ABSTRACT
The heat-labile toxin (LT) is one of the major virulence factors of enterotoxigenic Escherichia coli (ETEC). We recently described that 20 polymorphic LT variants are present in ETEC strains isolated globally. Two of the variants, LT1 and LT2, are particularly common and we found that they were associated with clonal ETEC lineages that express the colonization factors (CFs), CFA/I, CS1+CS3, CS2+CS3, and CS5+CS6. ETEC expressing these CFs are frequently found among ETEC strains isolated from cases with diarrhea. ETEC expressing the colonization factors CS1+CS3, and CS2+CS3 are found in 2 discrete clonal lineages and express the LT1 variant and heat stable toxin (STh). Although they clearly are virulent they neither produce, nor secrete, high amounts of LT toxin. On the other hand ETEC strains expressing LT, STh, CFA/I and LT, STh, CS5+CS6, carry the LT2 variant and produce and secrete significantly more LT toxin. Despite differences in toxin production, LT1 and LT2 are found in ETEC lineages that have managed to spread globally confirming that these variants are important for ETEC virulence.
Enterotoxigenic Escherichia coli heat labile toxin, and its natural variants
Enterotoxigenic Escherichia coli ETEC continues to be a major cause of diarrheal disease in children and adults and may also cause food-borne outbreaks in developed countries.Citation1 ETEC cause watery diarrhea through the actions of 2 toxins, the heat-stable toxin (ST), and the heat-labile toxin (LT). ETEC strains either express ST, LT, or a combination of ST/LT.Citation2,3 ETEC strains also show a large heterogeneity in expression of colonization factors (CFs) that mediate adherence to the human epithelium.Citation4 The ST toxin is divided into 2 subgroups, the STh and STp toxins. STh and STp are small non-immunogenic peptides containing 19 and 18 amino acids, respectively. The heat labile toxin, LT, encoded by the eltAB operon, is composed of an A subunit (LTA) and 5 B subunits (LTB) and forms a typical AB5 toxin. The pentameric B subunit binds to host receptors i.e GM1 and blood groups in the human intestine causing the toxic A subunit to be internalized.Citation5 This causes an increase in intracellular cyclic AMP (cAMP), which leads to deregulation of the cystic fibrosis conductance regulator (CFTR) and sub-sequent secretion of water and ions from the epithelium.Citation6
The LT toxin is polymorphic and different ETEC strains express various allele variants (in the present article, we discuss only the toxin that are neutralizable by anti-cholera toxin sera, i.e., LT-I toxins, and not the more distantly related LT-II toxins rarely found in human ETEC that are not neutralizable by anti-cholera toxin seraCitation6). Lasaro et al.Citation7 first demonstrated LT polymorphisms in a set of 51 human-derived ETEC strains from Brazil. Such a high level of polymorphism found in strains isolated in a restricted geographical area, indicated that the LT toxin was more variable than previously anticipated. The finding prompted us to question whether the other variants existed globally, and also if LT toxin could evolve into more virulent variants over time. To address this, we characterized the eltAB sequence in 192 LT-expressing ETEC strains, collected 1980 to 2011, from different parts of the world.Citation8,9 We could confirm that the eltAB operon is polymorphic in ETEC strains isolated globally.Citation9 We found in total 20 different amino acid variants of LT in our strain collection, 8 were identical to variants described by Lasaro et al. and we also found 12 novel variants.Citation9 A substantial genetic heterogeneity was mainly found in the A subunit with 22 amino acid changes, specifically in the A2 domain, while the B subunit was largely conserved with only 2 amino acid changes.Citation9 The B subunit pentamers are responsible for the recognition and binding to the host´s epithelial cells. The binding sites to GM1, blood sugar and, lipopolysaccharides located in the sequences of the B subunit, were conserved.Citation5,9 In addition, the ADP-ribosylation active site at amino acid residues 47-56 in LTA was conserved in all variants. This might indicate that all natural variants had intact binding, and virulence capacity.
