ABSTRACT
Methane is generated in the foregut of all ruminant animals by the microorganisms present. Dietary manipulation is regarded as the most effective and most convenient way to reduce methane emissions (and in turn energy loss in the animal) and increase nitrogen utilization efficiency. This review examines the impact of diet on bovine rumen function and outlines what is known about the rumen microbiome. Our understanding of this area has increased significantly in recent years due to the application of omics technologies to determine microbial composition and functionality patterns in the rumen. This information can be combined with data on nutrition, rumen physiology, nitrogen excretion and/or methane emission to provide comprehensive insights into the relationship between rumen microbial activity, nitrogen utilisation efficiency and methane emission, with an ultimate view to the development of new and improved intervention strategies.
Introduction
Global dairy sector
The global agri-food industry employs over 1 billion people, representing 1 in 3 of all workers.Citation1 In 2013, the largest producers of dairy milk were the US, EU and India, with global production levels at 617 billion litres of milk.Citation2 According to,Citation3 the EU is expected to become one of the world’s leading exporters of milk, with sales anticipated to rise to 19.2 million tonnes of milk equivalent. Since 2005, milk supply and demand has increased by 26%, at 2.4% per annum, respectively, with a further growth of 25% expected over the next 10 years. According to the FAO, the aggregate meat consumption of the industrial countries grew at 1.3% per annum in the last 10 years. With the global population set to increase to 9.1 billion by 2050, food production will need to increase by 70% to meet demands for adequate nutrition.Citation4
However, with an increase in milk and meat production comes an increase in greenhouse gases (GHG). On a global scale, it has been estimated that livestock production contributes between 9% and 11% of total anthropogenic GHG emissionsCitation5 with approximately 44% of livestock emissions in the form of methane (CH4).Citation6 Measuring and mitigation of methane emissions from livestock is increasing in significance. There are currently a number of methane mitigation strategies being investigated to reduce emissions from ruminants, including better feeds and feeding systems i.e. dietary manipulation of the rumen microbiome, improved genetics and overall health of animals (potentially leading to reduced herd sizes while maintaining milk yields), use of supplements that reduce methane emissions and immunisation against methanogens.Citation5
The bovine rumen microbiome
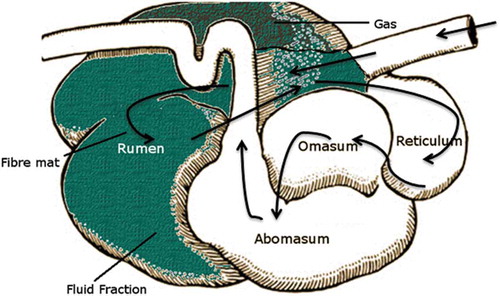
The rumen can be viewed as an anaerobic and methanogenic fermentation chamber that contains microorganisms that have the ability to utilise, and increase the productivity of, cellulolytic feeds (i.e. straw, hay, silage and grass). There are considerable benefits associated with understanding rumen function, as rumen dynamics are almost solely responsible for providing nutrients to the host animal.Citation7 shows the gastrointestinal tract of the bovine animal, where the rumen and its microbiota play a particularly important role in the degradation of feedstuffs. As a result of fermenting feedstuffs, carbon dioxide (CO2) and hydrogen (H2), which are the main electron acceptors and donors of the ecosystem, are produced in the rumen.Citation8 Recent developments relating to compounds used to shift fermentation towards more efficient microbial pathways and targeting of specific microorganisms will be discussed later in this review. The rumen microbiome, i.e., the community of microorganisms that inhabits the rumen, is characterised by its high population density, extensive diversity (encompassing bacteria, archaea, protozoa and fungi) and complexity of interactions.Citation9,Citation10 The continuous fermentation carried out by these microorganisms leads to ingested compounds being broken down into their subcomponents. There are three intersecting micro-environments found in the rumen that contain these microbes; the liquid phase making up 25% of the microbial mass, the solid phase making up 70% of the microbial mass, and the rumen epithelial cells and protozoa, containing 5% of the microbial mass.Citation11 A nutritionally balanced diet is important as it provides an environment that maximises the growth and activity of these microbes.Citation11 A rumen microorganism is anaerobic or facultatively anaerobic, and produces end products that are either utilised directly by the host or by other microorganisms as energy. The pH of the rumen is kept relatively constant, typically 6–7, but may vary depending on diet.Citation12 Such variations can result in a change in the microbial populations, and the levels of volatile fatty acids (VFAs) produced by them. These fatty acids are of interest as some, such as propionate and butyrate, can be absorbed across the gut wall to serve as an energy source for the ruminant. Buffering of the rumen to maintain a relatively constant pH is facilitated by the large quantity of saliva produced by the ruminant, which is high in sodium and potassium bicarbonate and urea. The saliva is swallowed into the rumen and then absorbed through the rumen walls. Further buffering is provided by ammonia produced during fermentation, which can then be used for microbial growth in the rumen.
Ruminants, through the action of their microbiota, can utilize components that the human body cannot break down, namely lignocellulose. Lignocellulose is the most abundant carbon polymer on the planet, with the rumen having a central role in releasing this vast energy store. The rumen ultimately uses lignocellulose to make products (i.e. milk and meat) that are then available to humans to consume as a nutrient dense food source. The interaction between the host and microbes in the rumen is synergistic, in that the host provides heat, moisture and food, while the microorganisms produce protein and by-products of digestion, such as the aforementioned VFAs, for use by the host.Citation13 The degradation of microbes by the host has also been described in literature, with microbes utilised for their protein, lipid and starch content.Citation14 The complex rumen ecosystem consists of bacteria, archaea, ciliate protozoa, fungi, bacteriophage and viruses, which will be discussed below.
Rumen bacteria
Bacteria are the most abundant microbes in the foregut of ruminant animals, with approximately 1010 – 1011 cells/ml and over 200 species.Citation16 The composition of the bacteria found in the rumen is dictated by a number of factors including preference for certain substrates (i.e. diet), energy requirements, and resistance to certain metabolic end-products that may be toxic to some species.Citation17 In a study by Hungate,Citation18 in which rumen microbial populations were studied using a culture-based approach, it was determined that rumen bacterial composition is mainly Gram negative when animals are being fed high forage diets, with more Gram positive bacteria, such as Lactobacillus, present in animals fed high grain diets, with ruminal pH levels dropping after the consumption of easily fermented carbohydrates.Citation19
The use of molecular techniques has become critical for the analysis of rumen microbiology. Such techniques can be used to determine the composition of the population present (using, for example, the conserved 16S rRNA gene to determine bacterial composition), predict their functionalities or enumerate targeted microbes within a complex ecosystem without the need for culturing.Citation20 This is important, as it is estimated that only 20% of the rumen microbiota can be cultured using standard techniques.Citation21 Since sequencing of the 16S rRNA gene was first used to study rumen microbial ecosystems, extensive coverage of low abundance species has enabled the analysis of rare microbial communities. Sequencing also showed that Prevotella, Butyrivibrio and Ruminococcus were the most dominant bacteria in the rumen, and that community structure is affected by changes in the diet of the host.Citation22 In particular, it has been shown that complexity in the diet favours increased microbial diversity.
