ABSTRACT
Previous studies indicated that caloric restricted diet enables to lower significantly the risk of cardiovascular and metabolic diseases. In experimental animal models, life-long lasting caloric restriction (CR) was demonstrated to induce changes of the intestinal microbiota composition, regardless of fat content and/or exercise. To explore the potential impact of short and long-term CR treatment on the gut microbiota, we conducted an analysis of fecal microbiota composition in young and adult Fisher 344 rats treated with a low fat feed under ad libitum (AL) or CR conditions (70%). We report here significant changes of the rat fecal microbiota that arise rapidly in young growing animals after short-term administration of a CR diet. In particular, Lactobacillus increased significantly after 8 weeks of CR treatment and its relative abundance was significantly higher in CR vs AL fed animals after 36 weeks of dietary intervention. Taken together, our data suggest that Lactobacillus intestinal colonization is hampered in AL fed young rats compared to CR fed ones, while health-promoting CR diet intervention enables the expansion of this genus rapidly and persistently up to adulthood.
Introduction
Diet is a major contributor to the diversity of the gut microbiota, exerting a strong impact on its composition since breast feeding in early life and throughout adulthood and advanced age. An overfeeding diet, rich in fat and free sugars, is a potent cause of gut microbiota dysbiosis,Citation1 with a variation of the phylogenetic microbial structure and a selection of pathobionts which affect negatively the gut mucosa homeostasis. Gut microbiota changes have been also vastly reported as a consequence of bariatric surgery, fasting, use of laxatives,Citation2 or low fat dietary intervention to counteract the metabolic damages associated to obesity and high fat diet. Recently, Tomas et al. demonstrated that both composition and spatial distribution of the small intestinal microbiota are rapidly affected by high fat diet in mice.Citation3 After only 30 days, mice under high fat diet showed an increased amount of Firmicutes, Proteobacteria and Verrucomicrobia, and a decrease of Bacteroidetes. In addition, high fat diet led to a massive spatial redistribution of the microbiota with a dense colonization of the intervillous zone. Noteworthy, changes in microbiota composition and segregation were reversed to the initial conditions after restoration of normal diet.
The ultimate reason of such great interest in the effect of diet on the gut microbial communities relies on the paramount importance of assessing the microbiota role in pathogenesis of metabolic disorders, age-related diseases and other conditions that depend on a correct balance between food intake and health status. In this respect, caloric restriction (CR), defined as decreased nutrient intake without causing malnutrition, has been reported to increase both healthspan and lifespan in animal models, including primates.Citation4-6
Following a previous study assessing the health benefits of CR treatment in a mouse model of aging,Citation6 Zhang et al. reported that life-long CR - regardless of diet composition – significantly changes the overall structure of the gut microbiota at mid-life (62–83 weeks) and aged-life (141 weeks).Citation7 All together, these pioneering results demonstrated that, as for other dietary intervention, long-lasting CR treatment enables changes of the microbiota structure, reduction of diet-associated metabolic disorders and a prolonged and healthy lifespan.Citation6, 7 Clearly, the development of diet-dependent metabolic disorders and accelerated aging processes in ad libitum (AL) fed animals might exert, in turn, a negative impact on gut physiology, leading to modifications of gut microbiota composition. However, it remains unclear whether (i) the microbiota variation is itself a direct consequence of diet-dependent metabolic disorders, further contributing to poor health; or else, (ii) specific microbiota changes occur as a direct consequence of overfeeding per se (AL regimen), setting the stage for altered host physiology. In both cases such early microbiota changes could play a pathogenetic role in intestinal and systemic diseases. If so, early microbial biomarkers might become predictive of clinical benefits of CR treatment, and/or might serve as diagnostic tools to identify overfeeding-related dysbiosis.
In this work, we investigated whether fecal microbiota variation is induced by short-term CR and whether such change persists in long-lasting CR, in a rat model of aging.Citation8 As for the majority of murine experimental models, animals were fed AL, a condition that might be assimilated to overfeeding, given the poor energy expenditure in the animal house settings, compared to wild rats in their natural environments.Citation9 To shed light on timing and nature of microbial composition changes, fecal microbiota was evaluated in young growing and adult Fisher 344 rats, comparing animals fed AL and animals fed with 70% of the AL ratio (CR).
