Abstract
Aflatoxin M1 contamination in dairy products is a risk when feedstuff contaminated with aflatoxin B1 produced by moulds is consumed by milk-producing animals. Milk can be screened for aflatoxin M1 at the European Union maximum limit of 50 ng l−1 by a lateral flow test, the MRLAFMQ (Aflatoxin M1) Test. The method takes 15 min with no milk dilution or a sample preparation step. The lateral flow assay was validated at the Technology and Food Science Unit of the Institute for Agricultural and Fisheries Research (ILVO-T&V) according to European Union guidelines using fortified raw milk samples. A detection capability of 50 ng l−1 was demonstrated with a false negative rate lower than 2% at 50 ng l−1 and a false positive rate of less than 0.3%. Quantitative readings had a mean bias of +2 to 6 ng l−1 at 50 ng l−1 with a standard deviation of 5–8 ng l−1. Based on the validation results, the test could be considered appropriate for milk screening prior to milk unload at dairies.
Introduction
Aflatoxins are natural toxins produced as a secondary product of Aspergillus flavus and/or Aspergillus parasiticus mould growth (Klich Citation2007). When mould growth occurs on feeds or grains, aflatoxin B1 could be produced. Aflatoxin B1 is the most toxic aflatoxin and a potent hepatocarcinogen (Wogan Citation1966; Carnaghan Citation1967; Wong & Hsieh Citation1976). Feeds and grains are generally screened in a range of 2 µg kg–1 aflatoxin B1 for European Union (EU) Regulation No. 165/2010 and 20 µg kg–1 aflatoxin B1 for the USFDA action level for human consumption and for dairy animals (USDA Citation2002).
When feed contaminated with aflatoxin B1 is consumed by dairy cows, usually some 1–3% of the aflatoxin B1 is excreted as aflatoxin M1 into the milk (Veldman et al. Citation1992; Masoero et al. Citation2007). Aflatoxin M1, which is the aflatoxin B1 hydroxylated metabolite, is a less potent carcinogen but still classified as a group 1 carcinogen by the IARC (Wogan & Paglialunga Citation1974; Hsieh et al. Citation1986; Cullen et al. Citation1987; IARC Citation2012). There are no strategies for removal of aflatoxin M1 from milk, hence attention has to be focused on methods to prevent or reduce mycotoxin formation at all stages in the milk production chain (van Egmond et al. Citation1997; Prandini et al. Citation2009). Pre-screening of feeds and grain prior to consumption could be a first control strategy to limit the amount of aflatoxin M1 in milk. Because grain and feed screening is not always performed, screening of the milk supply on aflatoxin M1 at the raw milk level prior to dairy intake is an important control check since the occurrence of mould growth and toxin production in grains and feeds is increasing due to the changing climatic conditions (Driehuis et al. Citation2010).
In some countries like Iran, Turkey, Pakistan and India, the levels of aflatoxin M1 in milk often (30.8%, 47%, 81%, and 86%, respectively) exceeds the EU permissible level of 0.05 µg l−1 (Muhammad et al. Citation2010) due to the poor storage conditions along the animal feed chain which exacerbates the growth of moulds and consequently increases the concentration of mycotoxins in cow feed.
The established CODEX health level for aflatoxin M1 in milk, and the US action level, is 0.5 µg l–1 or 500 ng l−1 (Codex Citation2001). In Europe, a maximum limit (ML) based on the ALARA (as low as reasonably achievable) principle is set for aflatoxin M1 in raw milk at 0.05 µg l−1 or 50 ng l−1 (Commission Regulation (EU) No. 165/2010).
A variety of methods exist for evaluating aflatoxin M1 in milk. One AOAC method is also the ISO method validated at the 80 ng l−1 level for reconstituted powdered milks (ISO/IDF Citation2007). This uses an immunoaffinity purification column followed by derivatisation and fluorescent HPLC detection. The repeatability at the lowest study concentration of 80 ng aflatoxin M1 l−1 was 50 ng l−1 (CV = 62.5%). Due to the equipment costs and complexity of the method, the reference method has limited to no field applicability for the rapid detection of aflatoxin M1. More rapid methods with high throughput using ELISA and lateral flow methods are employed for screening purposes. While many methods have been validated for the detection of aflatoxin B1 in feeds and grain, only a few have been validated for aflatoxin M1 in milk (Anfossi et al. Citation2010). Most of these methods are designed to screen aflatoxin M1 in milk at USFDA-established action level and are hence not suitable for the European market. Other rapid tests are not available as a single test or the test protocol is too complex to be used for screening at the entry of the dairy. In other tests, e.g. Afla M1-V (Vicam), a filtration step is involved as sample pre-treatment. To our knowledge there are actually only two rapid tests on the market based on the lateral flow principle with no sample dilution or preparation for raw milk screening on aflatoxin M1 at the 50 ng l−1 level, namely Aflasensor (Unisensor s.a.) and MRLAFMQ (Aflatoxin M1) Test (Charm Sciences Inc., Lawrence, MA, USA).
