion of the variation was explained by differences between NC and the other Atlantic populations (1.8%; FCT = 0.018, P > 0.05), but neither of these differences was significant ().
TABLE 4. Results from analyses of molecular variance based on mitochondrial DNA control region sequences in Southern Flounder. Populations are grouped at two different geographic scales: (1) ocean basin (Gulf of Mexico versus U.S. South Atlantic) and (2) U.S. South Atlantic (North Carolina versus South Carolina, Georgia, and Florida; *P < 0.05; **P < 0.0001).
DISCUSSION
We observed divergence between Gulf and Atlantic Southern Flounder populations based on both mtDNA control region sequences and AFLP fingerprints. All population pairwise FST and AMOVA FCT values were significant when comparing between ocean basins, regardless of the molecular marker used (). The same result was observed with AFLP population assignments () and in the mtDNA haplotype network (). These findings are in agreement with previous Southern Flounder studies that examined allozymes (Blandon et al. Citation2001), mtDNA (Anderson et al. Citation2012), and microsatellites (Anderson and Karel Citation2012), and they correspond with a well-known biogeographic break in marine fauna between the Gulf and Atlantic (Avise Citation1992). Southern Flounder are absent from the southern tip of Florida (Ginsburg Citation1952; Gilbert Citation1986), so a transition in genetic composition between ocean basins appears to correspond with the habitat discontinuity that is responsible for the disjunct distribution of Southern Flounder across south Florida.
We predicted that both markers would perform well for evaluating deep divergence, but we also wanted to assess how AFLPs and mtDNA would compare when examining subtle structure, as we expected to see in the Atlantic Southern Flounder populations. Although mtDNA markers are commonly used in population genetic studies because of their high mutation rates and purportedly neutral evolution, the mitochondrial genome is also haploid (maternally inherited, effectively as a single locus) and is very small in comparison with the nuclear genome. Thus, studies that rely solely on mtDNA markers may draw incomplete conclusions when attempting to make inferences about organisms and populations as a whole (reviewed by Ballard and Whitlock Citation2004). Amplified fragment length polymorphism loci are scattered across the genome and may better reflect demographic, historical, and selective events for which influences on diversity patterns can vary across loci, so we expected to obtain increased genetic resolution with AFLP analysis.
The AFLP and mtDNA results were generally in agreement when assessing Southern Flounder differentiation between the two ocean basins, but the results began to diverge when we examined structure within ocean basins. The magnitude of differentiation for pairwise comparisons between ocean basins was much higher with mtDNA (FST = 0.358–0.456) than with AFLPs (FST = 0.065–0.133), although both were highly significant. The opposite was seen for population comparisons within the Atlantic, as mtDNA differentiation values (FST = −0.010 to 0.052) were nonsignificant and lower than AFLP values (FST = 0.006–0.072), which showed significant differentiation for nearly all within-Atlantic comparisons. If we were to interpret the degree of population structure as proposed by Wright Citation(1978), genetic differentiation among Southern Flounder populations when comparing ocean basins would be characterized as “very great” (mtDNA) or “moderate” (AFLPs), whereas comparisons within the Atlantic basin showed “little” to “moderate” genetic differentiation based on AFLPs.
The levels of genetic differentiation detected in this study were consistent with previously reported FST values for Southern Flounder mtDNA (Anderson et al. Citation2012), but we observed markedly higher differentiation using AFLPs than was previously found with microsatellites (within-basin FST = −0.002 to 0.007; between-basin FST = 0.025–0.046; Anderson and Karel Citation2012). Similar discrepancies between markers have also been observed in recent studies of other species, for which differentiation was both higher (Mock et al. Citation2002; Whitehead et al. Citation2003; Gruenthal et al. Citation2007) and lower (Maguire et al. Citation2002) when comparing AFLPs to microsatellites. For analyses of highly variable multi-allelic loci, as in the Anderson et al. Citation(2012) study where there were up to 53 alleles per microsatellite locus, very high levels of within-population heterozygosity can result in reduced magnitudes of differentiation between populations (Hedrick Citation1999). Different molecular markers are subject to different evolutionary processes, which can affect levels of diversity within and between populations (reviewed by Zink and Barrowclough Citation2008; Toews and Brelsford Citation2012). Therefore, the lack of correspondence in the magnitude of differentiation estimated with the different markers used to assess Southern Flounder genetic structure thus far is not surprising.