Clonally related lineages of ETEC are persistent over time and have global distribution
ETEC strains that have the same serotype, toxin, and CF profile are often closely related and clonal ETEC lineages have been proposed by studies using randomly amplified polymorphic DNA analysis (RAPD), and multi locus sequence typing (MLST)Citation2,10-13. Using whole genome sequencing, we recently described that certain clonal lineages of ETEC with conserved toxin and CF profiles, have emerged in modern time and spread globally.Citation8 The sequences of eltAB described in Joffre et al.Citation9 were extracted from the same set of whole genome sequenced strains, and allowed us to superimpose the LT variants onto the genetic background of the corresponding ETEC strain. As previously described, we found associations between LT variants and CF profiles,Citation9 but we also found that the LT toxin variants were usually stable over time in the same clonal lineage. Hence, there was no evidence that the LT toxin changed into more virulent variants over time in the same lineage. Our results rather seem to suggest that conserved expression of certain LT variants provided certain ETEC lineages with an advantage. We found that some ETEC clonal lineages, that were frequently derived from diarrheal cases globally, expressed the same LT variants over time. This was particularly evident for the 2 most prevalent LT variants, previously described as LT1 and LT2.Citation14
The LT1 and LT2 variants are expressed by several globally distributed ETEC lineages
The LT1 and LT2 variants were initially described by Lasaro et al.,Citation7 who found that these variants were common in ETEC isolates from Brazil. In addition, they also noticed that ETEC isolates that were clonally distinct could express these LT variants. The LT1- and LT2-expressing ETEC isolates also harbor the most prevalent CFs according to epidemiological studies.Citation15-17 By exploring the virulence diversity carried by the ETEC strains that expressed the LT1 and LT2 variants, and mapping the CF profiles onto the eltAB operon amino acid-based phylogenetic 3,Citation9 we found a correlation between CF profile, toxin variants, and clonal lineages (). Of the identified ETEC lineages several of the largest lineages contained strains that expressed either LT1 or LT2. For instance, the LT1 variant was found in 2 major clonal linages that co-expressed the heat stable toxin STh and the CFs CS1+CS3 and CS2+CS3, respectively. The two lineages, L1, (LT1, STh, CS1+CS3), and L2, (LT1, STh, CS2+CS3), are closely related and both emerged approximately 60 years ago.Citation8 These 2 lineages have spread globally and are repeatedly isolated worldwide.Citation15
Figure 1. Distribution of LT1 and LT2 expressing strains throughout ETEC linages. The ETEC phylogenetic tree is derived from von Mentzer et al. Citation8. The E. coli pylogroups A, B1, B2 and D/E are indicated and the 5 major ETEC lineages are indicated in bold. LT1 expressing strains are found in lineages L1, L2, L3, L5, L10, L16 and L17 that express CFs CS1-CS3, CS7, CS17 and CS19. LT2 expressing strains are found in L3, L5, L11 and L17 that express CS5+CS6, CFA/I and CS14. The color of the bubbles represents the respective CF profile and the area is proportional to the number of ETEC isolates.
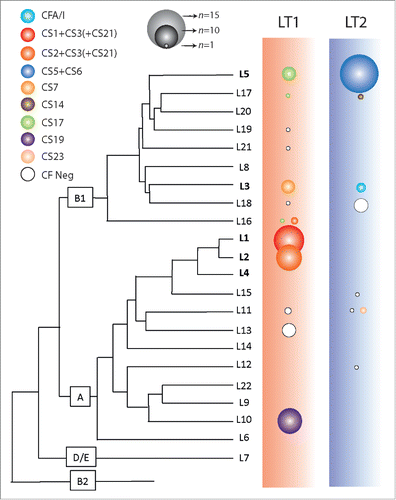
LT1 was also found in lineages harboring strains that expressed e.g CS7, CS17, and CS19. Although several strains that express LT1 were CF negative it seems to be a correlation between LT1 variant and the CFs CS1-CS3, CS7, CS17 and CS19. Strains with these CFs are frequently found in ETEC isolated from diarrheal cases. LT2 was expressed by strains from the major lineage 5 that contain ETEC expressing LT2, STh, and the CFs CS5+CS6.Citation8 Lineage 5 also contains a subset of strains that express CS17, all the CS17 positive ETEC co-express the LT1 variant confirming the close relationship between LT variants and CFs (). In addition all CFA/I expressing ETEC (in lineage 3), that co-expressed LT, had the LT2 variant. Strains expressing CS14 in linage 17 also had the LT2 variant. Numerous studies have found that CFA/I, CS1-CS3, CS5 and CS6 are the most frequently detected CFs, followed by CS7, CS17 and CS14.Citation3,15 Seven CF negative isolates that expressed LT2 clustered together in lineage 18 (). This lineage circulated in Guatemala and Mexico between 1998-2003, and its persistence might indicate that it had colonization advantages although no known CF could be identified. The presence of an important proportion of LT1 and LT2 expressing strains in separate and stable linages of ETEC may indicate that acquisition of plasmid encoded LT1 or LT2 in combination with certain CFs and/or other virulence characteristics conferred traits that allowed successful dissemination through clonal expansion of these strains.