This study describes a dairy stakeholder requested independent laboratory evaluation of the MRLAFMQ (Aflatoxin M1) Test for detecting 50 ng l−1 of aflatoxin M1 in milk and quantifying in a 15–75 ng l−1 range. The validation design performed at the Technology and Food Science Unit of the Institute for Agricultural and Fisheries Research (ILVO-T&V) is based on its applicability as a qualitative screening test according to Commission Decision Citation2002/657/EC and CRL Guidelines (CRL Citation2010) in order to check if the test is suitable as a raw milk screening test at 50 ng aflatoxin M1 l−1, the European ML. Additionally, because the method provides a semi-quantitative result, the data may be used to evaluate quantitative test parameters defined in Commission Decision Citation2002/657/EC to compare with other semi-quantitative methods. The test is typically used for testing farm tanks and truck loads of milk before the receipt of milk into dairies.
Materials and methods
Tests and equipment
MRLAFMQ tests (kit lot 009 (Exp. 09/2013) and lot 010 (Exp. 10/2013)) (Charm Sciences Inc., Lawrence, MA, USA), a 45°C ROSA incubator, and reader models ROSA Pearl and EZ were provided to ILVO-T&V. The MRLAFMQ Test for milk is a lateral flow method that works in 15 min with a single milk addition step. The test principle is similar to the optimised system described by Anfossi et al. (Citation2010). However, it is different in the way it employs two competitive binding lines as well as a control line. This test is based on the competitive binding of aflatoxin M1 in the milk with colloidal gold-antibody construct contained within a lateral flow device and solubilised by milk as it flows through the strip. When complete, two test lines and a control line are visible that are read by a reader using a line refractance algorithm to provide a quantitative test result in ng aflatoxin M1 l−1, also referred to as parts per trillion (ppt). The readers also employ a 40 ng l−1 qualitative negative/positive control limit to provide confidence that milk containing aflatoxin M1 at 50 ng l−1 or greater is not accepted. Two reader models, the ROSA Pearl Reader and the EZ Reader, were evaluated in this study because both readers models are currently in use in dairies worldwide.
Reported values
Values reported in tables for spiked concentrations include mean, standard deviation (SD) and minimum and maximum values of the n-replicates.
Clean-up by immunoaffinity chromatography and determination by HPLC with fluorescence detection based on ISO 14501:2007 (ISO/IDF Citation2007). LOD = 1.5 ng aflatoxin M1 l−1 and LOQ = 3 ng aflatoxin M1 l−1.
Reagents
Aflatoxin M1 analytical standard of 10 µg ml−1 in acetronitrile (Supelco-46319-U Sigma-Aldrich, Bornem, Belgium) and further used for the preparation of an 1 µg aflatoxin M1 ml−1 stock solution in phosphate buffer stored refrigerated for spiking raw and pasteurised milk.
Milk supply
The blank milk for spiking was raw commingled milk from local Belgian farms with a low aflatoxin M1 background originating from at least four healthy cows in mid-lactation that were not treated with antibiotics or chemotherapeutics for at least 3 months and stored refrigerated for a maximum of 3 days. Pasteurised milk was whole milk from the Netherlands purchased at the supermarket. Farm and truck milk samples were from the Flemish milk control station Melkcontrolecentrum-Vlaanderen. Incurred individual farm silo milk samples were obtained from two Belgian farms with aflatoxin M1 problems in the milk.
Results and discussion
Detection capability
Raw milk was spiked with aflatoxin M1 at three levels: 25, 50 and 75 ng l−1 in seven different blank raw milks. Sixty replicates tested at each concentration were used based on the closeness of the predicted 95% sensitivity (concentration capabilities, CCβ) to the European maximum limit (ML) according to CRL Screening Test Guidelines (CRL Citation2010). The testing was performed over at least 7 days with the use of at least two different stock solutions for spiking. Two different lots of MRLAFMQ reagents were used for this validation and the results were measured using both the ROSA Pearl Reader and the EZ Reader. To verify the detection capability at 50 ng aflatoxin M1 l−1, the test and reader must provide a positive interpretation above the positive/negative cut-off, with a quantitative result of 40 ng aflatoxin M1 l−1 or higher in at least 57 upon 60 tests. The performance of the test, for each reader, and at each concentration are given in and .