In this study, nearly all Southern Flounder geographic populations within the Atlantic were significantly differentiated (after Bonferroni correction for multiple comparisons) when using the AFLP data set (), thus contradicting the mtDNA results. The power to detect differentiation increases with the number of loci, and even very small values of FST can become statistically significant when enough markers are used (Ryman et al. Citation2006). We compared a single mtDNA marker to an AFLP profile comprising 671 loci, so we expected increased resolution. However, the levels of within-Atlantic AFLP differentiation we detected are not considered low for marine populations. Even very weak but significant levels of genetic differentiation can be biologically relevant; for example, Knutsen et al. Citation(2011) used empirical mark–recapture data to confirm the existence of distinct, localized populations of coastal Atlantic Cod Gadus morhua that produced low levels of genetic differentiation. Based on AFLP differentiation, genetic structure appears to be present among geographic populations of Southern Flounder in the Atlantic, although this does not necessarily mean that each locale represents a discrete stock.
Waples et al. Citation(2008) cautioned that statistical significance is not always correlated with biological significance and that the life history and biology of an organism must be taken into consideration when determining stocks. Thus far, we have no evidence that Southern Flounder from different locales within the Atlantic are biologically distinct. Based on otolith morphometric analysis, Midway et al. Citation(2014) also detected only weak evidence for differentiation among Southern Flounder populations within the Atlantic, including within NC. These results are consistent with our findings. Despite the possibility that many AFLP loci should allow for greater power in the detection of population subdivision, AMOVA did not find significant genetic structure when comparing NC (or portions of NC) to the remaining Atlantic locales. The same lack of NC regional structure was found with AMOVA based on mtDNA. These results do not support the consideration of NC Southern Flounder as a distinct stock within the Atlantic.
In addition to evaluating patterns of differentiation, we also assessed the genetic composition of each Southern Flounder population. When sufficient numbers of loci are used, AFLPs are highly accurate for population assignment tests and can be particularly informative for systems with weak genetic structure (Campbell et al. Citation2003). We saw similar patterns in individual assignments to inferred genetic clusters (i.e., K) across the most likely values of K for the AFLP profiles (). Although we examined patterns of variation at K-values of 2 and 5, the major genetic patterns remained the same. The two summary plots both showed a strong genetic distinction between Gulf and Atlantic Southern Flounder. Within NC, the NCN and NCC samples appeared to be homogeneous, whereas NCS displayed a small difference in assignment probabilities, suggesting that a mild barrier to gene flow may exist between these regions. The most unexpected observation was the shared clustering of SC samples—obtained near the center of the Atlantic populations—with the Gulf samples, particularly those from SBTX.
Overall, the AFLP genetic cluster analyses suggested high gene flow within the Atlantic, with the possible exception of SC. Of the three mtDNA haplotypes shared between ocean basins, one consisted of individuals from SC and both Gulf locales, although this was a low-frequency haplotype (). In addition, the SC samples were differentiated from one of the NC geographic populations in the mtDNA FST analysis, but this subdivision was no longer significant after Bonferroni adjustment. In contrast, SC was always highly significantly differentiated from both Gulf locales in pairwise FST comparisons, whether based on mtDNA or AFLPs. With these varying results, the potential distinction of SC genotypes from other Atlantic samples remains inconclusive.
By examining pairwise differences in AFLP profiles between geographic populations, we were able to identify a single primer pair (EcoRI-AGC–MseI-CTT) as the likely cause for the assignment of SBTX and SC individuals to the same genetic cluster. This was confirmed by a genetic cluster analysis that excluded loci from the primer pair in question, although the similarity between SC and Gulf samples was still present to a smaller degree when the number of assumed genetic clusters was increased (). Because one primer pair appears to be responsible for this grouping, it is possible that the pattern may be an artifact of the AFLP procedure. Errors in peak scoring are a common source of AFLP genotyping error (Bonin et al. Citation2004), so we used an established method to reduce scoring subjectivity and error in our AFLP data by setting peak height thresholds for inclusion of loci and phenotype calling (Whitlock et al. Citation2008). This resulted in a conservative data set that performed well in our tests for reproducibility (98.8%).