Material and methods
Animal intervention and sampling
A total of 22 Fisher 344 rats were used from a colony available at the Department of Biomedical Sciences, University of Cagliari. Animals were fed with Purina Rodent Lab Chow diet with 3% of fat (Mucedola srl, Settimo Milanese, Italy) until the age of 8 weeks. Then two groups of 6 rats each were either continued on the AL diet or given 70% of the AL ratio (CR). Food to CR group was delivered nightly at 1AM through a computer assisted automated food dispenser. Fecal samples were collected after 3 and 8 weeks. In a parallel experiment, 10 rats were fed as above since the age of 8 weeks and up to their mid-life. Fecal samples from adult rats were collected after 36 weeks of CR treatment (6 rats) and from controls (4 rats) fed AL. To evaluate the abundance of Lactobacillus spp. in fecal samples, bacterial cultures were performed on Man-Rogosa-Sharpe Agar (MRS, Oxoid, Basingstoke, UK) supplemented with the fungicide cycloheximide (7 µg/mL, Sigma-Aldrich, Saint Louis, MO) at 37°C in anaerobic atmosphere. Lactobacillus colonies were identified according to their morphology, followed by a genus-specific PCR amplification.Citation10
Animal studies were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Cagliari.
Sequencing of the 16S rRNA gene
A total of 46 fecal samples were collected and immediately stored at −80°C. Each sample was subjected to DNA extraction according to QIAamp Fast Stool Kit protocol (QIAGEN, Hilden, Germany). The extracted DNA was purified according to E.Z.N.A.® Soil DNA Kit (Omega Bio-Tek, Norcross, GA) and quantified on a Qubit 2.0 Fluorometer (Life Technologies, Grand Island, NY), using the Qubit ds DNA High Sensitivity Assay Kit (Life Technologies). Full length 16S rRNA genes were amplified from the adult rats samples using the universal primers 27F-1492R.Citation11 The V4 region of 16S rDNA was amplified from the young rats samples according to the protocol published by Caporaso and co-workers.Citation12 Two separate gene amplification reactions were performed for each experiment, pooled together, cleaned up using AMPure XP (Beckman Coulter, Brea, CA) magnetic beads and quantified with Qubit HS assay (Life Technologies). Libraries were constructed according to the Illumina Nextera XT sample preparation protocol (Illumina, San Diego, CA). Normalized sample libraries were pooled and subjected to the cluster generation step using the cBOT cluster generation station, according to the Illumina TruSeq Paired-End Cluster Kit protocol (Illumina). DNA sequencing was performed with the Illumina HiScanSQ sequencer, using the paired-end method and 93 cycles of sequencing.
Bioinformatics and statistical analysis of sequencing data
After sequencing, all reads were subjected to a demultiplexing step using Casava software version 1.8.2 implemented in HiScan control software (Illumina). The Illumina demultiplexed paired-end reads were trimmed for the first 20 bp using FASTX and sequences with Nextera adapter contamination were identified using the UniVec database (ftp://ftp.ncbi.nlm.nih.gov/pub/UniVec/) and removed.
The paired-end reads with a minimum overlap of eight bases were merged using a specific script contained into QIIME (v.1.9.1).Citation13 OTUs generation was done using a QIIME pipeline based on USEARCH's OTU clustering recommendations (http://www.drive5.com/usearch/manual/otu_clustering.html) using the closed-reference OTU picking to allow clustering of shotgun 16S sequences, as previously described.Citation14 Reads were clustered at 97% identity using UCLUST to produce OTUs.Citation15 Taxonomy assignment of resulting OTUs was performed using the Greengenes 13_8 database.Citation16 A number of OTUs were assigned to taxa given in square brackets, including [Prevotella], [Paraprevotellaceae] [Ruminococcus] gnavus, and [Clostridium] difficile. These outputs from QIIME analyses refer to taxonomic assignments recommend by GreenGenes, mainly based on genome trees. As normalization step, rarefaction to equal sequencing depth was performed on all samples in each experiment. Richness was quantified as observed OTU counts. Alpha diversity was quantified as Shannon diversity index; the statistical significance of differences in microbial community composition index between sample categories was determined by non-parametric Monte Carlo permuted two-sided t-test (999 permutation).
PCA plots were generated using MetaboAnalyst v.3.0 (http://www.metaboanalyst.ca/)Citation17 from even OTU tables; boxplots and scatter plots were created using GraphPad Prism (v.5.03), while heatmap was produced using ClustVis (http://biit.cs.ut.ee/clustvis)Citation18 and edited using Inkscape (https://inkscape.org).
Sequence similarity search was performed via the BLAST [nucleotide] tool available in the European Nucleotide Archive (ENA) website, using default search parameters (http://www.ebi.ac.uk/Tools/sss/ncbiblast/nucleotide.html).
OTUs with differential abundance between the sample groups were detected by applying a negative binomial generalized linear model (Wald test within the DESeq2 module available in QIIME 1.9.1) to the raw OTU counts. Differential abundance analysis of families and genera was performed between groups using t-test after log transformation and pareto scaling, while analysis within groups on paired data was performed with the inverted beta binomial testCitation19 available in ibb R package (http://www.oncoproteomics.nl/software/BetaBinomial.html), as detailed elsewhere.Citation14 P-values were corrected for multiple inference using the Benjamini-Hochberg false discovery rate procedure using an adjusted alpha cutoff value of 0.05.