Table 1. Spike milk sample results of MRLAFMQ by means of the ROSA Pearl Reader.
Table 2. Spike milk sample results of MRLAFMQ by means of the EZ Reader.
Using a value of 40 ng l−1 or greater as the cut-off for a qualitative positive result, 58 of the 59 fortified 50 ng aflatoxin M1 l−1 samples for both the ROSA Pearl Reader and the EZ Reader were found as positive, which qualifies the MRLAFMQ for ML detection at 50 ng aflatoxin M1 l−1. There were 60 upon 60 positives at 75 ng l−1. At 25 ng l−1 or 0.5 × ML, there were zero positives upon 58 replicates with the ROSA Pearl Reader and two positives upon 58 replicates with the EZ Reader, a false violative rate of 3.4%. These results indicate a high degree of discrimination between 0.5 × ML and ML. Blank milk tests demonstrated no positives of the 59 replicates with a maximum read of the ROSA Pearl Reader of 26 ng l−1 and of the EZ Reader of 20 ng l−1.
Quantitative detection parameters
Quantitative aspects of the data support the qualitative test results. SDs of blank milk of 4 ng l−1 and 3× ±SD support an LOD between 14 and 19 ng l−1 for the ROSA Pearl Reader and the EZ Reader, respectively. LOQ and blank mean + 10× SD are between 42 and 47 ng l−1, supporting a detection capability of 50 ng l−1. The 50 ng l−1 mean values minus 2 × SD are 40 ng l−1 for the ROSA Pearl Reader and 36 ng l−1 for the EZ Reader and demonstrate that the 40 ng l−1 limit is providing about a 95% confidence in detecting 50 ng aflatoxin M1 l−1 samples as positive.
The method LOD and LOQ as well as the SDs of quantification are consistent with semi-quantitative interpretation of test results. The estimated precision of a quantitative determination would be about 3 × SD or 15 ng l−1, which is consistent with the 30% relative standard deviation (RSD %) of Anfossi et al. (Citation2013) in the optimised lateral flow system. It is interesting to note that the SDs of lateral flow systems of about 5–8 ng kg–1 at 75 ng ml−1 are comparable with the ISO method repeatability, Sr = 0.005–0.008 µg kg−1, at the lowest study concentration of 0.08 µg kg−1 (ISO/IDF Citation2007). Additionally HorRat values less than 1.0 at the 50 µg kg−1 level are an indicator of the acceptable repeatability of the method for raw milk analysis. These results indicate that the lateral flow method might be useful as a dairy screening method for quantitative determination consistent with subsequent official methods for legal action and milk rejection. Further collaborative study according to international protocol to establish inter-laboratory repeatability and intra-laboratory reproducibility at 50 ng aflatoxin M1 l−1 and lower are needed to compare different semi-quantitative methods.
Selectivity
The selectivity of the method is its ability to distinguish the target analyte, aflatoxin M1, from other unrelated compounds, such as antibiotics; other unrelated mycotoxins, such as ochratoxin A, zearalenone, deoxynivalenol and fumonisin B1; and analogous compounds such as aflatoxin M2. Different compounds were evaluated at 10 × MRL to determine if there was any interference. If interference did occur, the levels were adjusted to determine the percentage of cross reactivity.
The results in indicate no interference in interpretation or reading with any unrelated antibiotic or mycotoxin. Aflatoxin M2 did show about 20% cross-reactivity in quantification and began to produce positive interpretation at about 500–600 ng l−1 in both readers. The MRLAFMQ is highly selective for aflatoxin M-related compounds.
Table 3. Test selectivity of MRLAFMQ test for aflatoxin M1.
Repeatability
The repeatability of the reader was evaluated by measuring in duplicate (removing and replacing into the reader) 20 different strips obtained after the testing of blank, low positive 25 ng aflatoxin M1 l−1, and high positive 75 ng aflatoxin M1 l−1. The test repeatability was checked by randomly analysing 15 samples in duplicate for blank, low positive 25 ng aflatoxin M1 l−1, and high positive 75 ng aflatoxin M1 l−1 milk. Differences between duplicate reader and test results were squared and SD are presented in and for the reader and test, respectively. Results indicate that reader variation is less than the test or assay variation. Reader variation of the ROSA Pearl Reader has SD of differences less than 1 at all concentrations, while the EZ Reader had a slightly higher variation with SD less than 2 at all concentrations. The test or assay variation is higher than the reader variation and the SD are similar to the SD of fortified sample experiments, suggesting that the test-to-test variation is the major contributing factor affecting result variance. Both the ROSA Pearl Reader and the EZ Reader had similar test variance SD at all test concentrations. The two readers can be considered to give equivalent test variation. This result supports the data of the fortified experiment that the EZ Reader is calibrated with a 5 ng l−1 higher positive bias and that this bias is consistently reflected at all four study concentrations.