Due to the high forage diets of ruminants, particularly those in receipt of grass-based diets, cellulose digesters are an important part of supplying the animal with vital nutrients. These bacteria degrade cellulose and hemicellulose, the main components of plant fibre.Citation23 The ability to degrade cellulose is strongly dependant on the type of forage, crop maturity and the accessibility of the cellulolytic bacterial communities.Citation17 The matrix of plant fibre is complex, composed of β-1, 4 linked glucose residues for cellulose and β-1, 4 linked xylose for hemicellulose, requiring the coordination of a number of hydrolytic enzymes in order to break it down.Citation23 Although there are many cellulose degrading bacteria, Fibrobacter succinogenes and Ruminococcus albus are the most desirable cellulose degraders. Their ability to digest cellulose is much higher than that of other cellulolytic bacteria,Citation23 which is probably due to the fact that they possess a number of genes encoding enzymes involved in fibre degradation. Fermentation end products of cellulolytic bacteria include acetate, butyrate, propionate and CO2. Hydrogen, ethanol, succinic acid, formic acid and lactic acid are also formed but are quickly used by other bacteria.Citation24
Starch is also an important constituent of the ruminant diet, in particular for highly productive dairy cows. High grain diets result in an increase in the amount of starch in the rumen. Streptococcus bovis, an amylolytic bacterium, is normally present in low numbers in cows fed high forage diets or cows adapted to grain diets over a course of time and in high abundance in un-adapted cows that consume high grain diets.Citation25 S. bovis has a lower pH optimum for growth than many other bacteria, and its high abundance following consumption of high grain diets is attributed to a sudden increase in glucose levels in the rumen and the loss of protozoa due to the more acidic environment created by high grain diets. More specifically, lactic acid is produced from starch and, as lactic acid is not metabolised by the animal, it is instead absorbed through the rumen wall causing an increase in lactic acid in the blood and reduced blood pH. If the diet of the animal is changed too quickly, there is also an accumulation of VFAs found in the rumen, having a detrimental effect on the microbiota and the host animal. These severe and sudden changes lead to a decrease in rumen pH and an increase in S. bovis and Lactobacillus species.Citation26
Some anaerobic bacteria acquire energy from the degradation of pectin, with the most important pectinolytic species, Lachnospira multiparus, Prevotella ruminicola and Butyrivibrio fibrisolvens, being capable of reducing pectin to oligogalacturonides, yielding large quantities of acetate, a VFA important in bovine metabolism.Citation27 Citrus by-products such as citrus pulp are used widely in ruminant feeding systems and contain a high percentage of pectin substances.Citation28 These by-products can be used as an alternative to highly fermentable grains, preventing the excessive growth of S. bovis, and associated ruminal acidosis, and some studies suggest that they may even improve the efficiency of feed utilisation for milk production.Citation29 With regards to microorganisms associated with milk-fat yield, Jami (2014) identified Firmicutes:Bacteroidetes to be strongly correlated to milk-fat yield.Citation30 Higher percentages of Firmicutes compensated for the lower abundances of Bacteroidetes. A decreased abundance of Bacteroidetes in comparison to Firmicutes was resulted in increased milk-fat percentages. Similar results were found by Bainbridge et al. (2016), where moderate correlations were found between milk yield, protein percentage, fat yield and bacterial communities. This alone indicates the role of the rumen microbiome in shaping host physiological parameters.Citation31
Methanogenic archaea
Archaea, in general, have a broad spectrum of unusual and distinctive metabolisms, enabling them to survive in a variety of different environments. Rumen archaea are strictly anaerobic and are the only known microorganisms present in the rumen capable of producing methane.Citation32 Such archaea are referred to as methanogens. Archaea are found in the rumen in the range of 106 to 108 cells per ml, accounting for less than 4% of the microbial community.Citation33 Archaea are found at the bottom of the trophic chain due to their need to use the end products of fermentation as substrates.Citation8 The domain Archaea is broken into two different kingdoms; Euryarchaeota, consisting of methanogens and extreme halophiles, and Crenarchaeota, consisting of hyperthermophiles and nonthermophiles.Citation34 Methanogens found in the kingdom Euryarchaeota require a very low redox potential and are among the strictest anaerobes known.Citation35 According to meta-analysis of global data, 90% of rumen methanogens belong to the following generaCitation36,Citation37; Methanobrevibacter (63.2% of methanogen population), Methanomicrobium (7.7% of methanogen population) Methanosphaera (9.8%) “Rumen Cluster C”, now referred to as Thermoplasma (7.4%) and Methanobacterium (1.2%). Most methanogens remove hydrogen gas by reducing CO2 with hydrogen gas to form methane. In contrast, Methanosphaera stadtmanae only produces methane through the reduction of methanol with H2, having one of the strictest energy metabolisms of all methanogenic archaea. Producing methane keeps hydrogen concentrations in the rumen low, allowing methanogens to promote the growth of other species, and enabling a more efficient fermentation.Citation11 However, methane produced in the rumen is eructated, leading to atmospheric pollution. Efforts to mitigate rumen methane emissions include vaccines (targeting rumen methanogens through the generation of antibodies to selected methanogen antigens that enter via saliva, binding to targets on the methanogens),Citation38 small-molecule inhibitors (targets enzymes essential for the growth of methanogens), additives and breeding approaches. In a study carried out by Goopy et al.,Citation39 it was found that sheep that emitted low methane levels had a smaller rumen in comparison to high methane-emitting sheep. There was no difference in dry matter intake or digestibility between the two groups. The study also found that low methane-emitting animals had a shorter mean retention time for both solid and liquid phase. This may be the basis for breeding animals with a smaller rumen size to reduce methane emissions. However, dietary manipulation is regarded the most effective and straightforward method of lowering rumen methane emissions,Citation40 as selective breeding is slow and selection of specific traits may affect favourable variants.