Results
Growth curves and serum lipids profile in rats fed AL or CR diet
Growth curves of AL and CR animals were already well separated after 3 weeks from the beginning of the CR diet (). Rats in AL group showed initially a rapid body weight increase and continued to gain weight until week 36, when their weight remained relatively stable until the end of the experiment. In contrast, CR rats showed a slower growth compared to AL until 21 weeks of treatment, after which their weight remained stable. These data are consistent with those reported in the literature for the Fisher 344 rat strain.Citation20 While absolute food consumption was obviously 30% less in CR animals, the amount of diet eaten by AL and CR groups relative to their body weight became comparable after 28 weeks of feeding, i.e. CR rats were in fact fed an equal amount of diet compared to AL group in relation to their actual size (Fig. S1).
Figure 1. Growth curves of young growing (A) and adult (B) rats during ad libitum (AL, red) and caloric restriction (CR, green) treatment. Arrows indicate the different weeks at which stools were collected, while the first point in which the two groups exhibit a statistically significant difference in weight is marked with an asterisk.
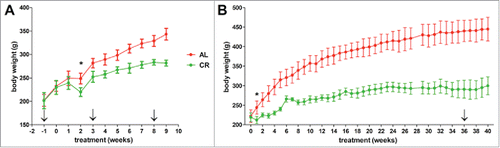
It is well established that both short-term and long-term CR result in significant changes in serum lipids profile.Citation21 Interestingly, we observed that significant decreases in total cholesterol and triglycerides were already evident after 8 weeks of CR regimen (Fig. S2).
Sequencing of 16S rDNA from the gut microbiota of young and adult Fisher 344 rats
To investigate the gut microbiota composition in the Fisher 344 rats subjected to either CR or AL chow diet, we collected fecal samples from the 12 young rats one week before the beginning of dietary intervention (T0) and after 3 and 8 weeks (). Fecal samples were also collected from 10 adult rats after 36 weeks from the beginning of CR (). Sequencing of the amplicons, from a total of 46 fecal samples, enabled to collect 30,107,702 and 3,358,687 reads, respectively for the adult and the young rats experiments, after merging of paired-end reads. Richness, measured as the number of OTUs detected within each community, showed no significant difference comparing samples from animals of the same age with or without CR treatment (Table S1).
As reported above for richness, Shannon's index analyses indicated that the microbial diversity was consistent between the CR and AL groups of young growing rats, with the exception of samples obtained at 3 weeks of CR that showed a lower alpha-diversity compared to the AL control group (8.22 ± 0.16 and 8.49 ± 0.06, respectively; P = 0.015). Since the richness was not affected by CR treatment, this difference in alpha-diversity might be accounted for a reduced evenness of dispersion of the gut microbial community members after the first three weeks of dietary intervention. It is noteworthy that this coincided with the time point when CR animals experienced a sudden change in average food intake per body weight (Fig. S1). Similarly, when comparing adult rats, alpha-diversity index did not differ between animals with or without CR treatment (Table S1).
CR treatment rapidly affects gut microbial community composition in young rats
To measure the similarity between microbial communities in young rats with or without CR treatment, principal component analysis (PCA) was performed (). PCA showed that as early as after 3 weeks there was a clear clustering of fecal microbiota CR samples compared to AL controls. Indeed, at this time point, rats fecal microbiota showed a very large number of differentially abundant OTUs (n = 1,703), of which 1,175 were higher in AL fecal samples and 528 were higher in CR fecal samples (Dataset S1). Analysis of the most abundant genera show significant differences comparing AL and CR treated animals, with [Ruminococcus] gnavus (Lachnospiraceae) more abundant in AL rats and 5–7N15 (Bacteroidaceae) more abundant in CR rats (). At family level, 3 weeks of CR treatment showed a significant impact on the relative abundance of Lachnospiraceae and Desulfovibrionaceae (higher in AL rats) and of S24-7 and Bacteroidaceae (higher in CR rats). This suggests that the dramatic reduction of average food intake per body weight, in the first 3 weeks of the experiment, induces a wide variation along the spectrum of microbial competition (Fig. S3).
Figure 2. Beta-diversity at OTU level between ad libitum (AL, red) and caloric restriction (CR, green) groups before (T0, A) and after 3 weeks of treatment (3W, B) in young rats. Principal component analysis (PCA) was carried out on even OTU table data.
