?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Database systems are pervasive components in the current big data era. However, efficiently managing and querying grid-based or array-based multidimensional climate data are still beyond the capabilities of most databases. The mismatch between the array data model and relational data model limited the performance to query multidimensional data in a traditional database when data volume hits a cap. Even a trivial data retrieval on large multidimensional datasets in a relational database is time-consuming and requires enormous storage space. Given the scientific interests and application demands on time-sensitive spatiotemporal data query and analysis, there is an urgent need for efficient data storage and fast data retrieval solutions on large multidimensional datasets. In this paper, we introduce a method for multidimensional data storing and accessing, which includes a new hash function algorithm that works on a unified data storage structure and couples with the memory-mapping technology. A prototype database library, LotDB developed as an implementation, is described in this paper, which shows promising results on data query performance compared with SciDB, MongoDB, and PostgreSQL.
1. Introduction
In climate earth community, with the growth of both data volume and complexity of spatiotemporal gridded data, the importance of efficient data storage, fast access, and analysis are well recognized by both data producers and data users. Using new technologies to enhance big earth data discovery, storage, and retrieval is attractive and demanded not only in the climate earth community but also in all science domains and data-related industries (Yang et al. Citation2017). As the nature of many natural phenomena, they can be modelled as array data sets with some dimensionality and known lengths (Baumann Citation1999). Grid-based data sets, raster structured data sets, array-based data sets or data cubes are all referring to the same concept of a data model, array data model. It is a multidimensional (n-D) array with values in the computer science context. In geoscience, it is an n-D gridded data set. The Earth Observation data is one of the main gridded data sources in climate earth community, according to Earth Observing System Data and Information System (EOSDIS)’s metrics of 2014, it manages over 9 PB of data and adds 6.4 TB of data to its archives every day (Blumenfeld Citation2015). Its data distribution is often in forms of flat files, and almost all the data are array based. Traditional file storage for multidimensional arrays involves using various formats such as the popular GeoTiff, NetCDF, and HDF. The data in those formats are suitable for data transfer because of their size advantages but require programming skills to perform further analysis. In addition to the original formats, data are often needed to be stored using DBMSs, and the mainstream technologies for sensing image management are relational database and array database technologies (Tan and Yue Citation2016). For DBMSs, a well-developed relational database management system (RDMS) is based on the relational data model, and the database storage structures on lower storage level are often trees (B+ trees). Historically, since traditional databases are not designed in the climate earth domain, nor are they built to store array data sets, RDMS does not directly support the array data model to the same extent as sets and tables. Mapping attempts have been made to utilize the mature relational model as the backend to support array datasets, like in van Ballegooij’s work (Citation2004), and theories have been developed to support array query (Libkin, Machlin, and Wong Citation1996; Marathe and Salem Citation1997); however, with the growth in data volume, storing, accessing, and analysing multidimensional arrays in RDMS became problematic.
Meanwhile, NoSQL databases have been developed to meet the demand to store and query the increasing amount of data from various sources, in which, array data could also be mapped to different data models, like using key-value pairs to store cells of the array. However, the data model mapping brings longer data pre-processing time and larger data storage volume. At the same time, as the data volume and complexity increase, there is no promising performance to be expected because of the corresponding increase in unit computer resource consumption. Recently, the array database is drawing increasing attention because of its array data model’s support on the database level, which provides a more native solution for array data storage. Tree data structures, which are usually used in relational databases’ storage level on disks, have lower computational complexities for value search when comparing with an array data structure. In contrast, the array data structure offers the lowest computational complexity for data access, perfect for static data retrieve if indexes could be presented as integers. Popular array databases like Rasdaman and SciDB are widely used in many practical projects (Baumann et al. Citation1998; Stonebraker et al. Citation2013b), showing promising performance gain comparing to traditional methods. However, existing array databases are still suffering from the problems including 1) low performance on non-cluster environment: the standalone version of array databases usually have limited performance compare to their clustered setup (Hu et al. Citation2018), 2) long data pre-processing time: current array databases does not provide direct support for multidimensional climate gridded data, data pre-processing is therefore necessary and cannot be avoided (Das et al. Citation2015), and 3) high data expansion rate compare to raw data: because of the data pre-processing and data chunking inside databases, data volume could be expanded multiply times compare to the raw data.
As the multidimensional array datasets are growing in both size and complexity, we are becoming limited in our abilities to handle those datasets. Our objective is to develop a solution that performs fast data retrieve queries on grid-based climate data and minimizes the data pre-processing time and data expansion rate at the same time. In this paper, we present LotDB, a prototype array database library, developed by utilizing memory-mapping technology and a new n-dimensional hash function algorithm. The query run-time and storage consumption are assessed and compared against PostgreSQL, MongoDB, and SciDB for performance evaluation by using the Modern-Era Retrospective analysis for Research and Applications, Version 2 (MERRA-2) data set.