Another concern in AFLP studies is fragment size homoplasy, which can occur as a result of the anonymous nature of AFLP loci. Equal-length DNA fragments are scored as a presence on the same locus, regardless of whether the fragments are homologous in genomic origin. Amplified fragment length polymorphism homoplasy can result in the underestimation of differentiation between populations and may provide one potential explanation for the unforeseen similarity we observed between SC and SBTX Southern Flounder. Homoplasy bias increases with the number of loci scored per primer pair and with smaller fragment sizes (<125 bp; Vekemans et al. Citation2002; Caballero et al. Citation2008). We reduced the numbers of AFLP loci analyzed per primer pair by applying peak height thresholds as previously described (only 38 of 139 loci initially detected were analyzed for EcoRI-AGC–MseI-CTT), but small fragment sizes were not excluded in this study. The potential influence of AFLP homoplasy on genetic diversity estimates must be considered, but experimental quantification of the degree of homoplasy is not within the scope of most AFLP studies (Meudt and Clarke Citation2007).
Alternatively, it may be possible for the geographically disjunct individuals from SC and the Gulf to have similar molecular profiles based on shared adaptive traits. Like other genome-scale molecular data sets, AFLP loci are anonymous and consist mostly of neutral loci, but they often include loci that are linked to genes under selection (Luikart et al. Citation2003). To investigate any patterns of distribution that may be related to adaptive evolution, future studies would benefit from a genome scan, which detects AFLP loci that show abnormally high among-population differentiation; such outliers are a signature of selection (Beaumont and Balding Citation2004).
The results of this study confirmed a clear divergence between Gulf and Atlantic Southern Flounder. There was also evidence of genetic structure among geographic populations of Southern Flounder at a finer spatial scale, which was only revealed upon analysis of AFLPs. We found intriguing patterns of differentiation and genetic clustering that did not correspond with obvious geographic patterns. However, there was no differentiation between samples from NC and those from the other Atlantic states when examining hierarchical population structure. Southern Flounder in NC are currently managed as a unit stock. The present molecular analysis suggests that a single-stock designation within NC should be adequate, as there is no clear evidence for additional subdivisions at a finer spatial scale (i.e., between regions within the state). However, the potential for high levels of mixing throughout the Atlantic basin, as evidenced by the presence of only weak genetic structure within the basin, suggests that interstate cooperation will be necessary to achieve a comprehensive population assessment of Southern Flounder at an appropriate spatial scale. To sustain fishery yields and meet conservation objectives for Southern Flounder, the spatial scale of stock assessment will need to be more closely aligned with the spatial scale of important ecological processes that determine stock dynamics. Our present findings suggest that these processes likely operate over broad spatial scales and that future management of Southern Flounder could be improved through a synthesis of biological information encompassing the species' range.
ACKNOWLEDGMENTS
This research was funded by the North Carolina Division of Marine Fisheries (NCDMF) through the Marine Resources Fund. Several individuals provided help with tissue sample collection and genetic data. Southern Flounder from NC were collected by L. Paramore, K. West, J. Rock, M. Seward, and C. Collier as part of NCDMF's fishery-independent sampling program. The SC fish were provided by B. Roumillat and S. Arnott (South Carolina Department of Natural Resources); GA fish were provided by J. Page and E. Robillard (Georgia Department of Natural Resources); and FL fish were provided by R. Brodie (Florida Fish and Wildlife Conservation Commission). S. Midway collected additional fish from Florida and was essential in providing the organizational framework for this study. J. Anderson (Texas Parks and Wildlife Department) graciously provided fish tissue samples and genomic DNA from the Gulf. L. Jarvis helped with initial development of molecular methods and generated a preliminary data set. We also thank three anonymous reviewers for providing comments that helped us to improve the presentation of our findings. The interpretation of the data and the conclusions expressed in this paper are those of the authors and do not necessarily represent the views of the NCDMF.
REFERENCES
- Anderson, J. D., and W. J. Karel. 2012. Population genetics of Southern Flounder with implications for management. North American Journal of Fisheries Management 32:656–663.
- Anderson, J. D., W. J. Karel, and A. C. S. Mione. 2012. Population structure and evolutionary history of Southern Flounder in the Gulf of Mexico and western Atlantic Ocean. Transactions of the American Fisheries Society 141:46–55.
- Avise, J. C. 1992. Molecular population structure and the biogeographic history of a regional fauna: a case history with lessons for conservation biology. Oikos 63:62–76.
- Ballard, J. W. O., and M. C. Whitlock. 2004. The incomplete natural history of mitochondria. Molecular Ecology 13:729–744.
- Bandelt, H. J., P. Forster, and A. Röhl. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16:37–48.
- Beaumont, M. A., and D. J. Balding. 2004. Identifying adaptive genetic divergence among populations from genome scans. Molecular Ecology 13:969–980.
- Begg, G. A., K. D. Friedland, and J. B. Pearce. 1999. Stock identification and its role in stock assessment and fisheries management: an overview. Fisheries Research 43:1–8.