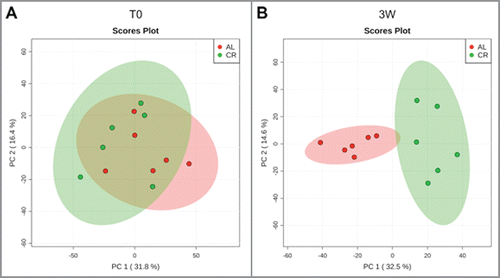
Figure 3. Top 10 genera within the young rats gut microbiota fed ad libitum (AL, left) and with caloric restriction (CR, right). Genera are ordered by decreasing median of the relative abundance among subjects at 1 week before treatment (A), 3 weeks (B), and 8 weeks of treatment (C). Boxes are colored based on the phylum membership (blue, Firmicutes; purple, Bacteroidetes). Asterisks indicate significant difference (P < 0.05) between AL and CR rats at same time of treatment, with the asterisk corresponding to the group in which a given genus was more abundant.
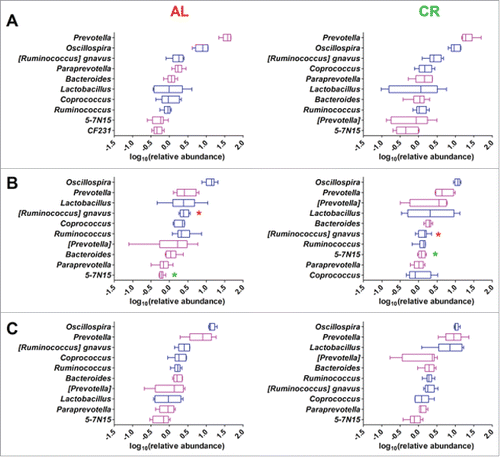
After 8 weeks of CR treatment, a lower number of differentially abundant OTUs were recorded (708), of which 364 were higher in AL samples and 344 were higher in CR treated samples. Differential analysis of the most abundant genera and family did not show significant differences comparing AL and CR treated animals at 8 weeks ( and Fig. S3). This result was in keeping with the behavior of differential OTUs belonging to the same taxon but with an opposite correlation to diet (Dataset S1).
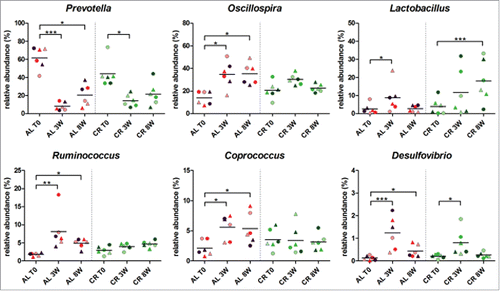
Taking into account that these rats were in a fast growing phase, gut microbiota composition at 8 weeks could be ascribed to physiological changes depending on both diet (CR vs AL) and rate of growth from 7 to 16 weeks of age (). Thus, to evaluate whether CR and AL diets induced specific changes during gut microbiota maturation in growing rats, the relative abundance of genera at different time points within the same group of animals was analyzed. Specifically, rats maintaining the AL diet for 8 weeks showed a decrease of the relative abundance of genus Prevotella and a parallel increase of genera Oscillospira, Ruminococcus, Coprococcus, and Desulfovibrio (). These variations were already evident and significant after 3 weeks. Interestingly, fecal samples from CR treated rats showed similar changes only for Prevotella and Desulfovibrio (to a lesser extent) but none of these genera was significantly varied after 8 weeks of CR. In the CR-fed group, on the contrary, Lactobacillus showed a significant increase in young growing rats after 8 weeks of treatment (). Therefore, CR appears to attenuate the variations observed in AL rats and to promote, instead, Lactobacillus abundance in the young growing rats.
Figure 4. Relative abundance variation of genera in young rats fecal microbiota after 8 weeks of ad libitum (AL) or caloric restriction (CR) diets. Only genera with a relative abundance > 1% in at least one time point and significant variation at 8 weeks are represented. Each dot represents a different sample, with the same combination shape-color indicating the same rat. Samples were evaluated 1 week before treatment (T0) and after 3 weeks (3W) and 8 weeks of treatment (8W). Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001) within AL and CR groups between different time points.
Composition of gut microbiota in adult rats
To evaluate the effect of long-term dietary intervention on the rat fecal microbiota, PCA was again used to describe beta-diversity in CR- and AL-fed animals. We again observed a clear clustering of the gut bacterial communities according to the CR or AL diet (Fig. S4).
However, comparing the relative abundance of each of the most prevalent genera and families, values were similar in the two experimental groups ().
Figure 5. Fecal microbiota of ad libitum (AL, left) and caloric restriction (CR, right) treated adult rats. Boxplots show the top 10 microbial families (A) and genera (B). Features are ordered by decreasing median of the relative abundance among subjects. Boxes are colored based on the phylum membership (blue, Firmicutes; purple, Bacteroidetes; orange, Proteobacteria). Asterisks indicate significant difference (P < 0.05) between AL and CR rats at same time of treatment, with the asterisk color corresponding to the group in which a given genus was more abundant.