It should also be noted that methane production by archaea represents an energy loss of about 2 – 12% of gross energy intake,Citation41,Citation42 meaning this energy is no longer available for animal growth, lactation, maintenance or pregnancy. Manipulating the diet of ruminants to reduce the number of methanogens would therefore both help reduce the negative impact on the environment, and also improve the efficiency of livestock production.Citation41 Methanogenesis is the only mechanism of ATP synthesis available to methanogenic archaea.Citation42 The methyl coenzyme A reductase (McrA, encoded by mcrA) catalyses the final step in methanogenesis, which means that this enzyme and the genes associated with it may be good markers for the presence of methanogens.Citation36 Furthermore, anti-methanogens, such as bromochloromethane, can be used to inhibit methane production. Bromochloromethane reacts with reduced vitamin B12 and inhibits the cobamide-dependant methyl transferase step,Citation43 which is responsible for the synthesis of McrA. Targeting this step may break the pathway and inhibit production. A recent study, carried out by Kinley et al.,Citation44 investigated the use of an invasive species of macroalgae as a means of reducing methane emissions in vitro. The results showed a decrease in methane production as a result of a secondary metabolite found in the macroalgae Asparagopsis taxiformis, bromoform, which is similar to bromochloromethane in its ability to decrease methanogenic activity.
Ciliate protozoa
Ciliate protozoa are found in the range of 104 – 106 cells/ml in rumen fluid and are responsible for 30 to 40% of overall fibre digestion.Citation16 They are also relatively active in lipid hydrolysis and can produce hydrogen via their hydrosomes.Citation45 The Entodinium genus is the most dominant protozoan in high grain diets. This genus rapidly degrades starch, engulfing it and converting it to an iodophilic storage polymer.Citation16 Degradation occurs through a combination of debranching, amylase and glucosidase enzymes. More research may be needed in order to determine their immediate role in methanogenesis and therefore create a better understanding of the value of manipulating this population as a means of reducing methane emissions in ruminants.
Defaunation is a mechanism used to prevent the growth of, or remove, protozoa from the rumen. This can be carried out by several different means such as through the isolation of calves after birth, chemical defaunation through the use of copper sulphate, calcium peroxide, alcohol ethoxylate, coconut oil, linseed oil or soya oil hydrolysate.Citation46 Another method of defaunation, reviewed by Hook et al. Citation32 involves removal of rumen contents from the host, an invasive method whereby the mucosa is carefully washed and the rumen contents are treated by either heating or freezing to eliminate protozoa. The rumen contents are then returned to the rumen. This method has been used to remove protozoa from the rumen to investigate the role they play in rumen function.Citation32,Citation47 suggested that protozoa-associated methanogens (methanogens sequestered within rumen protozoa) which account for approximately 37% of methanogenesis in ruminants,Citation48 may be the most active methanogen communities in the rumen, and as defaunation would also eliminate them, this would account for the decrease in methane production. Conversely, in a study carried out by Morgavi et al.,Citation8 it was concluded that there was no direct correlation between the number of methanogens and amount of methane produced, but there was a correlation between the number of protozoa and methane production (Wallace et al.).Citation42 showed that methane emissions increased as protozoal abundance increased and stated that protozoa showed the strongest link with methane emissions (in comparison to other domains) after qPCR using DNA extracted from rumen contents. Despite the conflicting reports on their roles in methanogenesis, it is agreed that, ultimately, the presence of protozoa can probably have both beneficial and negative effects on the rumen microbiome,Citation49 as protozoa engulf bacteria and other smaller microbes and particles in the rumen, but also larger molecules including proteins and carbohydrates.Citation50 They actively ingest the bacteria as a source of protein and also act as a stabilising factor for fermentation end products.Citation11
Amoeba
Amoebae can represent an important reservoir for bacteria in the environment, but their role in the rumen is unclear. In the vegetative cycle (multiplication by binary fission), amoeba, similar to ciliate protozoa, survive by ingesting bacteria through phagocytosis.Citation51 While further research is necessary in order to ascertain the role of amoeba in the rumen, it is known that some bacteria can survive phagocytosis by protozoa and live as endosymbionts.Citation52 For instance, Campylobacter jejuni has been shown to invade Acanthamoeba polyphaga and can replicate in vacuoles.Citation53 C. jejuni and C. fetus can have large effects on cow fertility, immunity and overall health.Citation54 Amoeba therefore may be important in rumen and general bovine health.
Fungi
Rumen fungi (103 – 106 zoospores/ml) are anaerobic, falling into the class Neocallimastigomycetes, consisting of 6 previously recognised genera (Anaeromyces, Caecomyces, Cyllamyces, Neocallimastix, Orpinomyces and Piromyces) with 21 known species and, using molecular techniques, 2 recently discovered genera OontomycesCitation55 and Buwchfawromyces.Citation56 Their discovery required a re-evaluation of the central dogma within microbiology that all fungi were aerobic.Citation21 It has been proposed that these fungi evolved through horizontal transfer of genes from bacteria.Citation57 Anaerobic fungi may influence the rest of the microbial community as they produce H2 during initial degradation of plant tissue and this H2 can be used as fuel for the degradation mechanisms of other communities. Fungi colonize plant cell walls and account for approximately 8–12% of total microbial biomass in the rumen, but this figure varies widely depending on diet.Citation21 Working with both the liquid and solid phase of rumen contents is important for the detection of fungal species as, while most plant fibre-associated fungi are retained in the solid phase, the liquid phase may contain smaller particulate matter that fungi may have attached to.