1.1. Outline
The rest of this paper is organized as follows. In Section 2, the studies of storing arrays in databases are reviewed. Section 3 introduces the n-dimensional hash function as an efficient method to execute data retrieve for a unified data storage structure. LotDB is introduced and compared with PostgreSQL, MongoDB, and SciDB in Section 4. Section 5 concludes this research with a summary and an outlook for future works.
2. Related work
There are many attempts to store multidimensional arrays in a database system, studies could be generalized as 1) array data model mapping in relational databases, and 2) developing native array databases. Meanwhile, recent studies that involve using big data frameworks like Hadoop systems to store and retrieve multidimensional climate data sets have some excellent performance results on data analysis queries (Li et al. Citation2017; Cuzzocrea, Song, and Davis Citation2011; Song et al. Citation2015). However, most of the solutions were developed as customized solutions for specific data formats or data sets and could not be applied to arbitrary multidimensional arrays. The related works in this section focus on the database solutions for multidimensional array storage and retrieval, and efforts done by researchers to enhance array data query.
2.1. Relational database
The data model in relational databases are tables, most of the relational database systems do not support storing multidimensional arrays natively. A natural way to store arrays in tables is to split arrays cell into rows, which is straightforward for 1D/2D datasets, however, problematic for higher dimensional data. Operational speaking, array orientated analysis is often done on platforms like MATLAB, which is limited by memory capacity and may have error-prone developing process. To close this gap, in 2004, van Ballegooij introduced array data structure in a relational database environment, developed a multidimensional array DBMS called RAM (Relational Array Mapping) which maps array onto a traditional relational schema. The multiple index columns in a multidimensional array were compressed into a single column in their approach. Data retrieve process was based on existing database system with bulk array processing. They provided an intellectual framework and ease the use for map arrays to relational models in a distributed application as the implementation (van Ballegooij et al. Citation2005). However, mapping the array model to relational could only be treated as the compromised solution rather than the ideal solution.
In 2005, the prototype of MonetDB was introduced as a main memory database system uses a column-at-a time execution model for the data warehouse. Although MonetDB is not a native array database but a full-fledged relational DBMS (Idreos et al. Citation2012), it provided useful thoughts and ideas for processing array models. It is a column-oriented database and each column, or BAT (Binary Association Table), in the database is implemented as a C-array on storage level. (Boncz, Zukowski, and Nes Citation2005) MonetDB has focused on optimizing the major components of traditional database architecture to make better use of modern hardware in database applications that support analyse massive data volumes (Boncz, Kersten, and Manegold Citation2008). In 2008, Cornacchia et al. also introduced an example of using a matrix framework with a Sparse Relational Array Mapping (SRAM) by Information Retrieval (IR) researchers, they used MonetDB/X100 as the relational backend, which provided fast response and good precision. The matrix framework is based on the array abstraction, and by mapping them onto the relational model and develop array queries, MonetDB allows them to optimize the performance and developed a high-performance IR application.
2.2. Array database
Research on array DBMS usually includes two parts, the storage of multidimensional array and query language (Baumann et al. Citation1999). While online analytical processing (OLAP) focuses on business array data, scientific studies on computing and imaging have been developed formal concepts on array data manipulation, like AQL (Libkin, Machlin, and Wong Citation1996) and the array manipulation language AML (Marathe and Salem Citation1997), but not been implemented and evaluated as real-life applications before Rasdaman (Baumann Citation1999). A declarative query language suitable for multiple dimensions was firstly introduced by Libkin, Machlin, and Wong (Citation1996), Rasdaman introduced its algebraic framework in 1999 for express cross-dimensional queries, for which consists of only three operations: an array constructor, a generalized aggregation, and a multidimensional sorter. On data model level, (Baumann et al. Citation1999) Rasdaman integrates array model into existing overall model, which is based on a formal algebraic framework and developed with declarative multidimensional discrete data (MDD). MDD is the data storage unit in Rasdaman. On storage level, binary large objects (BLOBs) are the smallest units of data access (Reiner et al. Citation2002), and size stored in Rasdaman is from 32KB to 640 KB. MDD object is divided into arbitrary tiles or subarrays and combined with a spatial index to access the tile object used by a query (Baumann et al. Citation1998).
Two years later, SciDB was first debuted in 2009 by Cudré-Mauroux et al. It provides a native array data structure on storage level and supports for clustering (Brown Citation2010). Different from Rasdaman (which store chunking information in the relational model), the metadata, location information of chunks are also stored within the logical array. The size of a chunk could be an order of 64 MB, which is much larger than Rasdaman and MonetDB. Since the chunks are potentially overlapped, this characteristic increases the total data volume stored in the database (Brown Citation2010). SciDB also provided a new way of dealing will ‘not valid’ cells, which no space is allocated in the array data chunk for such cells (Stonebraker et al. Citation2013a). At the same time, SciDB optimizes CPU performance by performing vector processing, which was from the contribution of MonetDB. The version 1 of SciDB was released in 2010, same year, a science benchmark paper (SS-DB) was published which included a comparison between SciDB and MySQL (Cudre-Mauroux et al. Citation2010), SciDB presented a high-performance potential. In 2012, Planthaber, Stonebraker, and Frew (Citation2012) provided an integrated system of using SciDB to analyse MODIS level 1B data. They pre-processed MODIS data from HDF format to CSV, then CSV to SciDB DLF files as a system. The SciDB engine showed a promising performance and being the most powerful competitor of Rasdaman. Three years after SciDB firstly announced, a SQL-based query language called SciQL was introduced in the relational model for scientific applications with both tables and arrays as first-class citizens (Kersten et al. Citation2011; Zhang et al. Citation2011). In nature, it is an array extension to MonetDB and makes MonetDB effectively function as an array database. However, it still involves data model mapping and is not a fully operational solution.