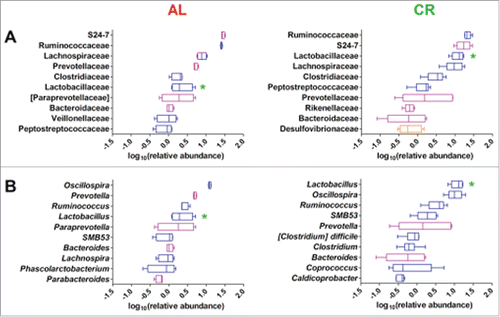
As discussed previously for the young rat gut microbiota, this result can be easily explained given the common occurrence of OTUs belonging to the same family or genus but with opposite correlation with dietary intervention. Most interestingly, the relative abundance of Lactobacillus was an exception, showing significantly higher relative abundance in CR fecal microbiota than in samples from AL-fed (“overfed”) rats (). Therefore, the high number (n = 32, Dataset S1) of differential Lactobacillus OTUs that correlate with the 36 weeks long CR treatment also corresponded to an overall higher relative abundance of Lactobacillus genus in these animals. The differential abundance of Lactobacillus was also confirmed by bacterial cultures. Following fecal culture in MRS agar plates, a significantly higher number of CFUs per gram of feces was detected in CR samples compared to AL samples (respectively 2.1 ± 0.31 × 109 vs 5.8 ± 0.38 × 108; p value < 0.03).
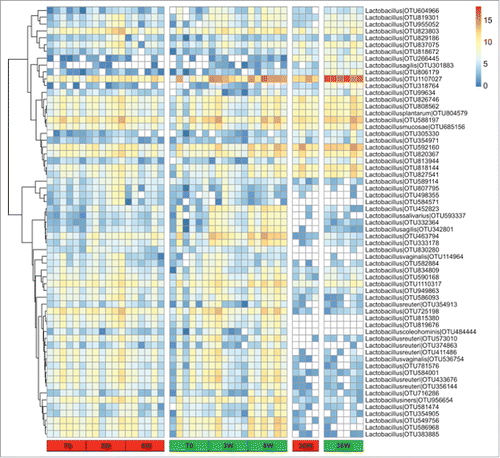
Finally, we focused on the assortments of Lactobacillus OTUs identified in the young CR rats (n = 446) and in the adult CR rats (n = 320). All the OTUs recorded in the latter group of animals were also present in the former group, suggesting the persistence in aged animals of the very same Lactobacillus strains identified in young animals. To highlight the variation of relative abundance of Lactobacillus from young growing rats to adulthood, OTUs assigned to the genus Lactobacillus and present with >0.01% of relative abundance in at least one group at one time point were selected and their relative abundance compared in all animals (). Few OTUs showed either low or high relative abundance in both groups at all time points, whereas a larger group was less represented in adult animals compared to the young ones, again regardless of the diet fed. On the other hand, a number of OTUs were more represented in CR rats at 8 and at 36 weeks of treatment compared to matched AL controls. Among these, OTU 1107027 was the most abundant in the fecal microbiota of CR samples collected at 8 weeks (mean = 0.17% and 2.39% in AL and CR fed rats, respectively) as well as in samples from adult animals (mean = 0.72% and 8.79% in AL and CR fed rats, respectively). To further refine the taxonomic assignment, we performed a BLAST search against the European Nucleotide Archive (ENA) database. OTU 1107027 showed 100% of sequence identity with an uncultured bacterial clone (HM124081) and 99.7-99.8% of identities with three strains of L. faecis, a recently assigned species described as obligately homofermentative.Citation22
Figure 6. Heatmap showing the relative log-transformed abundance distribution of Lactobacillus spp. OTUs among all samples. OTUs with >0.01% of relative abundance in at least one group at one time point were selected. Columns are hierarchically ordered based on the experiment, treatment (ad libitum, red; caloric restriction, green), and time point.
Discussion
The gut microbiota is recognized as central to gut health, local immune function and nutrient uptake. Gut health and microbiota are key to gut-liver and gut-brain signaling, since prolonged mucosal immune activation (gut inflammation) is a potential factor contributing to both liver injury and behavioral patterns. Besides, microbial communities inhabiting the gut environment are continuously challenged by the host health conditions, the food taken and its digestion. Such environmental challenges trigger the continuous adaptation of each member of the microbial communities, inducing a variation in microbiota composition and, occasionally, dysbiosis. For example, high fat diet (HFD) is among the most powerful drivers of gut microbiota dysbiosis.Citation1 Recently, Tomas et al. have shown how rapid (30 days) could be the occurrence of a microbiota structure reassortment induced by HFD, together with the on-site colonization under the intestinal mucus in the intervilli area.Citation3
Less is known about the effects of caloric restriction on the gut microbiota, despite the fact that dietary restriction is the most studied and reproducible non-genetic intervention known to extend healthspan and/or lifespan in organisms ranging from unicellular yeast to primates.Citation23 Indeed, CR exerts a beneficial effect on the incidence of chronic diseases related to old age, including cancer, consistent with the notion that changes occurring during the aging process may bear direct relevance to the pathogenesis of chronic disease. For example, we have obtained evidence indicating that CR is able to modulate the age-associated, neoplastic prone tissue landscape in rat liver, thereby decreasing the growth of transplanted pre-neoplastic cells.Citation24 Even short term CR protects from metabolic heart disease, reduces the rate of renal senescence, proteinuria and triglyceridemia.Citation25-27 However, while the effects of HFD have been well characterized after short and long term exposure, the timing of the microbiota changes induced by CR has not been carefully evaluated so far.