Although bacteria are the most prominent microorganisms in the rumen, fungi are the best degraders. They produce high levels of cellulases and hemicellulases, as well as possessing the ability to break down xylan due to the production of xylanases.Citation58 Fungi appear to initiate the feed breakdown process, indicating that anaerobic fungi may be pivotal for feed utilization efficiency and animal growth and production in pasture-fed ruminants.Citation59 As mentioned, anaerobic fungi are believed to have evolved through horizontal gene transfer from bacteria, and are the only known fungi to retain cellulosomes, which are cell-wall associated multienzyme complexes.Citation57 Metagenomic approaches may be able to indicate the degree of horizontal gene transfer and where these genes were transferred from. By breaking down carbohydrates, fungi produce metabolites used for nutritional purposes by the host.Citation59 They have many features which make them unique in comparison to fungi found outside the rumen, including polyflagellate zoospores (the reason why rumen fungi were originally mistaken for ciliate protozoa), hydrogenosomes, cellulosomes and a wide enzymatic spectra.Citation60
Bacteriophage
Bacteriophage are obligate pathogens of bacteria and occur in dense populations of approximately 107 – 109 particles per gram of digesta in the rumen.Citation61 As is the case for other populations, bacteriophage abundances are also influenced by external sources, meaning they may also be controlled through different strategies. The bacteriophage and virus population found in a sample is referred to as the virome.Citation62 The high number of rumen bacteriophage suggests that they may have an important function in the balance of the rumen system, but there is little known about the effect of the rumen virome on the system it inhabits. Viruses, however, have been shown to be a driving factor for evolution of many microbial systems in different environments, often facilitating horizontal gene transfer.Citation63 Indeed Garcia-Vallve et al.,Citation57 showed that this was important in the transfer of glycoside hydrolase between bacteria and rumen fungi (Ricard et al.).Citation64 proposed that this may also be important in the case of rumen ciliate protozoa, whereby horizontal gene transfer occurs from rumen bacteria in order to aid in protozoa adaption to the carbohydrate-rich environment of the rumen, transferring genes encoding plant cell wall degradation. This information indicates that bacteriophage may represent a shared gene pool for the rumen ecosystem. Again, metagenomics allows us to determine the origin of these genes. In a study carried out by,Citation63 it was found that most viruses found in the rumen were associated with the most dominant phyla, namely Firmicutes, Bacteroidetes and Proteobacteria. Bacteria-phage interaction can be identified by the presence of clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins in the microbial population.Citation63 These genes are fundamental in adaptive immunity in some bacteria and archaea, which enable the organism to respond and eliminate invading genetic material. Once infected by these invading genetic materials, the new DNA is integrated into the host CRISPR locus as new spacers, ultimately encoding a unique spacer sequence.
Techniques for analysis of the rumen microbiome
Culture-dependant approaches
The gastrointestinal tract of ruminant animals has a wide range of extremities, making it difficult to replicate conditions for optimal growth. While specific microbes can grow and can be characterised, a large percentage are unculturable in vitro and cannot be grown on laboratory media. Culture-dependant techniques rely on various selective and enrichment culture conditions in order to replicate the microbes’ natural environment. Culturing anaerobes is quite difficult due to the need to exclude oxygen, the slow growth of the microbes and the complexity of other growth requirements (Rufener et al.).Citation65 used a continuous culture system in order to replicate the rumen environment. This technique, along with similar techniques, was used for the enumeration and identification of rumen microbes.
Using the solid phase of rumen contents can pose many problems when attempting to culture microbes. Many microbes adhere to particulate matter and are thus difficult to separate. Methylcellulose solution can be used to encourage detachment of bacteria from feed particles by providing a readily available feed source as described by Fessenden.Citation66 Traditional methods of classifying rumen bacteria were based on the standard bacterial identification methods; morphology, shape and Gram stain. Nutritional requirements and fermentation end products were also used as a means of classification. Roll tubes came to be employed to grow and isolate anaerobic species, and were used instead of conventional agar plates. The method was developed by Hungate,Citation18 whereby agar medium was distributed in a thin layer over the internal surface of a test tube, which was then flushed with an oxygen-free gas of choice. According to Hungate, the tube required no particular incubation and could be examined with no anaerobic precautions necessary. The Hungate 1000 project, named after the aforementioned Robert Hungate, one of the first rumen microbiologists, seeks to combine culturing and high throughput sequencing approaches. More specifically, it is a catalogue of reference genomes from the rumen microbiome, which aims to produce a reference set of 1000 rumen genome sequences of cultivated rumen bacteria, ciliate protozoa, archaea and fungi. The project focuses on rumen bacteria and archaea, with a similar project, The Fungal 1000, focusing on fungal cultivation. The aim of the project is to support international efforts to develop methane mitigation strategies and rumen adaption technologies, as well as to initiate genome enabled research into understanding rumen function, feed conversion efficiency, methanogenesis and plant cell wall degradation in order to find a balance between food production and GHG. Along with this, the project will aid in discovering genes encoding fibre degradation that will enhance animal production and other functional genes such as those involved in biofuel production.Citation20
Creevey et al.Citation67 assessed several culture collections and databases, and found that members of the phylum Firmicutes and family Lachnospiraceae were the most commonly cultured bacteria from the rumen. Bacteriodetes represented just a small number with the strains identified belonging to just 2 genera. It was concluded that although the culture collections identified all of the major taxonomic groups that are widely known to be key players in rumen function, they do not represent the full diversity of the rumen microbiome. As stated previously Krause et al.,Citation21 estimated that only 20% of the rumen microbiota can be cultured using standard techniques, and, more recently McCabe et al.,Citation68 suggested that culturable microbes in the rumen account for less than 1% of the total microbial species.
Culture-independent approaches
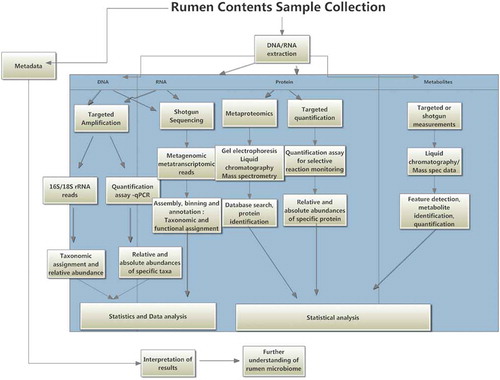
Culture-independent methods, or, more specifically DNA-based methods of identification and detection of microorganisms, allow the examination of microbial communities at a molecular level. Metagenomic analysis allows the description of a microbial community by high throughput sequencing technology. Methods associated with this type of analysis include 16S rRNA and Internal Transcribed Spacer (ITS) amplicon sequencing, used for bacterial and fungal communities, respectively, or, shotgun sequencing, where DNA fragments are sequenced randomly, regardless of the microbe from which they came. Targeting of the mcrA gene has also been suggested in recent studies as a means of identifying methanogens.Citation69 All of these methods use bioinformatics as a means of analysis and can be used in order to compare different microorganisms within a diverse ecosystem. Although these methods identify unculturable microbes, they do not provide a strain for further study. They may, however, provide insights that would allow the culturing of species that, thus far, have not been cultured. Understanding community structure is an important part in the recognition of how microorganisms are affected by the environment which they live in and how they in turn affect the host organism. DNA sequencing technologies have transformed research of microbial and animal ecosystems. They have completely changed the approach involved in identifying microorganisms and the limitations, outlined above, associated with culture-dependant studies. Several platforms are available to achieve results; these differ only in small details, and all follow the same basic protocol of template preparation and clonal amplification, followed by rounds of parallel sequencing.Citation70 Strategies used by each platform determine quality, quantity and bias of sequenced data. , identifies the steps which may be included for metagenomic, metabolomic and proteomic studies.