In 2013, Rasdaman added the in-situ support for tiff files and NetCDF files (Baumann, Dumitru, and Merticariu Citation2013). Same year, Dr. Stonebraker et al. (Citation2013b) pointed out the SciDB research directions are now in the areas of elasticity, query processing, and visualization, also presented an example of using SciDB for MODIS processing pipeline. Additional researches have been done in the recent years to customize SciDB. For example, Soroush and Balazinska (Citation2013) developed TimeArr as a new storage manager for SciDB, for which speed up the process of creating or selecting a specific version of arrays, allow researches explore how time changes affect the cell values in a specific array section.
To sum up, without the support of standard databases, scientists tend to employ customized programs or file-based implementations serving and analysing array data (Baumann, Dumitru, and Merticariu Citation2013). Array DBMSs like Rasdaman (Baumann 1998), SciDB (Cudré-Mauroux et al. Citation2009), and SciQL (Kersten et al. Citation2011) fill this demand by extending the supported data structure in the database with unlimited-size multidimensional arrays. However, mapping array data model to the relational model is a performance sacrifice, limited the data access ability. Also, although data tiling and chunking are good for clustering and mapping to the non-array data structure, it causes the problem of data volume increase and decreases the I/O performance on secondary storage by using random access instead of sequential access.
3. Methodology
Array databases have shown potentials to be valid solutions for storing and manipulating multidimensional array data. However, current solutions fail to provide good support for multidimensional climate gridded data and are facing issues related to overall performance. In this section, a new data management solution is presented as the foundation to build an array database. In general, a valid data management solution for data storage and retrieval includes 1) a data storage structure and 2) a data access and update method. Specifically, the solution should have the essential functions of data manipulating: create, read, update, and delete (CRUD). To feed the needs, a unified data storage structure is adopted for persistent storage of data on secondary-storage, and an n-dimensional hash function is proposed as the main algorithm for data manipulation.
3.1 A unified data storage structure
For array data structure on entity-level, the array data structure provides complexity for single data retrieval; this is the fastest way computers can achieve when retrieving something. Traditional ways involve tiling and chunking big arrays (Baumann et al. Citation1999; Boncz, Zukowski, and Nes Citation2005; Cudré-Mauroux et al. Citation2009). Typically, tiling and sub-setting are necessary for array database, like in Rasdaman, the size of its smallest element is from 32KB to 640KB, as the order of default page file size (4KB). However, arrays are not suitable for fragmenting because it will increase data access and search complexity. Since disks are designed to do the sequential reading, too small and too much tilling is not good for I/O performance. In 2002, Reiner developed a
tree for MDD tile node searching (Reiner et al. Citation2002), however, since the storage rectangles are duplicated,
tree increases the data volume and makes construction and maintenance more expansive. In computational complexity concern, data retrieve complexity will be
. Ideally, an multidimensional array will provide
complexity when retrieving data from it. However, because of tiling and sub-setting, the complexity for search increases considerably if the tiling unit is small and the whole dataset is large. Meanwhile, when a multidimensional array is stored in a secondary storage device, data files are often not sequentially stored on the device and the cost to access different byte address is different. Although the computational complexity is the same for each cell in an array, the cost of finding the index and let the disk head (in HDD) to retrieve is different. Even for SSDs, which does not have a physical head inside, random access and sequential access varies largely in speed. Since the ability to do random read/write and sequential read/write is different, storing data bytes closer to each other will have better performance for data read when they are retrieved together. Therefore, instead of chunking and tiling, putting a multidimensional array in a holonomic array form should have a performance gain for sequential data retrieval.