In this study, we showed that during the first 3 weeks of CR treatment of young growing rats (aged 8–11 weeks), the fecal microbiota showed significant changes, reaching a different structure compared to AL fed rats. Interestingly, these changes involved a highest number of OTUs (1,703 out of 33,910) accompanied by a significantly lower value of alpha-diversity in CR treated rats, without variation of OTUs richness. The sudden reduction of diet intake (down to 70%) appears to promote, thus, an important reassortment of the relative abundance of specific phylotypes. Following 5 more weeks of treatment (8 weeks total), the diversity between the 2 groups of rats was still evident, as shown by PCA analysis, yet the alpha-diversity was similar and the number of statistically different OTUs (relative abundance) was lower than at 3 weeks (708 out of 32,801). This trend parallels the difference between the 2 diet groups in the average food intake per body weight (g/g%), that decreases rapidly from the first three weeks to the 8 weeks time points, given the lower weight gained by CR rats. Despite the overall similarity of the gut microbial taxonomic organization observed between AL and CR young rats after 8 weeks of treatment, the longitudinal analyses of paired samples from T0 showed a significant increase of the Lactobacillus in CR. This suggests that CR promotes changes in the gut milieu that selectively facilitate the growth of a number of OTUs belonging to this genus.
Our results obtained with long-term CR treatment of Fisher 344 rats are consistent with findings reported previously in a mouse model.Citation7 In mice, CR treatment to mid-life (62 weeks of age) led to very high abundance in Lactobacillus spp.Citation6 Furthermore, after 10 weeks of CR treatment in obese human adolescents, a PCR-based analysis of the gut microbiota revealed a significant increase of Lactobacillus, but only for those individuals that had lost > 4.0 kg of weight.Citation28
It should be noted that in our study the adult and the young growing animals belonged to two distinct experimental groups, yet as part of the same colony of rats. Not surprisingly, hence, the Lactobacillus OTUs identified in the group of the adult rats were also identified as a selection of those present in the 12 young growing animals. With aging, therefore, specific Lactobacillus species/strains might outcompete others for growth rate, due to different metabolic capabilities. In particular, OTU 1107027 was the most abundant OTU in young rats after 8 weeks of CR treatment and its relative abundance was even higher (8.7% of total OTUs) in aged CR rats, suggesting a striking adaptation capability to dietary restriction and, conversely, a poor fitness in the gut of AL “overfed” animals. This OTU has been previously reported to be significantly lowered in a mouse model of colitis-associated colorectal cancer, compared to wild-type healthy controls.Citation29 Regardless the noteworthy abundance of OTU 1107027, the overall blooming of Lactobacillus spp. observed in CR treated mice is of great interest, given the acknowledged role of this genus in promoting intestinal homeostasis by stimulating host signaling pathways and the immune system.Citation30 The anti-inflammatory effect of Lactobacillus has been attributed mostly to surface antigens with structural or enzymatic functions. Growing evidence also indicates that supplementation with probiotic Lactobacillus strains improves lipid metabolism.Citation31 However, different moieties have been identified in different strains/species, suggesting that a considerable diversity exists in their capability to promote intestinal health. In our model, 94 OTUs out of 437 belonging to Lactobacillus showed an increased abundance in subjects with a decrease in total cholesterol and triglycerides after 8 weeks of CR diet. Hence, an increased abundance of specific Lactobacillus spp. might concur to the diet-dependent effect on lipid metabolism in CR-treated rats.
In conclusion, Lactobacillus species appear to compete well with the other members of the gut microbiota under CR dietary conditions, while their fitness seems to be limited when excessive intake of food occurs, even with low fat diet (standard chow, in our study). This evidence has to be considered carefully when Lactobacillus bacteria are proposed as probiotic and/or part of fermented food in dietary interventions,Citation32-36 since its gut colonization capability might be strongly affected by the diet regimen itself. On the other hand, our data suggest that Lactobacillus might serve as a marker of dietary intervention in experimental animal models, since its increased relative abundance pairs well the lowering of cholesterol and triglycerides, shortly after dietary restriction, then persists highly abundant after long-term CR treatment.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KGMI_A_1371894_Supplemental.zip
Download Zip (629.3 KB)Acknowledgments
Authors are particularly grateful to Dr. Francesco Fancello, Department of Agricultural Sciences, University of Sassari, for his precious assistance in bacterial isolates identification.