- Bensch, S., and M. Åkesson. 2005. Ten years of AFLP in ecology and evolution: why so few animals? Molecular Ecology 14:2899–2914.
- Blandon, I. R., R. Ward, T. L. King, W. J. Karel, and J. P. Monaghan Jr. 2001. Preliminary genetic population structure of Southern Flounder, Paralichthys lethostigma, along the Atlantic coast and Gulf of Mexico. U.S. National Marine Fisheries Service Fishery Bulletin 99:671–678.
- Bonin, A., E. Bellemain, P. Bronken Eidesen, F. Pompanon, C. Brochmann, and P. Taberlet. 2004. How to track and assess genotyping errors in population genetics studies. Molecular Ecology 13:3261–3273.
- Caballero, A., H. Quesada, and E. Rolán-Alvarez. 2008. Impact of amplified fragment length polymorphism size homoplasy on the estimation of population genetic diversity and the detection of selective loci. Genetics 179:539–554.
- Campbell, D., P. Duchesne, and L. Bernatchez. 2003. AFLP utility for population assignment studies: analytical investigation and empirical comparison with microsatellites. Molecular Ecology 12:1979–1991.
- Conover, D. O., L. M. Clarke, S. B. Munch, and G. N. Wagner. 2006. Spatial and temporal scales of adaptive divergence in marine fishes and the implications for conservation. Journal of Fish Biology 69:21–47.
- Craig, J. K., and J. A. Rice. 2008. Estuarine residency, movements, and exploitation of Southern Flounder (Paralichthys lethostigma) in North Carolina. North Carolina Sea Grant, Final Report for Fishery Resource Grant 05-FEG-15, Raleigh.
- Crandall, K. A., O. R. P. Bininda-Emonds, G. M. Mace, and R. K. Wayne. 2000. Considering evolutionary processes in conservation biology. Trends in Ecology and Evolution 15:290–295.
- Earl, D., and B. vonHoldt. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4:359–361.
- Evanno, G., S. Regnaut, and J. Goudet. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14:2611–2620.
- Excoffier, L., and H. E. L. Lischer. 2010. Arlequin suite version 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10:564–567.
- Excoffier, L., P. E. Smouse, and J. M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491.
- Falush, D., M. Stephens, and J. K. Pritchard. 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes 7:574–578.
- Fu, Y.-X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925.
- Gilbert, C. R. 1986. Species profiles: life histories and environmental requirements of coastal fishes and invertebrates (south Florida)–Southern, Gulf, and Summer Flounders. U.S. Army Corps of Engineers, Coastal Ecology Group, Technical Report 82 (11.54), Vicksburg, Mississippi.
- Ginsburg, I. 1952. Flounders of the genus Paralichthys and related genera in American waters. U.S. National Marine Fisheries Service Fishery Bulletin 52:267–351.
- Gruenthal, K. M., L. K. Acheson, and R. S. Burton. 2007. Genetic structure of natural populations of California red abalone (Haliotis rufescens) using multiple genetic markers. Marine Biology 152:1237–1248.
- Hauser, L., and G. R. Carvalho. 2008. Paradigm shifts in marine fisheries genetics: ugly hypotheses slain by beautiful facts. Fish and Fisheries 9:333–362.
- Hedrick, P. W. 1999. Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution 53:313–318.
- Hutchinson, W. F., C. van Oosterhout, S. I. Rogers, and G. R. Carvalho. 2003. Temporal analysis of archived samples indicates marked genetic changes in declining North Sea Cod (Gadus morhua). Proceedings of the Royal Society of London Series B: Biological Sciences 270:2125–2132.
- Jakobsson, M., and N. A. Rosenberg. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806.
- Knutsen, H., P. E. Jorde, C. André, and N. C. Stenseth. 2003. Fine-scaled geographical population structuring in a highly mobile marine species: the Atlantic Cod. Molecular Ecology 12:385–394.
- Knutsen, H., E. M. Olsen, P. E. Jorde, S. H. Espeland, C. André, and N. C. Stenseth. 2011. Are low but statistically significant levels of genetic differentiation in marine fishes “biologically meaningful”? A case study of coastal Atlantic Cod. Molecular Ecology 20:768–783.
- Lee, W.-J., J. Conroy, W. H. Howell, and T. Kocher. 1995. Structure and evolution of teleost mitochondrial control regions. Journal of Molecular Evolution 41:54–66.