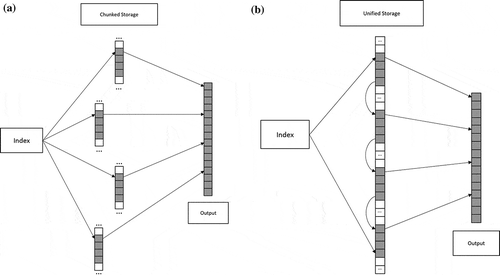
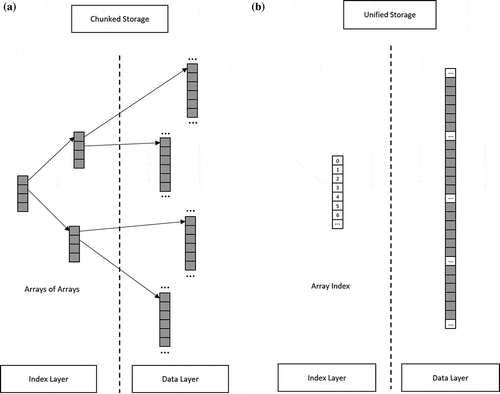
illustrates the difference in storage structure between a chunked storage and a unified storage. The chunked storage requires additional indexing for data storage and retrieval; however, a unified storage has the native array index as its storage index. Another benefit of using a unified storage is the possibility of saving storage space. Specifically, if a multidimensional array has
dimensions, and the size for the
dimension is
. Then when it stores in a unified form, it takes the
space in terms of bytes as expressed in the following:
Figure 1. Chunked storage and unified storage (a) chunked storage with multilayer array indexing, (b) unified storage with no extra indexes
It is the minimum space an array will cost without any compression. For example, if a 5-D array (32-bit float) with dimensions will cost about 33KB. The HDF and NetCDF file formats use similar structures to hold multidimensional array along with other information integrated. In practical use, it requires additional programming skills to access the data set, and a full data set load to memory space. Although a specific cell location could be calculated through its dimension sizes, there is no indexing function to map from the original multidimensional array to its unified form for subsetting.
3.2 An N-dimensional hash function for the unified storage structure
The unified storage structure provides a one-dimensional native index for each cell in it. Querying an n-dimensional array needs a hash function to map the n-dimensional index to the one-dimensional native index. In this section, we propose an n-dimensional Hash Function Algorithm to fill the demands for fast data retrieval based on the unified array data storage structure.
Definition:
is a multidimensional array with
dimensions.
The dimensions form an ordered set as
.
The data storing order follows the same order as
. The size of each axis forms another ordered set as
.
Query a subset of
is expressed as
‘Find)’
Name: Computation of 1-D storage index from n-D query index
Input:
Output:
Procedure:
(1) Compute the first element of
:
(2) Compute the last element of
:
(3) If (
) then {
Return }
(4) Else: index
(5) Compute array
, for
(6) For (
) do {
For () do {
For () do {
…
For () do {
Appending as the last element of
If () then {
}
}
…
If () then {
}
}
If () then {
}
}
If () then {
}
}
(7) Return
As a result, if the total number of data amount for retrieve is , the n-dimensional hash function will hold an
complexity. shows the difference between the idea of traditional indexing method and the idea of the proposed indexing method. In the traditional indexing method, executing a data retrieve query forces the index to locate the initial pointers to each chunk and return with the integrated output. In the proposed indexing method, the indexing system is only used to calculate the first and last elements and a jumping step set
and data are retrieved sequentially with specific jumping steps. Typically, algorithms are designed independently from hardware (H/W), which may lead to a diminishing return of actual performance. The intention to design this algorithm is to consider the H/W limit while performing data retrieve action. Specifically, by implementing a sequential-like data retrieval action, the n-dimensional hash function algorithm will utilize the sequential read ability in disks when it is possible. It is similar to the motivation of building MonetDB to break the memory wall (Boncz, Kersten, and Manegold Citation2008).
4. Implementation and experiments
4.1. System design
LotDB is a prototype array database library developed with C++ and the memory-mapping technology; it is an implementation of the n-dimensional hash function and unified data storage design. It follows a light-weight, standalone, and single-use design. LotDB acts as both a client-based database manager and a database library for applications on top, like Google’s LevelDB.
4.1.1. Memory-mapping technology
In LotDB, data are stored in the secondary storage system, and the access to it is done by utilizing memory-mapping technology and through page files. This technology is widely used in database systems like LMDB and MongoDB. Specifically, instead of loading the whole file into memory, the file handler maps the file to virtual memory as a big array and assign a virtual memory address to each page file without loading any actual data into the memory other than file’s metadata. When a data access call is made for a page file, it will cause a page fault and enable read/write of the secondary storage. In this way, bytes are copied to actual memory addresses directly, no need to go through disk caches as the standard open/write will do. In addition, by utilizing memory-mapping of arrays, LotDB could exceed the memory cap for accessing large data files and makes it possible for LotDB accessing big arrays without tiling. Meanwhile, when integrating with the n-dimensional hash function, the array indexes could be virtually calculated with low costs and could increase data retrieve speed exponentially when compared with traditional database solutions.