Table 4. Reader repeatability of MRLAFMQ.
Table 5. Test repeatability of MRLAFMQ.
Milk sample screening and false positive results
The incidence of aflatoxin M1 contamination of milk farm tanks and milk bulk tanks is of interest based on the discovery of contamination of feed originating from Eastern Europe with traces found in the milk supply (Epi South Citation2013). This contaminated feed was used in some Western European countries. This study found that the baseline levels of aflatoxin M1 in milk from Belgium and the Netherlands were below the LOD, 14–19 ng l−1. This evaluation tested the MRLAFMQ-positive rate with 123 frozen and thawed farm blank milk samples and 131 fresh blank truck bulk tank milk samples. The evaluation also evaluated 20 powder milk samples rehydrated to 10% solids, pH balanced and centrifuged prior to testing. Since these milk samples have an unknown history, if positive they were also tested with HPLC methods to determine method agreement and if the MRLAFMQ results were correct. All samples evaluated tested negative on both reader types; these results are summarised in . There was one truck sample that tested positive with the ROSA Pearl Reader, but this result was not confirmed on a duplicate retest. There were five farm milk samples that tested positive by the EZ Reader due to positioning errors corrected on reinsertion and excluded from .
Table 6. Summary of negative and positive results with different farm milk, tanker/truck milk and reconstituted milk powder samples.
These results indicate that the false positive incidence of the MRLAFMQ is about 1 upon 274 tests, or about 0.3%. Care should be taken to insert strips properly into the EZ Reader when used in the read-only mode. These results are consistent with antibiotic screening test validations and appropriate for farm and tanker/truck screening (Reybroeck & Ooghe Citation2012). The results also indicated that the majority of farm and tanker milk tested from Belgium and the Netherlands region is below the LOD by the MRLAFMQ Test.
Some incurred individual farm silo milk samples were tested by both the MRLAFMQ and reference HPLC-affinity fluorescence detection method (ISO/IDF Citation2007). In general there was a good agreement (compliant/non-compliant) between the results obtained with both test methods, indicating that the MRLAFMQ method is also working comparatively with the reference method with incurred samples.
Ruggedness
The test performance under assay variance conditions was evaluated using pipette variances of 330 µl (high) and 270 µl (low) and with different milk temperatures, 3, 10, 15 and 20°C. Three replicates were evaluated using negative and positive milk doped with aflatoxin M1 at 25 and 50 ng l−1. The minimum and maximum results and the mean reading are presented in and .
Table 7. Effect of change in milk volume on MRLAFMQ readings.
Table 8. Effect of the milk temperature on MRLAFMQ readings.
Milk volume did not significantly affect negative or positive results. One false negative result occurred with the ROSA Pearl Reader at low volume dispense, and one false violative result occurred with the high volume dispensed on the EZ Reader. Similarly, milk temperature did not have an effect on MRLAFMQ results. There was one false negative result with the ROSA Pearl Reader at 15°C. None of the results is outside the normal spiking population of 60 data points and therefore the results are not considered significant indicators of perturbation. Similar readings and positive biases were produced with both readers in comparison with the spike data.
Interferences
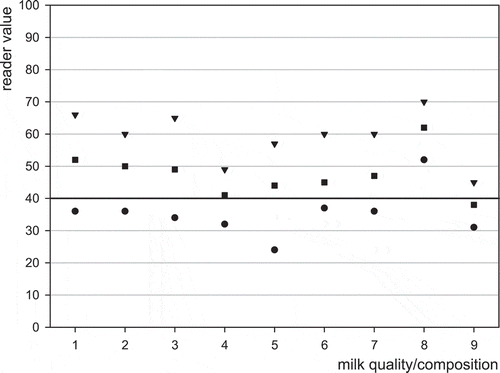
The influence of milk compositional components or milk quality, >106 somatic cells ml−1, >5 × 105 bacteria ml−1, low and high fat, low and high protein, and low and high pH were compared with blank milk of normal quality/composition, and spiked near CCβ with aflatoxin M1 at 50 ng l−1. Ten replicates of each milk type were performed. Mean, minimum and maximum values are shown graphically for the EZ Reader in and for blank milk and milk spiked with aflatoxin M1, respectively. The results for blank and spike milk for the ROSA Pearl reader are not shown. The results from the compositional/quality analysis show that compositional milk quality aspects did not influence the blank results but did have the effect of lowering the mean positive result in the cases of high pH, low and high fat, and low and high protein. These effects are likely due to slow flow of the milk through the test. The figures show that high somatic cell and bacteria have little effect on the results. The likely causes of false negative results with the abnormal milk compositional variants are a slight sensitivity shift due to flow differences.