- Lesack, K., and C. Naugler. 2011. An open-source software program for performing Bonferroni and related corrections for multiple comparisons. Journal of Pathology Informatics 2:52–52.
- Librado, P., and J. Rozas. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452.
- Liu, J.-Y., Z.-R. Lun, J.-B. Zhang, and T.-B. Yang. 2009. Population genetic structure of Striped Mullet, Mugil cephalus, along the coast of China, inferred by AFLP fingerprinting. Biochemical Systematics and Ecology 37:266–274.
- Liu, Y.-G., S.-L. Chen, B.-F. Li, Z.-J. Wang, and Z. Liu. 2005. Analysis of genetic variation in selected stocks of hatchery Flounder, Paralichthys olivaceus, using AFLP markers. Biochemical Systematics and Ecology 33:993–1005.
- Luikart, G., P. R. England, D. Tallmon, S. Jordan, and P. Taberlet. 2003. The power and promise of population genomics: from genotyping to genome typing. Nature Reviews Genetics 4:981–994.
- Maguire, T. L., R. Peakall, and P. Saenger. 2002. Comparative analysis of genetic diversity in the mangrove species Avicennia marina (Forsk.) Vierh. (Avicenniaceae) detected by AFLPs and SSRs. Theoretical and Applied Genetics 104:388–398.
- McCairns, R. J. S., and L. Bernatchez. 2008. Landscape genetic analyses reveal cryptic population structure and putative selection gradients in a large-scale estuarine environment. Molecular Ecology 17:3901–3916.
- Meudt, H. M., and A. C. Clarke. 2007. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends in Plant Science 12:106–117.
- Meyer, A., J. M. Morrissey, and M. Schartl. 1994. Recurrent origin of a sexually selected trait in Xiphophorus fishes inferred from a molecular phylogeny. Nature 368:539–542.
- Mickett, K., C. Morton, J. Feng, P. Li, M. Simmons, D. Cao, R. A. Dunham, and Z. Liu. 2003. Assessing genetic diversity of domestic populations of Channel Catfish (Ictalurus punctatus) in Alabama using AFLP markers. Aquaculture 228:91–105.
- Midway, S. R., S. X. Cadrin, and F. S. Scharf. 2014. Southern Flounder (Paralichthys lethostigma) stock structure inferred from otolith shape analysis. U.S. National Marine Fisheries Service Fishery Bulletin 112:326–338.
- Mock, K. E., T. C. Theimer, O. E. Rhodes, D. L. Greenberg, and P. Keim. 2002. Genetic variation across the historical range of the wild turkey (Meleagris gallopavo). Molecular Ecology 11:643–657.
- Monaghan, J. P. Jr. 1996. Life history aspects of selected marine recreational fishes in North Carolina, study 2: migration of paralichthid flounders tagged in North Carolina. North Carolina Division of Marine Fisheries, Grant F-43, Segments 1–5, Completion Report, Morehead City.
- NCDMF (North Carolina Division of Marine Fisheries). 2005. North Carolina fishery management plan: Southern Flounder (Paralichthys lethostigma). North Carolina Department of Environment and Natural Resources, Technical Report, Morehead City.
- NCDMF (North Carolina Division of Marine Fisheries). 2013. North Carolina Southern Flounder (Paralichthys lethostigma) fishery management plan amendment 1. North Carolina Department of Environment and Natural Resources, Technical Report, Morehead City.
- O'Reilly, P. T., M. F. Canino, K. M. Bailey, and P. Bentzen. 2004. Inverse relationship between FST and microsatellite polymorphism in the marine fish, Walleye Pollock (Theragra chalcogramma): implications for resolving weak population structure. Molecular Ecology 13:1799–1814.
- Parchman, T. L., C. W. Benkman, and S. C. Britch. 2006. Patterns of genetic variation in the adaptive radiation of New World crossbills (Aves: Loxia). Molecular Ecology 15:1873–1887.
- Polzin, T., and S. V. Daneshmand. 2003. On Steiner trees and minimum spanning trees in hypergraphs. Operations Research Letters 31:12–20.
- Pritchard, J. K., M. Stephens, and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959.
- Rosenberg, N. A. 2004. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes 4:137–138.
- Ruzzante, D. E., C. T. Taggart, and D. Cook. 1999. A review of the evidence for genetic structure of cod (Gadus morhua) populations in the NW Atlantic and population affinities of larval cod off Newfoundland and the Gulf of St. Lawrence. Fisheries Research 43:79–97.