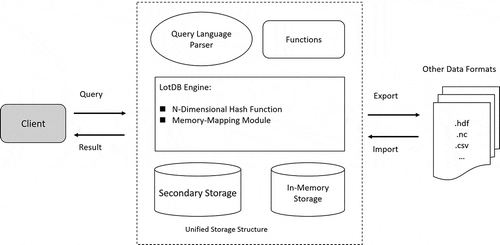
4.1.2. LotDB system architecture
The general architecture of LotDB is shown in . The system core is the LotDB engine, which consists of the n-dimensional hash function algorithm and memory-mapping module. Data are stored in files and separated from their metadata files; data retrieve queries are called through built-in functions, and results are stored in memory. Functions are designed to perform different calculations and services, including upload multidimensional arrays directly from their original formats like HDF or NetCDF, etc. Meanwhile, the query communications are done through a query language parser and a client. For spatiotemporal grid-based climate datasets, they are often produced and packaged without the demand of over-write. The empty cells in a grid-based climate data product do not require adjustments when storing, such cells or usually referred as noise will only be eliminated when the analysis is performed and will stay with the original data. LotDB is designed to meet this characteristic of climate datasets, it will be fast to store static array data which does not change once captured, and the drawback for this is that data updating will be very expensive if it involves changes in array shape or size and the empty cells cost same storage spaces as the non-empty cells. Although keeping the empty cells does waste storage space and not ideal for sparse data, it accelerates the data retrieval by simplifying the data storage architecture on disks.
4.2. Experiment design and performance evaluation
4.2.1. Experiment setup and design
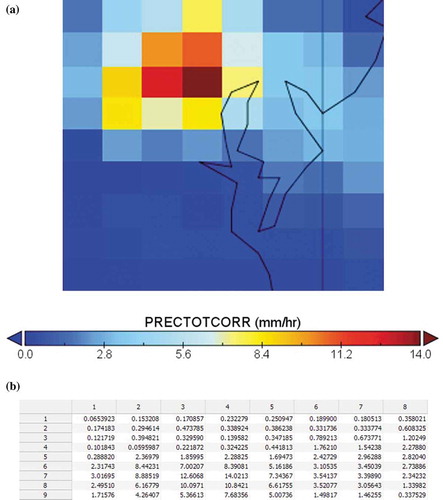
The MERRA-2 dataset is collected and selected as the experiment data, which is produced and provided by Global Modelling and Assimilation Office of NASA Goddard Space Flight Centre. MERRA-2 dataset is stored in NetCDF4 format and contains about 49 variables (e.g. Surface Wind Speed, Precipitation, Surface Air Temperature, etc.). PRECTOTCORR is chosen as the variable to use in this experiment, which is the bias-corrected precipitation output from an atmospheric model. The spatiotemporal resolution of this variable is 0.625 degree by 0.5 degree with hourly reads for each day of the year. The year selected for this experiment is 2017, and the dataset is a 4D array with dimensions , and its datatype is 32-bit float. This one-year dataset contains around 1.8 Billion grid points. As an example, shows the plot view and the array view of this dataset at one time step in Chesapeake Bay area.
To compare the performance of LotDB with other databases, three popular databases are chosen; specifically, they are PostgreSQL 9.3 (Relational Database), MongoDB 4.0.4 (NoSQL Database), and SciDB 18.1 (Array Database). PostgreSQL is an open source object-relational database management system, it has been launched for 30 years and maintained a very stable performance in different domains. MongoDB is a document-oriented NoSQL database system, which follows a schema-free design and is one of the most popular databases for modern applications. SciDB, as mentioned in the previous section, is a high-performance array database that designed specifically for storing and querying scientific datasets. All these databases are installed as standalone modes on individual servers with the same hardware configuration: Intel Xeon CPU X5660 @ 2.8 Ghz×24 with 24GB RAM size and 7200 rpm HDD, installed with CentOS 6.6 or Ubuntu 14.04. Data were uploaded to each database, and pre-processing work was done for databases that do not support NetCDF format. The databases are evaluated in the following aspects: (1) data uploading and pre-processing time, (2) data storage consumption, and (3) spatiotemporal query run-time. Different spatiotemporal queries are designed to evaluate the performance of selected databases () for the year 2017 with raw data size be 3.45 GB. Different raw data sizes were chosen to evaluate the data storage consumption in different databases in additional to 3.45 GB, specifically, they are 10 MB, 100 MB, 1 GB, and 10 GB. The number of grid points is the estimated number of array cells to be retrieved for the corresponding query. The query run-time refers to the elapsed real time or wall-clock time in this experiment.
Table 1. Spatiotemporal queries
4.2.2. LotDB system architecture
PostgreSQL and MongoDB do not have native support for NetCDF format, and SciDB does not have a stable and fully functional plugin for NetCDF data import. Therefore, data were converted into the CSV format and then uploaded to each database. Meanwhile, LotDB is developed with data import functions to upload multidimensional data directly from the NetCDF/HDF format. In order to accelerate the pre-processing process, SSDs are utilized to store the raw datasets and intermediate results. Then, the pre-processed results were uploaded to servers. lists the processing time and intermediate data size in CSV format, from 120 MB to 168 GB as the raw data increases from 10 MB to 10 GB.