Novel insights in the gene expression levels of the LT common LT variants, LT1 and LT2
The dominance of the LT1 and LT2 variants in successful ETEC lineages may suggest that they are highly virulent. To test whether these variants have higher expression levels than other toxin variants we analyzed toxin production of the different LT variants using GM1- ELISA.Citation9 We found high (LT2 and LT21), medium (LT11 and LT13), and low (LT1 and LT18), LT producers.Citation9 Hence, although LT2 was expressed at high levels, LT1 was not. Thus, increased levels of LT1 and LT2 compared to other toxin variants were not found.
When we quantified the production and secretion of LT in LT1 and LT2 strains by quantitative GM1-ELISA, the data showed that LT2 strains produced 5-fold more LT toxin than LT1.Citation9 In the original study we did not analyze the gene transcription levels of the toxin variants. Hence, in order to address if the difference in production between LT1 and LT2 was on the transcriptional level, we performed additional studies and analyzed the eltAB1 (LT1) and eltAB2 (LT2) gene expression using qPCR and primers previously described.Citation18
We analyzed the transcription levels in LT1 and LT2 ETEC, in media with, and without glucose, since several studies have shown that expression of LT is increased in the presence of glucose due to repression of the eltAB promoter by the cyclic AMP receptor protein (CRP).Citation5,18-21
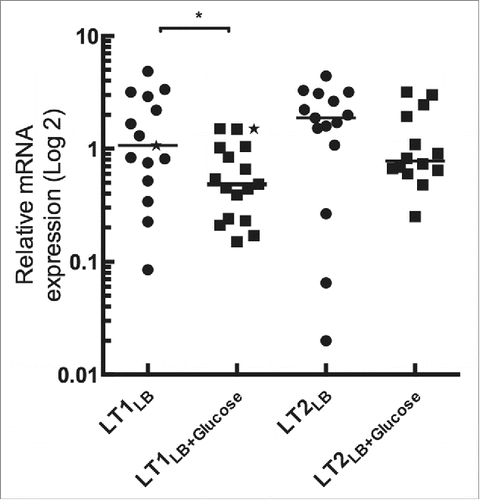
Surprisingly, in our hands, the expression of both eltAB1 and eltAB2 was reduced in the presence of glucose although expression of eltAB2 did not reach statistical significance (). The expression levels of eltAB2 were slightly, but not significantly, higher than the expression levels of eltAB1, both with, and without, presence of glucose in the medium.
Figure 2. Effect of the glucose on the transcriptional levels of the LT toxin in LT1 and LT2 expressing strains measured by qPCR. The LT1 and LT2 isolates were grown in either LB-only or supplemented with glucose (0,2% w/v). Comparative analysis of the expression of LT between LT1 and LT2 strains without glucose. Values of the relative mRNA expression of the isolate H10407 was labeled with a star (★). A Wilcoxon signed rank test was used to calculate P values using Prism version 6.0 (GraphPad Software, La Jolla Califoria USA). (P < 0,05)
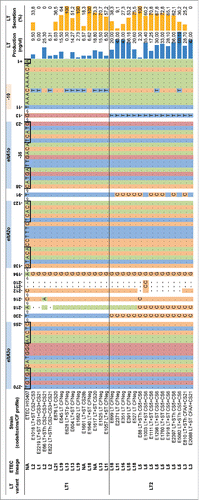
An analysis of the eltAB promoter sequence extracted from the LT1 and LT2 strains identified 3 CRP binding sites as reported by Bodero and MunsonCitation20 in all strains. No polymorphisms were found that could explain the differential gene expression in LT1 strains in presence of glucose, since the CRP binding sequences remain intact in all ETEC strains. The LT1 and LT2 promoters for a subset of ETEC strains are shown in .