Figure 2. Rumen microbiome project workflow including options for metabolomics and metaproteomics (Figure adapted from Deusch et al. (2014)).
16S rRNA amplicon sequencing requires the use of primers in order to identify the presence of specific bacteria and archaea. 18S target regions can also be used in identification of protozoa but may run the risk of amplifying bovine DNA, thereby affecting results. The ITS region found in fungi is the most widely used region to study the ecology of fungi. Various primers have been designed in order to yield the highest diversity. 16S sequencing is useful as it provides a good phylogenic marker and there is a large database of sequences to assist in data analysis. It can identify highly conserved regions that are identical in all bacteria, with the use of a single primer pair. However, primer specificity is key and lack thereof will contribute to bias in detection of a target organism. Often, it is difficult to choose or design a primer that is specific to diverse groups, such as Firmicutes.Citation20 Incorrect PCR conditions, such as improper annealing temperatures during the amplification step in 16S, may result in false amplification. Additionally, 16S rRNA sequencing only gives information on bacterial populations, not fungal, viral, protozoa or amoeba, all of which are believed to play an important role in the rumen, as discussed earlier.
Kunin et al.Citation71 defined metagenomics as the application of shotgun sequencing to DNA obtained directly from an environmental sample, producing at least 50Mbp of randomly sampled sequence data. Shotgun sequencing comprehensively samples all genes in all organisms, thus helping to explain what is there, what they are doing and to potentially examine why they are doing it. It evaluates total microbial diversity, detecting the abundance of microbes in a given environment. An important function is that it by-passes the need for obtaining pure cultures for sequencing, as this is often not possible for samples obtained from complex environments such as the rumen. It is a modern technology that is continuing to improve our ability to understand complex ecosystems, enabling the analysis of previously undetectable and unculturable microorganisms, otherwise difficult to analyse. It is also able to detect low abundance members of the microbial community, microbes which are dormant or inactive.
While the application of culture-based approaches to study ruminal contents has declined in recent years, our understanding of the rumen microbiome across different diets, species and geographical location has expanded due to the introduction of high throughput sequencing techniques.Citation67 Culture-independent approaches rely on bioinformatics tools to handle large datasets. Programmes such as Kraken, assign taxonomic labels to short DNA sequences achieved through metagenomic studies.Citation72 Superfocus, used in functional analysis of unannotated shotgun data (discussed below), classifies each sequence in the metagenome into a subsystem.Citation73 Other programmes and databases used include: Qiime, RDP and Humann2. Statistical packages are then used to analyse the data.
Dietary interaction with the rumen system
Feed conversion efficiency
Feed conversion efficiency can be defined as kg of milk of standardised composition with respect to protein and fat concentrations produced per kg of dry matter consumed.Citation74 It is an important measure of the performance of a farm feeding system. Residual feed intake (RFI) is a measure of feed efficiency, defined as the difference between the actual feed intake of an animal and its predicted feed intake based on maintenance energy requirement and growth rate.Citation75,Citation76 Many factors affect feed efficiency, with forages having the greatest effect due to making up the largest component of the slowly digestible portion of the diet.Citation77 The amount of feed fed, and the ingredients contained in the ration, can influence feed efficiency.Citation78 High quality, rather than poor quality, forage, also improves feed efficiency and is essential to farm profitability. Supplementary feeding in a grass-based system is necessary as nutritional needs cannot be met solely on a grass-based diet during times of the year when grass quantity and quality is not at its optimum. Improvements in feed efficiency would reduce on-farm costs and lead to production system efficiency.Citation76 Supplementing forage with concentrates, such as cereal grains, improves feed efficiency, due to grains and their by-products breaking down more effectively in the digestive tract. As our understanding of the rumen microbiota deepens, and it’s associated effect on the host, it is possible to make links between microbial communities and host phenotypes.Citation79 One of these being feed conversion efficiency. Metatranscriptomic analysis of the rumen microbial community has shown significant differences in both bacterial and archaeal community structure between high residual feed intake groups and low residual feed intake groups.Citation80 It was found that the microbiome of low RFI groups had the potential to adapt more quickly to environmental stresses, such as that of intense dietary changes therefore. Changes in the abundances of the family Lachnospiraceae were noted between groups. Members of this family are associated with butyrate production, a VFA produced through the digestion of carbohydrates.Citation81 The higher abundance of this family in high RFI groups may indicate an increase in butyrate metabolism, therefore, a resulting impact on feed efficiency. In contrast, the order Methanomassiliicoccales were more abundant in low RFI animals. Members of this order can use methylamine as an energy source, which may be harmful to the host, to produce additional ammonium during methanogenesis.
In high concentrate diets, methane reduction is induced via an increase in propionate which decreases H2. Enteric methane emissions are also influenced by concentrate composition.Citation82 Loss of energy in the ruminant digestive tract is a consequence of the production of methane. Therefore, including feeds which decrease methane production is important to reduce energy loss in the animal. Ruminal methane production represents approximately 2 – 12% of gross energy intake, which could otherwise be used for animal growth or milk production. Lowering emissions would benefit both the environment and the efficiency of livestock production. The interest in feed conversion efficiency is growing due to the need for more animal-derived protein and energy sources as well as minimizing the carbon footprint of the livestock sector.Citation83
Methane production through fermentation involves the conversion of almost all polymers to methane and carbon dioxide under anaerobic conditions. It is the result of a number of metabolic interactions among the microbial ecosystem described above. As reviewed by Sirohi et al.,Citation35 Wolfe began the first studies of CO2 reduction to methane in the early 1970s, discovering six new coenzymes in the following years. He found that the process of methanogenesis requires seven coenzymes and eight enzymes.Citation35 CO2 and methane are removed by eructation to the atmosphere while acetate, propionate and butyrate travel across the rumen wall to the blood stream. Methane is produced by utilising simple substrates at low reduction potential to produce cellular energy.Citation35 Methane synthesis contributes to the efficiency of the entire system and avoids the build-up of H2, which would inhibit the normal function of microbial enzymes involved in electron transfer.Citation8 Indeed, if ruminants did not produce any methane, the rumen pH would drop and fibre digestion would no longer be feasible.