Table 2. Pre-processing data (from NetCDF to CSV)
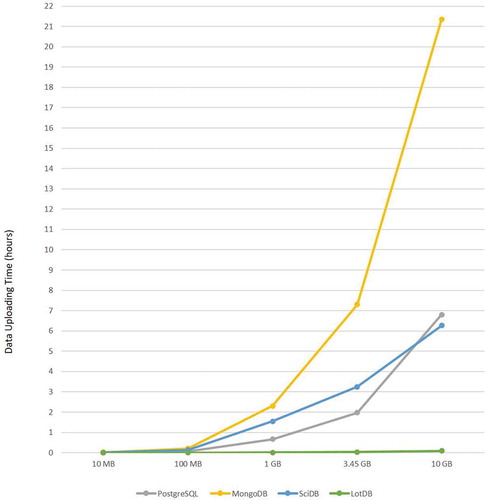
After the pre-processing, PostgreSQL, SciDB, and MongoDB all require additional time for data uploading. The detailed time variation is shown in . The vertical axis records the data uploading time to each database from CSV files, and the horizontal axis represents different uploading cases for different raw data size. The data uploading time in this figure is independent from the pre-processing time. As the figure shows, MongoDB took the longest time for data uploading in almost all the cases. Meanwhile, LotDB used the least time amount for data uploading because of its native support for NetCDF data files and unified data storage design. As a representative of relational database in this experiment, PostgreSQL shows a stable increase in uploading time as raw data size increases, it has an advantage when dealing with small amount of array datasets compare to MongoDB and SciDB. SciDB is increasing in its data uploading time with a decrease speed change, which implies a potential advantage on handling larger datasets. MongoDB has the highest rate of time complexity while LotDB has an almost linear rate for data uploading. As designed in LotDB data import function, data are dumped directly from the raw dataset if it was stored linearly in multidimensional array data formats like NetCDF. Therefore, it is not surprised that LotDB has significant advantages in multidimensional data uploading.
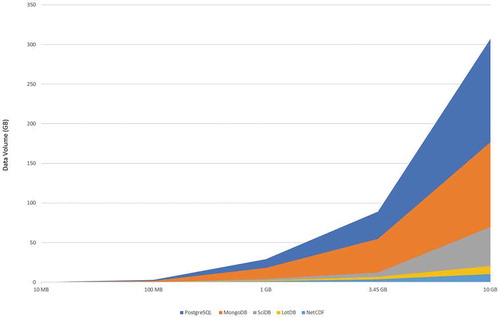
The corresponding data storage volume in different containers for different cases is also recorded and displayed in . The graph compares the difference in data expansion from raw data format to data storage volume in different databases. It is observed that although MongoDB took the longest time to upload data, it didn’t use the most storage space; instead, PostgreSQL consumed the largest storage space in all cases, and data volume increased about 20 to 30 times from the raw data size. SciDB used much less space and has a data expansion rate around 5. LotDB performed the best in all cases and has a 2 times data expansion rate; this meets the design of the unified storage structure and shows significant advantages compare to other tested databases. As data are stored in a unified storage structure in LotDB, no extra indexes or data chunks are needed. PostgreSQL and MongoDB are using non-array data model to store arrays, extra storage for indexes are expected. SciDB uses an over-lapped chunking design, which costs additional storage space for the redundant part.
Consider the performance for different database on spatiotemporal queries, MERRA-2 data set for the year 2017 was chosen. lists the detailed data pre-processing and uploading time for this specific case. Among these data containers, MongoDB took the longest time, and PostgreSQL required the largest space for data storage. By implementing the array as the fundamental data structure, SciDB and LotDB acquired much less storage. LotDB holds the top place both in data uploading time, and data storage size compare with the other three.
Table 3. Data pre-processing & uploading time and data size in different containers
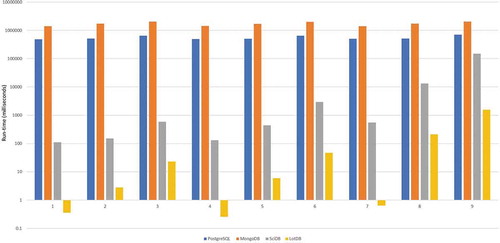
illustrates the spatiotemporal query run-time across four containers for nine queries. In terms of performance, LotDB used the shortest time in all the queries even compared with SciDB. The queries were designed to increase in its complexity both spatially and temporally; the general run-time patterns of PostgreSQL and MongoDB are similar to each other. They both tend to be stable in a certain range, the cost to do a simple query as Q1 is not much different from a complex query as Q9, although the number of data points retrieved varies largely. It implies that there exist some initial costs for each spatiotemporal query when executed in both databases. MongoDB is a document-based database, and PostgreSQL is a relational database. Both of them are not expected to have better performance than SciDB because their data models are mismatching with the array data model, which also agrees with the results from our previous studies on different data containers’ abilities for handling big multidimensional array dataset (Hu et al. Citation2018). As an insight for MongoDB, it tends to memory-map the whole collection into the physical memory of the machine during the query. When the total size of the dataset is enormously larger than the physical memory, a large number of memory page faults are observed and thus delayed query speed. As an array database, SciDB has much better performance outputs than PostgreSQL and MongoDB and proved to be one of the best databases on the market when handling multidimensional arrays. For LotDB, it has a significant time advantage among all selected databases. The reason behind the outstanding performance is due to the following: 1) LotDB implemented a native array data structure on the storage level, 2) memory-mapping technology is utilized for data retrieval on byte level, 3) query process is accelerated by the N-Dimensional hash function.