Figure 3. Promoter region of LT1 and LT2 including the regulatory elements, production level and secretion of LT. Alignment of 270 bp nucleotide sequence upstream from the start codon of eltA gene using the program MEGA6.0. The nucleotides are numbered above the sequence. The 3 CRP binding sites (eltAo1-3) proposed by Bodero & MunsonCitation20 were identified (blue boxes) along the sequence of eltAp. CRP binding sites are indicated by rectangles. The -35 and -10 hexamers are also labeled above the nucleotide sequence. The blue bars illustrate the amount of LT produced while the yellow bars represent the secretion rate (LT1: 50.29% and LT2: 50.91%). The black dashed vertical lines indicate the average of the production of LT per LT variant (LT1: 6.53 ng/ml and LT2: 30.77 ng/ml). The statistical analysis was performed by the Mann-Whitney test using Prism version 6.0 (GraphPad Software, La Jolla Califoria USA).
Expression of LT is suggested to be indirectly repressed by CRP
Recent data confirm our findings that the CRP/glucose regulation of LT is more complex than previously anticipated. Although we have shown CRP dependent repression of LT in Δcrp strains,Citation18 recent studies have revealed more complexity. Importantly, although the promoter of the eltAB operon contains 3 CRP binding sites upstream of the operon,Citation20 a search for CRP binding sites using Chip–seq failed to detect these sites as bona fide CRP binding sites.Citation19 The same authors showed that deletion of the crp binding site did not affect expression of eltAB in either the wt or Δcrp, compared to when the crp binding site was intact.Citation19 These results indicate that the CRP-mediated repression of LT expression is indirect. We analyzed the extracted eltAB1 and eltAB2 promoters for putative transcription factor binding sites but could not find evidence of other binding sites ().
Different toxin secretion ability in major ETEC lineages expressing LT1 and LT2
Several studies have shown that ETEC strains differ in their capacity to produce LT.Citation9,22,23 In addition, the ability to secrete the mature LT toxin is very variable and range from zero secretion to up to 70% of produced toxin.Citation24 The level of secretion of LT has been suggested to be linked to virulence. In a prior in vitro studyCitation22 a set of ETEC LT expressing strains where tested for production and secretion of the mature LT toxin. The results showed a 50-fold variation in secretion between strains and a correlation of high secretion capacity with fluid accumulation in rabbit ileal loops. We also reported differences in production and secretion of LT in our collection of ETEC isolates collected worldwide. Strains expressing either the LT1 or LT2 variants had no significant statistical difference in ability to secrete the toxin, although the individual secretion ranged form 0-100 % in both LT1 and LT2 isolates. This means that on average, LT1 and LT2 strains secrete 50% of the total amount of the mature toxin; however LT2 produce 5-fold more mature toxin and therefore a larger proportion of toxin is secreted into the external environment than in LT1 producers.
In the original study we did not look at the individual ETEC lineages in the context of LT secretion capacity. We therefore compared secretion capabilities between the most common LT1 and LT2 expressing lineages and we found that strains within lineages L1 and L2 not only produced low levels of LT toxin, but the toxin was also retained within the periplasm and not secreted during growth in LB medium (). These data are intriguing since LT1, STh CS1+CS3 and LT1, STh CS2+CS3 ETEC have been correlated to diarrhea in a number of studies globally.Citation3,25-27 On the other hand, strains from lineage 5, expressing LT2, STh and CS5+CS6, and lineage 3 expressing LT STh and CFA/I, had medium to high levels of toxin secretion ().
Our preliminary data indicate that polymorphisms found in the operon encoding the type II secretion system (T2SS) might provide heterogeneous efficiency of the secretion machinery which might affect LT toxin secretion (data not shown).
Conclusions and future directions
The frequent isolations of ETEC expressing CFA/I, CS1+CS3, CS2+CS3, and CS5+CS6 as well as CFA/I, confirm that these ETEC are very virulent and indicate that expression of either LT1 or LT2 can be advantageous for this pathogen. Although there are differences in toxin amount between the 2 major toxin variants in lab cultures, there is no indication to our knowledge, of differences in duration of diarrhea, or severity of disease, between strains expressing either of the variants during infection. To fully understand LT-induced diarrhea we need to study several ETEC strains and determine regulation in situ in the small intestine to unravel environmental cues that induce and modify virulence in ETEC. In addition, it would be interesting to address severity of disease in relation to infection with major ETEC lineages.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
The authors wish to thank Astrid von Mentzer, University of Gothenburg for bioinformatic help to extract the promoter sequences.