Figure 1. Effect of milk composition or quality effects on the screening of blank milk using MRLAFMQ and EZ Reader. Maximum reading (▾), average reading (■), minimum reading (), control point (40 ng l−1) dividing positive from negative (▬▬); 1 = reference: normal raw cows’ milk, 2 = somatic cell count > 106 ml−1, 3 = high bacterial count (>5 × 105 ml−1), 4 = low fat content (<2 g 100 ml−1), 5 = high fat content (>6 g 100 ml−1), 6 = low protein (<3 g 100 ml−1), 7 = high protein (>4 g 100 ml−1), 8 = low pH (6.0), 9 = high pH (7.5).
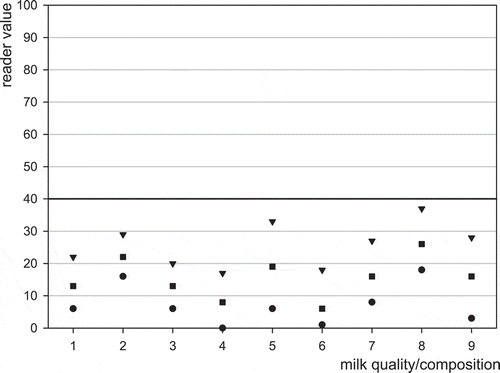
Figure 2. Effect of milk composition or quality on the detection of 50 ng l–1 aflatoxin M1 in milk using MRLAFMQ and EZ Reader. Maximum reading (▾), average reading (■), minimum reading (), control point (40 ng l−1) dividing positive from negative (▬▬); 1 = reference: normal raw cows’ milk, 2 = somatic cell count > 106 ml−1, 3 = high bacterial count (>5 × 105 ml−1), 4 = low fat content (<2 g 100 ml−1), 5 = high fat content (>6 g 100 ml−1), 6 = low protein (<3 g 100 ml−1), 7 = high protein (>4 g 100 ml−1), 8 = low pH (6.0), 9 = high pH (7.5).
The detection capability of the MRLAFMQ method was also evaluated in four different doped homogenised pasteurised whole-milk samples over 4 days. The performances of the test, for each reader, at each concentration are given in and . The results of pasteurised milk experiments are similar to the raw milk spike results, with a slightly higher positive bias on all the tested concentrations. There were no false negative results at 50 ng aflatoxin M1 l−1 and 75 ng aflatoxin M1 l−1. There were no false positive results with negative milk. At 25 ng aflatoxin M1 l−1 there were two false violative results using the ROSA Pearl Reader and five false violative results using the EZ Reader. It is important that non-raw matrices tested with the method are internally validated by the laboratory performing the method to assure the performance and reliability of the test results since these other dairy matrices can have a different flow rate as compared with raw milk.
Table 9. Pasteurised whole-milk spike sample results of MRLAFMQ by means of the ROSA Pearl Reader.
Table 10. Pasteurised whole-milk spike sample results of MRLAFMQ by means of the EZ Reader.
The MRLAFMQ Test is a test claimed for raw cows’ milk. It is not claimed and has not been validated with goats’ and ewes’ milk nor powdered, UHT or heat-treated milk. Additional testing of the effect of composition looked at the influence of these different types of milks: UHT, sterilised, reconstituted powder, frozen–thawed, goats’, ewes’ and mares’ milk. In this evaluation 10 negative raw milk and 10 negative heat-treated milk samples, and each sample spiked with aflatoxin M1 at 50 ng l−1 were tested and evaluated in each reader. Minimum, maximum and mean obtained in the ROSA Pearl Reader are plotted in and for blank milk and for milk spiked with aflatoxin M1 at 50 ng l−1, respectively. Comparable results were obtained in an EZ Reader (figures not shown).
Figure 3. Screening of different blank milk types using MRLAFMQ and ROSA Pearl Reader. Maximum reading (▾), average reading (■), minimum reading (), control point dividing positive from negative (▬▬) ; 1 = reference: normal raw cows’ milk, 2 = UHT milk, 3 = sterilised milk, 4 = reconstituted powder, 5 = frozen–thawed, 6 = goats’ milk, 7 = mares’ milk.