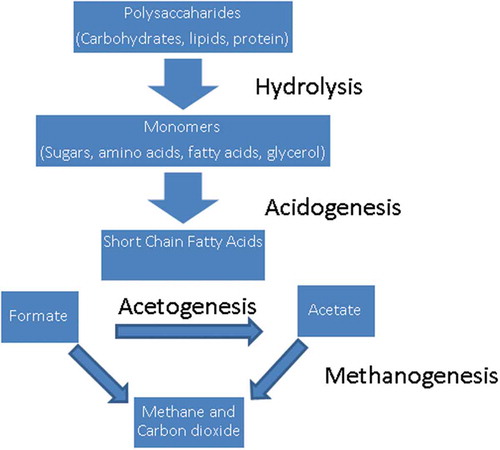
A recent study suggested that methane production can, however, be reduced substantially without adverse effects on fibre digestion.Citation84 The authors examined the effects of methane inhibition in the rumen of steers. The animals were fed either a hay:roughage diet or a hay:concentrate diet at a ratio of 60:40. An anti-methanogenic compound was also included, in this case, and in many others, chloroform. Results showed that with increasing levels of chloroform, there was an increase in H2 expelled and CH4 production decreased. illustrates the degradation of plant fibre from the hydrolysis of polysaccharides, releasing monomers. The monomers are then fermented through acidogenesis, resulting in the production of organic and short chain fatty acids. Hydrogen and CO2 are also formed and are used by methanogenic archaea to produce methane. Animals on the hay only diet showed a more efficient redirection of H2 into other microbial products in comparison to hay:concentrate diet. Metabolomic studies showed that there was an increase in the levels of amino acids, organic and nucleic acids found in the liquid phase of the rumen contents in both diets when methanogenesis was inhibited. This suggests that there may be enhanced microbial protein synthesis under these conditions. These changes showed an obvious alteration in the rumen microbiota, with an increase in the ratio of Bacteroidetes and Firmicutes and a decrease in Archaea and Synergistetes. However, there were no significant changes observed in the abundance of protozoa, fungi or fibrolytic bacteria, meaning that fibre degradation was not affected.Citation84 The conclusion was made that, although there was a 30% decrease in methane production, this did not negatively affect rumen fermentation and fibre degradation, and the microbes were able to adapt and redirect H2 to produce other end products. Further supplementation may be needed in order to drive excess H2 and improve the energy supply to the animal.Citation84 Although it had been believed that without the formation of methane, the rumen microbiota would be drastically affected, this study shows that it can adapt to changes in H2 flux and produce different end products without the need to produce methane, in turn reducing methane eructed by the host animal.
There have been many attempts to minimise energy loss as a result of methane production, some of which have been mentioned earlier in this review. Many studies have explored the application of compounds and natural products to feed, prior to consumption by the animals.Citation85–Citation87 A novel, promising approach, is the use of probiotics. Probiotics present health benefits for the host when provided in sufficient amounts.Citation88 Citation89 explored the use of Bacillus licheniformis as a novel probiotic to reduce enteric methane emissions (Deng et al.).Citation89 The study found that the use of the bacteria as a probiotic was sufficient in inhibiting methane emissions in sheep, increasing total tract digestibility and improve NUE. However, the subsequent effect on the overall rumen microbiome was not examined.
Emerging areas of interest
The usage of antibiotics in animal production systems has encouraged the abundance of antimicrobial resistance genes in farm environments to increase. It is a current threat to both human and animal health. Antibiotics are used therapeutically to treat or prevent specific diseases and those that are not entirely absorbed are excreted in the faeces and urine.Citation90 As animal manure is collected in large pits for use as a high nutrient fertiliser, antimicrobial compounds can then be transferred to the soil.Citation91 This may result in the possible uptake of these compounds by plants. Different land uses may require higher quantities of manure in order to provide higher concentrations of nutrients needed by specific plants. For example, land with which will be used for ensiling of grass may have larger quantities of manure applied in comparison to land used for seasonal grazing. This, in turn may result in varying abundances of AMR genes found in animals. In a study carried out by,Citation92 the influence of diet on the ruminal resistome and abundance of pathogenicity genes was examined. The animals used in the study were antibiotic-free beef cattle on two different diets consisting of different levels of concentrate. Over 200 genes associated with antimicrobial resistance were detected across 4966 metagenomic genes, with results showing a higher diversity and abundance of these genes in animals fed a higher level of concentrate than animals on lower levels of concentrate. These results suggest, therefore, that not only does diet impact the microbial ecosystem but it also has the potential to impact antimicrobial resistance in the gut.
Interestingly, a recent study on the human gut microbiota, suggest that even non-antibiotic compounds, such as anti-inflammatories, may alter the gut microbiota.Citation93 It was found that resistance mechanisms to antibiotics and human targeted drugs somewhat overlap. This then poses the question as to whether non-antibiotic compounds could have a comparable impact on the gut microbiota of cattle.
Emerging metagenomic techniques
The rumen microbiota is a topic that has been covered in great detail since the emergence of NGS technologies.Citation38,Citation94–Citation96 Most studies use reference databases such as Kraken (shotgun sequencing)Citation9 and Greengenes (16S rRNA),Citation10 among many others, to characterise the microbiota. The use of contigs or longer contiguous sequencesCitation97 for metagenomic analysis, may provide a deeper insight into the workings of the microbiome. Shorter reads, although having its advantages, do not represent complete genomes and may contain contamination. Studies have shown that the use of contigs may be an invaluable technique for the recovery of microbial genomes. Hess et al. (2011)Citation98 were the first to use metagenomic binning techniques for the analysis of the rumen microbiota. Fifteen uncultured microbial genomes were assembled to > 60% completeness. The technique was also used in a study of the moose rumen microbiome, which recovered 99 metagenome-assembled genomes.Citation99 The most recent study by Stewart et al. (2018),Citation100 assembled 913 microbial genomes from metagenomic data of the rumen of cattle. This resulted in over 69,000 proteins thought to be involved in carbohydrate metabolism, in addition to the expansion of the Erysipelotrichales order. The study has, therefore, contributed immensely to improved metagenomic classification.
This study also highlights the potential to use hybrid short-read and long-read sequencing approaches in order to obtain complete end to end assembly of microbial chromosomes direct from metagenomic data.Citation100 This approach may involve the use of both Illumina technologies for shorter reads and Pac-Bio or Oxford Nanopore MinIon for longer reads.Citation101 The advantages of this approach include providing data on within-species genomic variation,Citation102 identify pathogenic variants of species and determining the presence of pathogenicity islands and plasmids important for the detection of AMR.Citation101
Protein metabolism
Protein consumed by ruminants can be described as rumen degradable protein (non-protein and true protein N) or rumen un-degradable protein.Citation50 Dietary protein intake determines the outcome of several factors associated with both the usable by-products and non-usable by-products, namely milk nitrogen efficiency, urinary nitrogen losses and ammonia emissions for the ruminant faeces.