Funding
The study was supported by the Swedish Research Council grant no K2012-56X-22029-01-3, VINNOVA grant no 2011-03491, and the Swedish Foundation for Strategic Research (SSF) grant no SB12-0072 to ÅS. The project was performed as part of the UMSA-IBMB “Diarrheal Disease Project” supported by the Swedish Agency for Research Economic Cooperation (SIDA) to ÅS. EJ acknowledges the financial support from the Swedish Institute and the International Science Program (ISP).
References
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013; 382:209-22; PMID:23680352; http://dx.doi.org/10.1016/S0140-6736(13)60844-2
- Rodas C, Klena JD, Nicklasson M, Iniguez V, Sjoling A. Clonal relatedness of enterotoxigenic Escherichia coli (ETEC) strains expressing LT and CS17 isolated from children with diarrhoea in La Paz, Bolivia. PLoS One 2011; 6:e18313; PMID:22140423; http://dx.doi.org/10.1371/journal.pone.0018313
- Begum YA, Baby NI, Faruque AS, Jahan N, Cravioto A, Svennerholm AM, Qadri F. Shift in phenotypic characteristics of enterotoxigenic Escherichia coli (ETEC) isolated from diarrheal patients in Bangladesh. PLoS Negl Trop Dis 2014; 8:e3031; PMID:25032802; http://dx.doi.org/10.1371/journal.pntd.0003031
- Gaastra W, Svennerholm AM. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol 1996; 4:444-52; PMID:8950814; http://dx.doi.org/10.1016/0966-842X(96)10068-8
- Mudrak B, Kuehn MJ. Heat-labile enterotoxin: beyond G(m1) binding. Toxins (Basel) 2010; 2:1445-70; PMID:22069646; http://dx.doi.org/10.3390/toxins2061445
- Sears CL, Kaper JB. Enteric bacterial toxins: mechanisms of action and linkage to intestinal secretion. Microbiol Rev 1996; 60:167-215; PMID:8852900
- Lasaro MA, Rodrigues JF, Mathias-Santos C, Guth BE, Balan A, Sbrogio-Almeida ME, Ferreira LC. Genetic diversity of heat-labile toxin expressed by enterotoxigenic Escherichia coli strains isolated from humans. J Bacteriol 2008; 190:2400-10; PMID:18223074; http://dx.doi.org/10.1128/JB.00988-07
- von Mentzer A, Connor TR, Wieler LH, Semmler T, Iguchi A, Thomson NR, Rasko DA, Joffre E, Corander J, Pickard D, et al. Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution. Nat Genet 2014; 46:1321-6; PMID:25383970; http://dx.doi.org/10.1038/ng.3145
- Joffre E, von Mentzer A, Abd El Ghany M, Oezguen N, Savidge T, Dougan G, Svennerholm AM, Sjoling A. Allele variants of enterotoxigenic Escherichia coli heat-labile toxin are globally transmitted and associated with colonization factors. J Bacteriol 2015; 197:392-403; PMID:25404692; http://dx.doi.org/10.1128/JB.02050-14
- Pacheco AB, Guth BE, Soares KC, Nishimura L, de Almeida DF, Ferreira LC. Random amplification of polymorphic DNA reveals serotype-specific clonal clusters among enterotoxigenic Escherichia coli strains isolated from humans. J Clin Microbiol 1997; 35:1521-5; PMID:9163473
- Pacheco AB, Soares KC, de Almeida DF, Viboud GI, Binsztein N, Ferreira LC. Clonal nature of enterotoxigenic Escherichia coli serotype O6:H16 revealed by randomly amplified polymorphic DNA analysis. J Clin Microbiol 1998; 36:2099-102; PMID:9650973
- Nicklasson M, Klena J, Rodas C, Bourgeois AL, Torres O, Svennerholm AM, Sjoling A. Enterotoxigenic Escherichia coli multilocus sequence types in Guatemala and Mexico. Emerg Infect Dis 2010; 16:143-6; PMID:20031063; http://dx.doi.org/10.3201/eid1601.090979
- Steinsland H, Lacher DW, Sommerfelt H, Whittam TS. Ancestral lineages of human enterotoxigenic Escherichia coli. J Clin Microbiol 2010; 48:2916-24; PMID:20534806; http://dx.doi.org/10.1128/JCM.02432-09