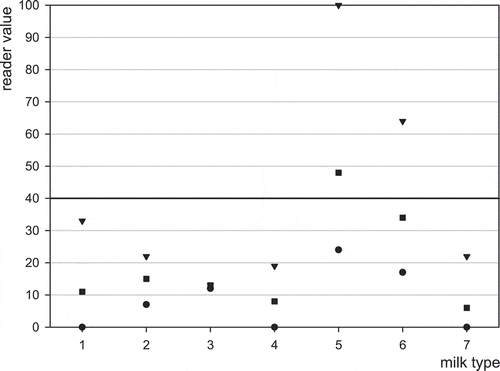
Figure 4. Detection of 50 ng l−1 aflatoxin M1 in different milk types using MRLAFMQ and ROSA Pearl Reader. Maximum reading (▾), average reading (■), minimum reading (), control point dividing positive from negative (▬▬) ; 1 = reference: normal raw cows’ milk, 2 = UHT milk, 3 = sterilised milk, 4 = reconstituted powder, 5 = frozen–thawed, 6 = goats’ milk, 7 = mares’ milk.
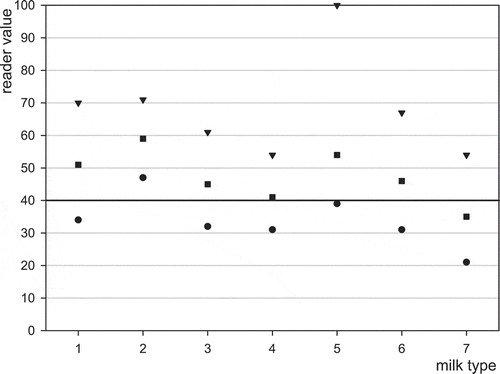
Results are generally biased low, or high, depending on milk type. UHT milk displayed a higher average result for both blank and spiked milk. Sterilised and powdered milk had lower spiked positive averages. Thawed milk had more erratic minimum and maximum compared with normal milk. Goats’ milk was biased positive, while mares’ milk was biased negative. Ewes’ milk is not reported as it did not produce valid results due to flow issues.
The MRLAFMQ Test is influenced by milk composition, and care should be taken to validate and calibrate instrumentation if other matrices than raw cows’ milk are tested.
Lot differences
The following samples were analysed at the same time with two different batches of MRLAFMQ reagents (Lot 010 (Exp. Sep. 2013) and Lot 011-EZ (Exp. Oct. 2013)):
Blank milk (antibiotic-free raw milk) (20 samples).
Raw milk spiked with 25 ng l–1 aflatoxin M1 (20 samples).
Raw milk spiked with 50 ng l−1 aflatoxin M1 (20 samples).
Raw milk doped with 75 ng l−1 aflatoxin M1 (20 samples).
The results are shown in . Both readers show similar performance for the two lots. In general comparable results were obtained for the two lots in that the differences between mean values were within 2–6 and 1–6 ng aflatoxin M1 l−1 for EZ and ROSA Pearl Reader, respectively. Likewise, the maximum and minimum extremes were within a few ng aflatoxin M1 l−1 of each other. The exception of this statement is with MRLAFMQ Lot 011 at 75 ng aflatoxin M1 l−1, which displayed a more negative minimum value, by about 10 ng aflatoxin M1 l−1, and in one case gave a false negative result with the ROSA Pearl Reader. There were no false negative results from any other of the positive spiked (50 and 75 ng aflatoxin M1 l−1) samples. There was one false positive result with blank milk analysed with the EZ Reader for both MRLAFMQ lots 010 and 011 and on reinsertion it was analysed as negative. This is likely a positional error discussed earlier in the false positive-selectivity paragraph.
Table 11. Testing of blank milk, milk spiked with 25 ng l−1 aflatoxin M1, 50 ng l−1 aflatoxin M1 and 75 ng l−1 aflatoxin M1 using two different lots of MRLAFMQ.
Stability of result
The stability in readings was also evaluated by control chart plotting daily control performance and calibration data over the 2-month evaluation period. Daily negative controls and 50 ng aflatoxin M1 l−1 Charm-positive controls for EZ Reader are depicted in . There were no false positive results and one false negative Charm-positive control with the EZ reader and three false negative Charm-positive controls with the ROSA Pearl Reader (figures not shown). In addition, milk spiked with aflatoxin M1 standard (diluted from Supelco 10 µg aflatoxin M1 ml−1) was monitored and gave results similar to the positive control standard, which are depicted for the EZ Reader in . There was one false negative with the 50 ng aflatoxin M1 l−1 spiked raw milk in each reader (the figure for the ROSA Pearl Reader is not shown). False-negative positive controls were followed up with true positive results to verify equipment operation before continuing evaluation.
Figure 5. Results for daily blank samples () and Charm-positive control (▾) by means of the EZ Reader.