- Ryman, N., S. Palm, C. André, G. R. Carvalho, T. G. Dahlgren, P. E. Jorde, L. Laikre, L. C. Larsson, A. Palmé, and D. E. Ruzzante. 2006. Power for detecting genetic divergence: differences between statistical methods and marker loci. Molecular Ecology 15:2031–2045.
- Sønstebø, J. H., R. Borgstrøm, and M. Heun. 2007. A comparison of AFLPs and microsatellites to identify the population structure of Brown Trout (Salmo trutta L.) populations from Hardangervidda, Norway. Molecular Ecology 16:1427–1438.
- Svensson, E. I., L. Kristoffersen, K. Oskarsson, and S. Bensch. 2004. Molecular population divergence and sexual selection on morphology in the banded demoiselle (Calopteryx splendens). Heredity 93:423–433.
- Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595.
- Takade-Heumacher, H., and C. Batsavage. 2009. Stock status of North Carolina Southern Flounder (Paralichthys lethostigma). North Carolina Division of Marine Fisheries, Technical Report, Morehead City.
- Tamura, K., G. Stecher, D. Peterson, A. Filipski, and S. Kumar. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30:2725–2729.
- Taylor, J. C., J. M. Miller, and D. Hilton. 2008. Inferring Southern Flounder migration from otolith microchemistry. North Carolina Sea Grant, Final Report for Fishery Resource Grant 05-FEG-06, Raleigh.
- Timmermans, M. J. T. N., J. Ellers, J. Mariën, S. C. Verhoef, E. B. Ferwerda, and N. M. Van Straalen. 2005. Genetic structure in Orchesella cincta (Collembola): strong subdivision of European populations inferred from mtDNA and AFLP markers. Molecular Ecology 14:2017–2024.
- Toews, D. P. L., and A. Brelsford. 2012. The biogeography of mitochondrial and nuclear discordance in animals. Molecular Ecology 21:3907–3930.
- Toews, D. P. L., and D. E. Irwin. 2008. Cryptic speciation in a Holarctic passerine revealed by genetic and bioacoustic analyses. Molecular Ecology 17:2691–2705.
- Vekemans, X., T. Beauwens, M. Lemaire, and I. Roldán-Ruiz. 2002. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Molecular Ecology 11:139–151.
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Friters, J. Pot, J. Paleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23:4407–4414.
- Waples, R. S. 1998. Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. Journal of Heredity 89:438–450.
- Waples, R. S., A. E. Punt, and J. M. Cope. 2008. Integrating genetic data into management of marine resources: how can we do it better? Fish and Fisheries 9:423–449.
- Ward, R. D. 2000. Genetics in fisheries management. Hydrobiologia 420:191–201.
- Ward, R. D., M. Woodwark, and D. O. F. Skibinski. 1994. A comparison of genetic diversity levels in marine, freshwater, and anadromous fishes. Journal of Fish Biology 44:213–232.
- Watterson, J. C., and J. L. Alexander. 2004. Southern Flounder escapement in North Carolina, July 2001–June 2004. North Carolina Division of Marine Fisheries, Final Performance Report F-73, Morehead City.
- Weetman, D., A. Ruggiero, S. Mariani, P. W. Shaw, A. R. Lawler, and L. Hauser. 2007. Hierarchical population genetic structure in the commercially exploited shrimp Crangon crangon identified by AFLP analysis. Marine Biology 151:565–575.
- Wenner, C. A., W. A. Roumillat, J. E. Moran Jr., M. B. Maddox, L. B. I. Daniel, and J. W. Smith. 1990. Investigations on the life history and population dynamics of marine recreational fishes in South Carolina: part 1. South Carolina Wildlife and Marine Resources Department, Marine Resources Research Institute, Final Report F-37, Charleston.
- Whitehead, A., S. L. Anderson, K. M. Kuivila, J. L. Roach, and B. May. 2003. Genetic variation among interconnected populations of Catostomus occidentalis: implications for distinguishing impacts of contaminants from biogeographical structuring. Molecular Ecology 12:2817–2833.
- Whitlock, R., H. Hipperson, M. Mannarelli, R. K. Butlin, and T. Burke. 2008. An objective, rapid and reproducible method for scoring AFLP peak-height data that minimizes genotyping error. Molecular Ecology Resources 8:725–735.
- Wright, S. 1978. Evolution and the genetics of populations: variability within and among natural populations, volume 4. University of Chicago Press, Chicago.
- Zink, R. M., and G. F. Barrowclough. 2008. Mitochondrial DNA under siege in avian phylogeography. Molecular Ecology 17:2107–2121.