- Lasaro MA, Mathias-Santos C, Rodrigues JF, Ferreira LC. Functional and immunological characterization of a natural polymorphic variant of a heat-labile toxin (LT-I) produced by enterotoxigenic Escherichia coli (ETEC). FEMS Immunol Med Microbiol 2009; 55:93-9; PMID:19076225; http://dx.doi.org/10.1111/j.1574-695X.2008.00506.x
- Isidean SD, Riddle MS, Savarino SJ, Porter CK. A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine 2011; 29:6167-78; PMID:21723899; http://dx.doi.org/10.1016/j.vaccine.2011.06.084
- Qadri F, Svennerholm AM, Faruque AS, Sack RB. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev 2005; 18:465-83; PMID:16020685; http://dx.doi.org/10.1128/CMR.18.3.465-483.2005
- Svennerholm A-M, Lundgren A. Recent progress toward an enterotoxigenic Escherichia coli vaccine. Expert Review of Vaccines 2012; 11:495-507; PMID:22551034; http://dx.doi.org/10.1586/erv.12.12
- Gonzales L, Ali ZB, Nygren E, Wang Z, Karlsson S, Zhu B, Quiding-Jarbrink M, Sjoling A. Alkaline pH Is a signal for optimal production and secretion of the heat labile toxin, LT in enterotoxigenic Escherichia coli (ETEC). PLoS One 2013; 8:e74069; PMID:24058516; http://dx.doi.org/10.1371/journal.pone.0074069
- Haycocks JRJ, Sharma P, Stringer AM, Wade JT, Grainger DC. The Molecular Basis for Control of ETEC Enterotoxin Expression in Response to Environment and Host. PLoS Pathogens 2015; 11:e1004605; PMID:25569153; http://dx.doi.org/10.1371/journal.ppat.1004605
- Bodero MD, Munson GP. Cyclic AMP Receptor Protein-Dependent Repression of Heat-Labile Enterotoxin. Infection and Immunity 2009; 77:791-8; PMID:19075028; http://dx.doi.org/10.1128/IAI.00928-08
- Wijemanne P, Moxley RA. Glucose significantly enhances enterotoxigenic Escherichia coli adherence to intestinal epithelial cells through its effects on heat-labile enterotoxin production. PLoS One 2014; 9:e113230; PMID:25409235; http://dx.doi.org/10.1371/journal.pone.0113230
- Lasaro MAS, Rodrigues JF, Mathias-Santos C, Guth BEC, Régua-Mangia A, Ferreira AJP, Takagi M, Cabrera-Crespo J, Sbrogio-Almeida ME, Ferreira LCdS. Production and release of heat-labile toxin by wild-type human-derived enterotoxigenic Escherichia coli. FEMS Immunol Med Microbiol 2006; 48:123-31; PMID:16965360; http://dx.doi.org/10.1111/j.1574-695X.2006.00134.x
- Rocha LB, Ozaki CY, Horton DS, Menezes CA, Silva A, Fernandes I, Magnoli FC, Vaz TM, Guth BE, Piazza RM. Different assay conditions for detecting the production and release of heat-labile and heat-stable toxins in enterotoxigenic Escherichia coli isolates. Toxins (Basel) 2013; 5:2384-402; PMID:24316604; http://dx.doi.org/10.3390/toxins5122384
- Hirst TR, Hardy SJ, Randall LL. Assembly in vivo of enterotoxin from Escherichia coli: formation of the B subunit oligomer. J Bacteriol 1983; 153:21-6; PMID:6336733
- Qadri F, Das SK, Faruque AS, Fuchs GJ, Albert MJ, Sack RB, Svennerholm AM. Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. J Clin Microbiol 2000; 38:27-31; PMID:10618058
- Oh KH, Kim DW, Jung SM, Cho SH. Molecular characterization of Enterotoxigenic Escherichia coli strains isolated from diarrheal patients in Korea during 2003-2011. PLoS One 2014; 9:e96896; PMID:24841334; http://dx.doi.org/10.1371/journal.pone.0096896
- Mansour A, Shaheen HI, Amine M, Hassan K, Sanders JW, Riddle MS, Armstrong AW, Svennerholm AM, Sebeny PJ, Klena JD, et al. Pathogenicity and phenotypic characterization of enterotoxigenic Escherichia coli isolates from a birth cohort of children in rural Egypt. J Clin Microbiol 2014; 52:587-91; PMID:24478492; http://dx.doi.org/10.1128/JCM.01639-13