5. Conclusion and future works
Although array is one of the oldest data structures, the study of storing and retrieving large multidimensional array datasets are limited. As earth observations and climate model simulations are producing larger and larger outputs in forms of multidimensional arrays due to increases in model resolution and remote sensing technologies, it is challenging to provide solutions to handle big multidimensional datasets. The primary goal of this research was to design and implement a solution for efficient gridded data storage and faster data retrieval. We reviewed past attempts for storing multidimensional arrays in both relational databases and array databases, although array databases are more native and advanced solutions than relational databases, current solutions still have their own limitations regarding query performance and data volume expansion problem. Therefore, an n-dimensional hash function algorithm was proposed to perform a fast data retrieval action on a unified data storage structure, and a prototype database library (LotDB) was developed as an implementation, which integrated memory-mapping technology and this algorithm. PostgreSQL, MongoDB, and SciDB were selected to compare the performance with LotDB on MERRA-2 data storage and retrieval. The preliminary experimental results have shown promising potentials of LotDB for efficient multidimensional gridded data storage, and abilities for fast data retrieval. The run-time results are validated by using multiple timers and repeating the same experiments several times. In addition, to avoid the effect of using in-memory cache, physical memory is cleaned each time before the query execution and cold-run time is recorded instead of hot-run. Therefore, the results are credible for general analysis. However, the standalone mode for NoSQL databases are far less potent than the clustered mode, MongoDB and SciDB would have better results if they were deployed in a cluster.
The work presented in this paper shows a potential for seeking fast gridded data retrieve and efficient storage solutions using existing technologies; however, it is challenging to provide up-to-date solutions as the data size is also growing in an increasing speed. There are many directions for related future works, include but not limited to: (1) design a strategy for big data store and retrieve, (2) design and develop LotDB into a complete database system, (3) extend current storage structure and algorithm to a distributed system. Current version of LotDB acts as a fast implementation of the N-Dimensional hash function algorithm, further implementations could be developed to enhance the data retrieve performance in large multidimensional arrays in different scenarios.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Baumann, P. 1999. “A Database Array Algebra for Spatio-temporal Data and Beyond.” In International Workshop on Next Generation Information Technologies and Systems, 76–93. Berlin, Heidelberg: Springer.
- Baumann, P., A. Dehmel, P. Furtado, R. Ritsch, and N. Widmann. 1998. “The Multidimensional Database System RasDaMan.” Acm Sigmod Record 27 (2): 575–577. doi:10.1145/276305.
- Baumann, P., A. Dehmel, P. Furtado, R. Ritsch, and N. Widmann. 1999. “Spatio-temporal Retrieval with RasDaMan.” In VLDB, 746–749. The 25th VLDB Conference, Edinburgh, Scotland.
- Baumann, P., A. M. Dumitru, and V. Merticariu. 2013. “The Array Database that Is Not a Database: File Based Array Query Answering in Rasdaman.” In International Symposium on Spatial and Temporal Databases, 478–483. Berlin, Heidelberg: Springer.
- Blumenfeld, J., 2015. Getting Petabytes To People: How EOSDIS Facilitates Earth Observing Data Discovery And Use | Earthdata. [online] Earthdata.nasa.gov
- Boncz, P. A., M. Zukowski, and N. Nes. 2005. “MonetDB/X100: Hyper-Pipelining Query Execution.” In Cidr 5: 225–237.
- Boncz, P. A., M. L. Kersten, and S. Manegold. 2008. “Breaking the Memory Wall in MonetDB.” Communications of the ACM 51 (12): 77–85. doi:10.1145/1409360.1409380.
- Brown, P. G. 2010. “Overview of SciDB: Large Scale Array Storage, Processing and Analysis.” In Proceedings of the 2010 ACM SIGMOD International Conference on Management of Data, 963–968. Indianapolis, Indiana: ACM.
- Cudre-Mauroux, P., H. Kimura, K.-T. Lim, J. Rogers, S. Madden, M. Stonebraker, S. B. Zdonik, and P. G. Brown. 2010. “Ss-db: A Standard Science Dbms Benchmark.” Under submission.
- Cudré-Mauroux, P., K.-T. Hideaki Kimura, J. R. Lim, R. Simakov, E. Soroush, P. Velikhov et al. 2009. “A Demonstration of SciDB: A Science-oriented DBMS.” Proceedings of the VLDB Endowment 2 (2), 1534–1537.