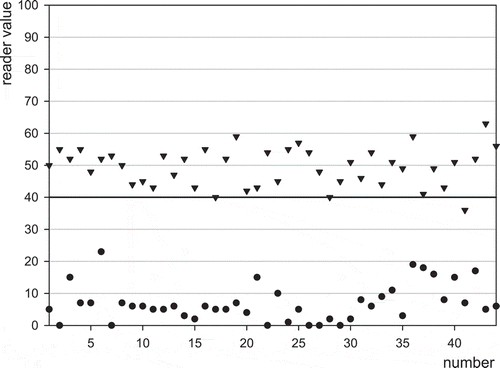
Figure 6. Results for daily blank samples () and 50 ng l−1 aflatoxin M1 spiked standard (▾) by means of the EZ Reader.
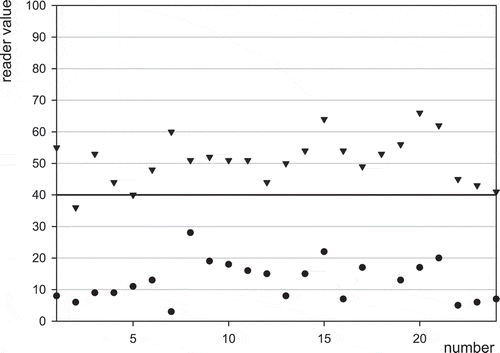
In summary, the MRLAFMQ is a very selective lateral flow test for commingled raw milk that detects aflatoxin M1 at 50 ng l−1 in 15 min. The method is qualitative using a 40 ng l−1 limit and demonstrated detection capability at 50 ng l−1, a low <3.4% false violative rate at 25 ng l−1, and a false positive rate of 0.3% While the method did demonstrate detection capability at EU ML with <2% false negative results, by lowering the cut-off to 38 ng l−1 no false-negatives could be obtained providing additional confidence and robustness at detecting 50 ng l−1 while not increasing false positive incidence. The method was also tested and is applicable to pasteurised milk, but based on a testing anomaly with a particular type of pasteurised milk it is recommended that laboratories test and specifically qualify non-raw milk type matrices before routine use of the assay.
The MRLAFMQ method provides a quantitative value with an LOD of 14–19 ng aflatoxin M1 l−1, an LOQ of 45–50 ng aflatoxin M1 l−1, and SDs of readings of about 5–10 ng l−1. The method was evaluated for influences from compositional components or milk quality and could result in false positive results when testing frozen–thawed samples and goats’ milk. The method is applicable to normal raw cows’ milk as abnormal fat, protein and pH levels caused loss of detection capability.
The method was easily performed using the existing ROSA Pearl Reader and the new EZ Reader equipment in use at dairy laboratories. The method was robust to milk temperature variations and to the correct amount of milk to within ±10% of the target level.
The MRLAFMQ method meets screening test specifications of low false negatives and positives, low false violatives, and detection capability of aflatoxin M1 at 50 ng ml−1 which is the EU ML. This indicates the method can reliably be used at dairy milk receipt and in trade to verify milk is free of aflatoxin M1 below levels of concern. Based on the validation data and the results for the incurred samples, the MRLAFMQ was accepted by the Belgian Federal Agency for the Safety of the Food Chain as a method that could be used for testing farm silo milk to re-allow milk collection after the farm was put in ‘quarantine’ due to the delivery of milk containing aflatoxin M1 above the ML. Routine use of the method can allow for quantification above the LOD and below the LOQ; this is useful for farm feed remedial action before milk becomes actionable and proactively maintain ALARA levels. An additional international collaborative study to determine method precision parameters is warranted based on these study results.
Acknowledgements
The authors appreciate the valuable work performed by Eline De Wispelaere and Daan Blomme and thank Vzw Melkcontrolecentrum-Vlaanderen for providing part of the raw cows’ milk samples.
References
- Anfossi L, Baggiani C, Giovannoli C, Biagioli F, D’Arco G, Giraudi G. 2013. Optimization of a lateral flow immunoassay for the ultrasensitive detection of aflatoxin M1 in milk. Anal Chim Acta. 772:75–80.
- Anfossi L, Baggiani C, Giovannoli C, Giraudi G. 2010. Mycotoxins in food and feed: extraction, analysis and emerging technologies for rapid and on-field detection. Recent Pat Food Nutri Agri. 2:140–153.
- Carnaghan RBA. 1967. Hepatic tumours and other chronic liver changes in rats following a single oral administration of aflatoxin. Brit J Cancer. 21:811–814.
- Codex. 2001. Comments submitted on the draft maximum level for aflatoxin M1 in milk. CX/FAC 01/20. Codex Alimentarius Commission. Codex Committee on Food Additives and Contaminants 33rd session, The Hague, the Netherlands.