Nitrate/nitrite cycle
The protein profile of feeds ingested by ruminants is extensively altered before reaching the small intestine due to fermentation within the rumen. This is primarily due to the conversion of nitrogenous compounds to microbial protein. The entire process is controlled by the fermentation of carbohydrates with recycling of nitrogen between the host and the gut.
The rumen has the ability to reduce nitrate to nitrite and nitrite to ammonia. Conversion of nitrate to ammonia leads to an increase of nitrogen available for use by the rumen microbes.Citation103 It is thought that feeding nitrate to dairy cows may be an alternative method of reducing methane emissions by suppressing methanogenesis in the rumen. This is because, thermodynamically, nitrate and nitrite reduction to ammonia is a preferred reaction compared to the formation of methane with CO2 as an electron acceptor, consuming electrons at the expense of methanogenesis. Recent studies have investigated the use of 3-Nitrooxypropanol (3NOP) to inhibit methane production.Citation104,Citation105 In a study carried out by,Citation105 rumen methane emissions were decreased linearly by 30% through the use of 3NOP, while increasing milk protein and lactose yields. Animal performance also increased, suggesting that energy previously wasted through the production of methane was instead used by the host to increase body weight. It is also important to note that the inhibitory effect continued over the 12 week period of the trial, thus suggesting this is a means of effective methane mitigation which can be applied to the livestock industry. Although evidently an exceptional means of reducing methane emissions, the safety of 3NOP is questionable in terms of milk processing due to fears that excess nitrate passing from the rumen to the host could result in trace amounts of nitrate being found in the milk. Trace amounts are often not a concern but, with most milk being processed for use as milk powders, nitrate may become concentrated after drying stages, and even more cause for concern is the use of these powders for baby formulas where nitrate can cause methemoglobinemia or “Blue Baby Syndrome”. Therefore, a need for more comprehensive studies on milk produced by animals on 3NOP will need to be carried out in order to certify the safety of end products.
Nitrogen fractions in milk
The composition of milk is important in milk processing as it affects the quality of the product produced and the economic output of the dairy industry.Citation106 Milk composition is an important factor with respect to milk processability. Key components are milk protein fractions, i.e., casein, whey and non-protein nitrogen (NPN), and minerals, with the most important being calcium, phosphorus and sodium. NPN refers to any compound that is not a true protein but can be converted into protein in the rumen following microbial synthesis.Citation107 NPN is currently wasteful with regard to the processing of milk, thus a push towards having a larger proportion of casein or whey, relative to NPN, in the milk would mean more profitability for the processors per kg of milk produced. A high milk protein quality is key to maintaining thermal stability and gelation during processing.Citation108 Urea is the most available NPN and is quickly broken down by bacterial ureases to form ammonia.Citation109 The ammonia that is formed is important for bacterial growth, as it is used for amino acid synthesis, required for optimum growth. However, when ammonia is produced in high concentrations in the rumen, a certain amount is reabsorbed back into the bloodstream, converted back into urea in the liver and excreted through the kidneys and passed out as urine. It should be noted that urea may play an important role in low protein diets, compensating for the low concentrations of dietary protein in the diet, making urea utilization higher in comparison to that of high protein diets.
Nitrogen utilization efficiency
The importance of reducing nitrogen loss is critical to agriculture from a nutritional, environmental and economic perspective.Citation110 Improving nitrogen utilization in dairy animals is fundamental to dairy product production and the environment. The agricultural industry contributes to approximately 40% of anthropogenic nitrous oxide emissions in Europe, with the dairy industry accounting for the most nitrogen loss in faeces and urine in comparison to any other ruminant animal in the agri-food sector. Not only does nitrogen loss have a huge impact on the environment, it also has huge consequences for the agricultural industry financially.Citation111 claimed that feeding nitrogen for optimum milk yields rather than maximum milk yields is the best way to reduce nitrogen pollution on dairy farms, without having consequences on productivity or profitability. As mentioned previously, dairy farms relying on grazed grass as their primary feed source may experience problems in improving efficiency of protein utilizationCitation110 due to the naturally high levels of protein in well-managed grass.
A means of improving nitrogen utilization efficiency (NUE) is by trying to synchronise the supply of energy and protein in the rumen through the use of dietary supplementation. Dietary protein and energy sources degrade in the rumen at different rates, available for use by the host at different times. Rumen synchrony refers to the provision of dietary protein and energy to the rumen so that they are available for use in proportions required or used by rumen microbes.Citation112
Reid et al.Citation106 examined the effect of dietary crude protein on grass-fed dairy cow production, nutrient status and milk heat stability. The study found that supplementing with high crude protein concentrates increased milk urea nitrogen concentration and milk non-protein nitrogen concentration, in comparison to supplementing with low crude protein concentrate. It was also found that increasing dietary crude protein lowered milk heat stability, which is undesirable in the processing of milk. Examining the nitrogen supply to the rumen based on the varying concentrations of protein consumed and how it effects the rumen environment and its microbial ecosystem is important in terms of determining what microbes can breakdown the feed stuff and use it efficiently, without wasting energy. Feeding animals more protein than is needed results in unnecessarily high feeding costs, reduced profits and further reductions in nitrogen utilization efficiency.Citation110 High levels of dietary crude protein have a positive effect on protein degradation in the rumen, owing to increased ammonia concentrations, and have been shown to reduce nitrogen utilization efficiency for milk production.Citation113 Reid et al.Citation106 found that supplementation improved NUE in comparison to a grass only diet. Supplementation is important as it aims to provide further nutrients for the animals that may not have been provided by grass alone.
Conclusion
In conclusion, the rumen microbial ecosystem is diverse and complex, consisting of microorganisms working symbiotically to break down feedstuffs consumed by ruminant animals. As addressed throughout the review, the microbiome controls the production efficiency of the animal, with certain pathways (such as those associated with methane production) resulting in energy loss in the animal. The microbiome also affects end-product quality (milk and meat) but also contributes to environmental pollution. High throughput sequencing-based techniques allow one to identify what microbes are there and, depending on the approach taken, what they are doing. Understanding the rumen microbiome and its connection to the ruminant itself is important for producing quality products, increasing profitability and reducing environmental impacts. Identifying certain metabolic pathways and further research into these pathways may determine the best diet for bovine ruminants in order to minimise energy loss, reduce methane production and increase nitrogen utilization efficiency. Examining the rumen microbiome can identify the effects of diet on the microbiome and in turn, the effects on milk yield, protein percentages, urea percentage (used as a NPN indicator) and milk protein yield. The composition of proteins found in milk can have a profound effect on milk processability, thus identifying pathways which may influence milk composition and improve processability is important.