- Cuzzocrea, A., I.-Y. Song, and K. C. Davis. 2011. “Analytics over Large-scale Multidimensional Data: The Big Data Revolution!” In Proceedings of the ACM 14th international workshop on Data Warehousing and OLAP, 101–104. Glasgow Scotland: ACM.
- Das, K., T. Clune, K. S. Kuo, C. A. Mattmann, T. Huang, D. Duffy, C. P. Yang, and T. Habermann, 2015. “Evaluation of Big Data Containers for Popular Storage, Retrieval, and Computation Primitives in Earth Science Analysis.” In AGU Fall Meeting Abstracts, December. San Francisco.
- Hu, F., X. Mengchao, J. Yang, Y. Liang, K. Cui, M. Little, C. Lynnes, D. Duffy, and C. Yang. 2018. “Evaluating the Open Source Data Containers for Handling Big Geospatial Raster Data.” ISPRS International Journal of Geo-Information 7 (4): 144. doi:10.3390/ijgi7040144.
- Idreos, S., F. Groffen, N. Nes, S. Manegold, S. Mullender, and M. Kersten. 2012. “MonetDB: Two Decades of Research in Column-oriented Database.”
- Kersten, M., Y. Zhang, M. Ivanova, and N. Nes. 2011. “SciQL, a Query Language for Science Applications.” In Proceedings of the EDBT/ICDT 2011 Workshop on Array Databases, 1–12. Uppsala, Sweden: ACM.
- Li, Z., H. Fei, J. L. Schnase, D. Q. Duffy, T. Lee, M. K. Bowen, and C. Yang. 2017. “A Spatiotemporal Indexing Approach for Efficient Processing of Big Array-based Climate Data with MapReduce.” International Journal of Geographical Information Science 31 (1): 17–35. doi:10.1080/13658816.2015.1131830.
- Libkin, L., R. Machlin, and L. Wong. 1996. “A Query Language for Multidimensional Arrays: Design, Implementation, and Optimization Techniques.” ACM SIGMOD Record 25 (2): 228–239. ACM. doi: 10.1145/235968.
- Marathe, A. P., and K. Salem. 1997. “A Language for Manipulating Arrays.” VLDB 97: 46–55.
- Planthaber, G., M. Stonebraker, and J. Frew. 2012. “EarthDB: Scalable Analysis of MODIS Data Using SciDB.” In Proceedings of the 1st ACM SIGSPATIAL International Workshop on Analytics for Big Geospatial Data, 11–19. Redondo Beach, California: ACM.
- Reiner, B., K. Hahn, G. Höfling, and P. Baumann. 2002. “Hierarchical Storage Support and Management for Large-scale Multidimensional Array Database Management Systems.” In International Conference on Database and Expert Systems Applications, 689–700. Berlin, Heidelberg: Springer.
- Song, J., C. Guo, Z. Wang, Y. Zhang, Y. Ge, and J.-M. Pierson. 2015. “HaoLap: A Hadoop Based OLAP System for Big Data.” Journal of Systems and Software 102: 167–181. doi:10.1016/j.jss.2014.09.024.
- Soroush, E., and M. Balazinska. 2013. “Time Travel in a Scientific Array Database.” In 2013 IEEE 29th International Conference on Data Engineering (ICDE), 98–109. Brisbane, QLD, Australia: IEEE.
- Stonebraker, M., J. Duggan, L. Battle, and O. Papaemmanouil. 2013b. “SciDB DBMS Research at MIT.” IEEE Data Engineering Bulletin 36 (4): 21–30.
- Stonebraker, M., P. Brown, D. Zhang, and J. Becla. 2013a. “SciDB: A Database Management System for Applications with Complex Analytics.” Computing in Science & Engineering 15 (3): 54. doi:10.1109/MCSE.2013.19.
- Tan, Z., and P. Yue. 2016. “A Comparative Analysis to the Array Database Technology and Its Use in Flexible VCI Derivation.” In 2016 Fifth International Conference on Agro-Geoinformatics (Agro-Geoinformatics), 1–5. Tianjin, China: IEEE.
- van Ballegooij, A., R. Cornacchia, A. P. de Vries, and M. Kersten. 2005. “Distribution Rules for Array Database Queries.” In International Conference on Database and Expert Systems Applications, 55–64. Berlin, Heidelberg: Springer.
- van Ballegooij, A. R. 2004. “RAM: A Multidimensional Array DBMS.” In International Conference on Extending Database Technology, 154–165. Berlin, Heidelberg: Springer.
- Yang, C., Q. Huang, L. Zhenlong, K. Liu, and H. Fei. 2017. “Big Data and Cloud Computing: Innovation Opportunities and Challenges.” International Journal of Digital Earth 10 (1): 13–53. doi:10.1080/17538947.2016.1239771.
- Zhang, Y., M. Kersten, M. Ivanova, and N. Nes. 2011. “SciQL: Bridging the Gap between Science and Relational DBMS.” In Proceedings of the 15th Symposium on International Database Engineering & Applications, 124–133. Lisboa, Portugal: ACM.