Funding
This work was supported by Sardegna Ricerche – Science and Technology Park of Sardinia, grant program “art. 26_2015” (to Porto Conte Ricerche), and by AIRC grant No. 14640 (to EL). VM and MA were supported by a Doctoral Fellowship from the International PhD Course in Life Sciences and Biotechnologies, University of Sassari.
References
- Murphy EA, Velazquez KT, Herbert KM. Influence of high-fat diet on gut microbiota: a driving force for chronic disease risk. Curr Opin Clin Nutr Metab Care. 2015;18:515-20. doi:10.1097/MCO.0000000000000209. PMID:26154278
- Remely M, Hippe B, Geretschlaeger I, Stegmayer S, Hoefinger I, Haslberger A. Increased gut microbiota diversity and abundance of Faecalibacterium prausnitzii and Akkermansia after fasting: a pilot study. Wien Klin Wochenschr. 2015;127:394-8. doi:10.1007/s00508-015-0755-1. PMID:25763563
- Tomas J, Mulet C, Saffarian A, Cavin JB, Ducroc R, Regnault B, Kun Tan C, Duszka K, Burcelin R, Wahli W, et al. High-fat diet modifies the PPAR-gamma pathway leading to disruption of microbial and physiological ecosystem in murine small intestine. Proc Natl Acad Sci U S A. 2016;113:E5934−E43. doi:10.1073/pnas.1612559113. PMID:27638207
- Lane MA, Black A, Handy A, Tilmont EM, Ingram DK, Roth GS. Caloric restriction in primates. Ann N Y Acad Sci. 2001;928:287-95. doi:10.1111/j.1749-6632.2001.tb05658.x. PMID:11795520
- Lettieri-Barbato D, Giovannetti E, Aquilano K. Effects of dietary restriction on adipose mass and biomarkers of healthy aging in human. Aging (Albany NY). 2016;8:3341-55. doi:10.18632/aging.101122. PMID:27899768
- Zhou B, Yang L, Li S, Huang J, Chen H, Hou L, Wang J, Green CD, Yan Z, Huang X, et al. Midlife gene expressions identify modulators of aging through dietary interventions. Proc Natl Acad Sci U S A. 2012;109:E1201−9. doi:10.1073/pnas.1119304109. PMID:22509016
- Zhang C, Li S, Yang L, Huang P, Li W, Wang S, Zhao G, Zhang M, Pang X, Yan Z, et al. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat Commun. 2013;4:2163. doi:10.1038/ncomms3163. PMID:23860099
- Marongiu F, Serra MP, Doratiotto S, Sini M, Fanti M, Cadoni E, Serra M, Laconi E. Aging promotes neoplastic disease through effects on the tissue microenvironment. Aging (Albany NY). 2016;8:3390-9. doi:10.18632/aging.101128. PMID:27929382
- Bischoff SC, Volynets V. Nutritional influences of overfeeding on experimental outcomes in laboratory mice: consequences for gut microbiota and other functional studies. Int J Med Microbiol. 2016;306:328-33. doi:10.1016/j.ijmm.2016.05.018. PMID:27432516
- Rinttila T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97:1166-77. doi:10.1111/j.1365-2672.2004.02409.x. PMID:15546407
- Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl Environ Microbiol. 2001;67:411-9. doi:10.1128/AEM.67.1.411-419.2001. PMID:11133473
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4516-22. doi:10.1073/pnas.1000080107. PMID:20534432
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335-6. doi:10.1038/nmeth.f.303. PMID:20383131
- Tanca A, Manghina V, Fraumene C, Palomba A, Abbondio M, Deligios M, Silverman M, Uzzau S. Metaproteogenomics reveals taxonomic and functional changes between cecal and fecal microbiota in mouse. Front Microbiol. 2017;8:391. doi:10.3389/fmicb.2017.00391. PMID:28352255
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460-1. doi:10.1093/bioinformatics/btq461. PMID:20709691
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069-72. doi:10.1128/AEM.03006-05. PMID:16820507
- Xia J, Wishart DS. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr Protoc Bioinformatics. 2016;55:14 0 1−0 91.
- Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566−70. doi:10.1093/nar/gkv468. PMID:25969447
- Pham TV, Jimenez CR. An accurate paired sample test for count data. Bioinformatics. 2012;28:i596−i602. doi:10.1093/bioinformatics/bts394. PMID:22962487
- Armbrecht HJ, Strong R, Boltz M, Rocco D, Wood WG, Richardson A. Modulation of age-related changes in serum 1,25-dihydroxyvitamin D and parathyroid hormone by dietary restriction of Fischer 344 rats. J Nutr. 1988;118:1360-5. PMID:3193253
- Choi YS, Goto S, Ikeda I, Sugano M. Age-related changes in lipid metabolism in rats: the consequence of moderate food restriction. Biochim Biophys Acta. 1988;963:237-42. doi:10.1016/0005-2760(88)90286-X. PMID:3196729
- Endo A, Irisawa T, Futagawa-Endo Y, Salminen S, Ohkuma M, Dicks L. Lactobacillus faecis sp. nov., isolated from animal faeces. Int J Syst Evol Microbiol. 2013;63:4502-7. doi:10.1099/ijs.0.052985-0. PMID:23907223
- Fontana L, Partridge L, Longo VD. Extending healthy life span–from yeast to humans. Science. 2010;328:321-6. doi:10.1126/science.1172539. PMID:20395504
- Cadoni E, Marongiu F, Fanti M, Serra M, Laconi E. Caloric restriction delays early phases of carcinogenesis via effects on the tissue microenvironment. Oncotarget. 2017;8:36020-32. PMID:28415598
- Xu XJ, Babo E, Qin F, Croteau D, Colucci WS. Short-term caloric restriction in db/db mice improves myocardial function and increases high molecular weight (HMW) adiponectin. IJC Metab Endocr. 2016;13:28-34. doi:10.1016/j.ijcme.2016.10.002. PMID:27942464
- Ning YC, Cai GY, Zhuo L, Gao JJ, Dong D, Cui S, Feng Z, Shi SZ, Bai XY, Sun XF, et al. Short-term calorie restriction protects against renal senescence of aged rats by increasing autophagic activity and reducing oxidative damage. Mech Ageing Dev. 2013;134:570-9. doi:10.1016/j.mad.2013.11.006. PMID:24291536
- Dong D, Cai GY, Ning YC, Wang JC, Lv Y, Hong Q, Cui SY, Fu B, Guo YN, Chen XM. Alleviation of senescence and epithelial-mesenchymal transition in aging kidney by short-term caloric restriction and caloric restriction mimetics via modulation of AMPK/mTOR signaling. Oncotarget. 2017;8:16109-21.
- Santacruz A, Marcos A, Warnberg J, Marti A, Martin-Matillas M, Campoy C, Moreno LA, Veiga O, Redondo-Figuero C, Garagorri JM, et al. Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity (Silver Spring). 2009;17:1906-15. doi:10.1038/oby.2009.112. PMID:19390523
- McFadden R-MT. The role of curcumin in modulating colonic microbiota during colitis and colon cancer prevention [Thesis dissertation]. Chapel Hill (NC, USA): The University of North Carolina, 2014.
- Ding YH, Qian LY, Pang J, Lin JY, Xu Q, Wang LH, et al. The regulation of immune cells by Lactobacilli: a potential therapeutic target for anti-atherosclerosis therapy. Oncotarget. 2017;8:59915-28.
- Sun J, Buys N. Effects of probiotics consumption on lowering lipids and CVD risk factors: a systematic review and meta-analysis of randomized controlled trials. Ann Med. 2015;47:430-40. doi:10.3109/07853890.2015.1071872. PMID:26340330
- Kamiya T, Watanabe Y, Makino S, Kano H, Tsuji NM. Improvement of Intestinal Immune Cell Function by Lactic Acid Bacteria for Dairy Products. Microorganisms. 2016;5:1.
- Adachi T, Kakuta S, Aihara Y, Kamiya T, Watanabe Y, Osakabe N, Hazato N, Miyawaki A, Yoshikawa S, Usami T, et al. Visualization of Probiotic-Mediated Ca2+ Signaling in Intestinal Epithelial Cells In Vivo. Front Immunol. 2016;7:601. doi:10.3389/fimmu.2016.00601. PMID:28018362
- Singh S, Sharma RK, Malhotra S, Pothuraju R, Shandilya UK. Lactobacillus rhamnosus NCDC17 ameliorates type-2 diabetes by improving gut function, oxidative stress and inflammation in high-fat-diet fed and streptozotocintreated rats. Benef Microbes. 2016:1-14.
- Ni Y, Wong VH, Tai WC, Li J, Wong WY, Lee MM, Fong FL, El-Nezami H, Panagiotou G. A metagenomic study of the preventive effect of Lactobacillus rhamnosus GG on intestinal polyp formation in ApcMin/+ mice. J Appl Microbiol. 2017;122:770-84. PMID:27782360
- Wang J, Tang H, Zhang C, Zhao Y, Derrien M, Rocher E, van-Hylckama Vlieg JE, Strissel K, Zhao L, Obin M, et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015;9:1-15. doi:10.1038/ismej.2014.99. PMID:24936764