- Commission Decision 2002/657/EC of 12 August 2002 implementing Council Directive 96/23/EC concerning the performance of analytical methods and the interpretation of results. Off J Eur Comm. L221:8–36.
- Commission Regulation (EU) No. 165/2010 of 26 Febuary 2010 amending Regulation (EC) No. 1881/2006 setting maximum levels for certain contaminants in foodstuffs as regards aflatoxins. Off J Eur Union. 2010, L50:8–12.
- CRL. 2010. Guidelines for the validation of screening methods for residues of veterinary medicines (initial validation and transfer). Community Reference Laboratories Residues (CRLs):1–18.
- Cullen JM, Ruebner BH, Hsieh LS, Hyde DM, Hsieh DP. 1987. Carcinogenity of dietary aflatoxin M1 in male Fischer rats compared to aflatoxin B1. Cancer Res. 47:1913–1927.
- Driehuis F, te Giffel MC, Van Edmond HP, Fremy JM, Bluthgen A. 2010. Feed–associated mycotoxins in the dairy chain: occurrence and control. Bull Intern Dairy Fed. 444/2010:1–25.
- Epi South. 2013. EpiSouth Weekly Epi Bulletin No:260. March 6–12, 2013 [Internet]. Report of new health events occurring inside the EpiSouth Area. [cited 2013 Sep]. Available from: http://www.episouthnetwork.org/sites/default/files/bulletin_file/eweb_260_14_03_13_1.pdf
- Hsieh DPH, Cullen JM, Hsieh LS, Shao Y, Ruebner BH. 1986. Cancer risks posed by aflatoxin M1. In: Hayashi Y, et al. editors. Diet, nutrition and cancer. Tokyo: Japan Science Society Press; Utrecht: VNU Science Press; p. 57–65.
- IARC. 2012. Aflatoxins: IARC monographs on the evaluation of carcinogenic risks to humans [Internet]. Chemical agents and related occupations. 100F:225–248. [cited 2014 Sep]. Available from: http://monographs.iarc.fr/ENG/Monographs/vol100F/index.php
- ISO/IDF. 2007. Milk and milk powder: determination of aflatoxin M1 content – cleanup by immunoaffinity chromatography and determination by high-performance liquid chromatography ISO/FDIS 14501 | IDF 171:2007 ISO TC 34/SC 5/WG:1–10.
- Klich MA. 2007. Aspergillus flavus: the major producer of aflatoxin. Mol Plant Pathol. 8:713–722.
- Masoero F, Gallo A, Moschini M, Piva G, Diaz D. 2007. Carryover of aflatoxin from feed to milk in dairy cows with low or high somatic cell counts. Animal. 1:1344–1350.
- Muhammad K, Tipu MY, Abbas M, Khan AM, Anjum AA. 2010. Monitoring of aflatoxin M1 in market raw milk in Lahore city, Pakistan. Pakistan J Zool. 42:697–700.
- Prandini A, Tansini G, Sigolo S, Filippi L, Laporta M, Piva G. 2009. On the occurrence of aflatoxin M1 in milk and dairy products. Food Chem Toxicol. 47:984–991.
- Reybroeck W, Ooghe S. 2012. Validation of the Charm Beta-lactam and Tetracycline 2 minute Test (MRLBLTET2) for raw commingled milk – Report of ILVO-T&V, Melle, Belgium: 1–16.
- USDA. 2002. Aflatoxin handbook [Internet]. Washington (DC): USDA, Grain Inspection, Packers and Stockyards Administration, Federal Grain Inspection Service; p. 1–9 [cited 2014 Jul]. Available from: http://www.gipsa.usda.gov/publications/fgis/handbooks/afl_insphb.html
- van Egmond HP, Svensson UK, Fremy JM. 1997. Mycotoxins. In: Residues and Contaminants in Milk and Milk Products (Special Issue 9701). Brussels: International Dairy; p. 79–88.
- Veldman A, Meijst JAC, Borggreve GJ, Heeres-van der Tol JJ. 1992. Carry-over of aflatoxin from cows’ food to milk. Anim Prod. 55:163–168.
- Wogan GN. 1966. Chemical nature and biological effects of the aflatoxins. Bact Rev. 30:460–470.
- Wogan GN, Paglialunga S. 1974. Carcinogenicity of synthetic aflatoxin M1 in rats. Food Cosmet Toxicol. 12:381–384.
- Wong JJ, Hsieh DPH. 1976. Mutagenicity of aflatoxins related to their metabolism and carcinogenic potential. Proc Natl Acad Sci. 73:2241–2244.