Acknowledgments
The authors wish to thank the Moorepark DNA sequencing and bioinformatics teams.
Additional information
Funding
References
- International Farm Comparison Network. Long-term dairy outlook. IFCN - the dairy research network. 2016.
- Agriculture and Horticulture Development Board. Dairy Statistics - An insider’s guide 2015 [Internet]. 2015. http://www.dairyco.org.uk/non_umbraco/download.aspx?media=1438.
- OECD-FAO. OECD - FAO agricultural outlook 2016-2025. 2016.
- Lagrange V, Whitsett D, Burris C. Global market for dairy proteins. J Food Sci. 2015;80(S1). DOI:10.1111/1750-3841.12801
- Pickering NK, Oddy VH, Basarab J, Cammack K, Hayes B, Hegarty RS, Lassen J, McEwan JC, Miller S, Pinares-Patiño CS, et al. Genetic possibilities to reduce enteric methane emissions from ruminants. Animal. 2015;9(9):1431–1440. doi:10.1017/S1751731114003280.
- Rojas-Downing MM, Nejadhashemi AP, Harrigan T, Woznicki SA. Climate change and livestock: impacts, adaptation, and mitigation. Clim Risk Manag [Internet]. 2017;16:145–163.
- Krehbiel CR. Invited Review: applied nutrition of ruminants: fermentation and digestive physiology. Prof Anim Sci [Internet]. 2014;30(2):129–139.
- Morgavi DP, Forano E, Martin C, Newbold CJ. Microbial ecosystem and methanogenesis in ruminants. Animal. 2012;6(5):871.
- Brulc JM, Antonopoulos DA, Miller MEB, Wilson MK, Yannarell AC, Dinsdale EA, Edwards RE, Frank ED, Emerson JB, Wacklin P, et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA. 2009;106(6):1948–1953.
- Mccann JC, Wickersham TA, Loor JJ. High-throughput methods redefine the rumen microbiome and its relationship with nutrition and metabolism. Bioinform Biol Insights. 2014;109–125.
- Ishler V, Heinrichs J, Varga G. From feed to milk : understanding rumen function. 1996.
- Byrant MP. Normal Flora - Rumen Bacteria. Am J Clin Nutr. 1970;23(11):1440–1450.
- Gordon GL, Phillips MW The role of anaerobic gut fungi in ruminants. Nutr Res Rev [Internet]. 1998;11(1):133–168. http://www.ncbi.nlm.nih.gov/pubmed/19087463.
- Husvéth F. Physiological and reproductional aspects of animal production. 2011.
- Depeters EJ, George LW. Rumen transfaunation. Immunol Lett [Internet]. 2014;162(2):69–76.
- McSweeney CS, Mackie R. Micro-organisms and ruminant digestion: state of knowledge, trends and future prospects. 2012.
- Castillo-Gonzalez AR, Burrola-Barraza ME, Dominguez-Viveros J, Chavez-Martinez A. Rumen microorganisms and fermentation Microorganismos y fermentación ruminal. Arch Med Vet. 2014;46:349–361.
- Hungate RE. The Rumen and its Microbes. New York: Academic Press; 1966.
- Oetzel GR. Introduction to Ruminal Acidosis in Dairy Cattle. Technology. 2003;15:307–317.
- Chaucheyras-Durand F, Ossa F. Review: the rumen microbiome: composition, abundance, diversity, and new investigative tools. Prof Anim Sci [Internet]. 2014;30(1):1–12.
- Krause DO, Nagaraja TG, Wright ADG, Callaway TR. Board-invited review: rumen microbiology: leading the way in microbial ecology. J Anim Sci. 2013;91(1):331–341.
- Henderson G, Cox F, Ganesh S, Jonker A, Young W, Janssen PH. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep [Internet]. 2015;5(April):14567. http://www.scopus.com/inward/record.url?eid=2-s2.0-84943655505&partnerID=tZOtx3y1.
- Koike S, Kobayashi Y. Fibrolytic rumen bacteria: their ecology and functions. Asian-Australasian J Anim Sci. 2009;22(1):131–138.
- Ratti RP, Botta LS, Sakamoto IK, Silva EL, Varesche MBA. Production of H2 from cellulose by rumen microorganisms: effects of inocula pre-treatment and enzymatic hydrolysis. Biotechnol Lett. 2014;36(3):537–546.
- Fernando SC, Ii HTP, Najar FZ, Sukharnikov LO, Krehbiel CR, Nagaraja TG, Roe BA, Desilva U. Rumen microbial population dynamics during adaptation to a high-grain diet. 2010;76(22):7482–7490.
- Chen Y, Oba M, Guan LL. Variation of bacterial communities and expression of Toll-like receptor genes in the rumen of steers differing in susceptibility to subacute ruminal acidosis. Vet Microbiol [Internet]. 2012;159(3–4):451–459.
- Duskova D, Marounek M. Fermentation of pectin and glucose and activity of pectin-degrading enzymes in the rumen bacterium Lachnospira multiparus. Lett Appl Microbiol. 2001;33:159–163.
- Santos GT, Lima LS, Schogor ALB, Romero JV, De Marchi FE, Grande PA, Santos NW, Santos FS, Kazama R. Citrus pulp as a dietary source of antioxidants for lactating holstein cows fed highly polyunsaturated fatty acid diets. Asian-Australasian J Anim Sci. 2014;27(8):1104–1113.
- Santos F, Menezes Junior M, Simas J, Pires A, Nussio C. Corn grain processing and its partial replacement by pelleted citrus pulp on performance, nutrient digestibility and blood parameters of dairy cows. Asian-Australasian J Anim Sci. 2001;23:923–931.
- Jami E, White BA, Mizrahi I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS One. 2014;9:1.
- Bainbridge ML, Cersosimo LM, Wright ADG, Kraft J. Rumen bacterial communities shift across a lactation in Holstein, Jersey and Holstein × Jersey dairy cows and correlate to rumen function, bacterial fatty acid composition and production parameters. FEMS Microbiol Ecol. 2016;92(5):1–14.
- Hook SE, Wright A-DG, McBride BW. Methanogens: methane producers of the rumen and mitigation strategies. Archaea [Internet]. 2010;2010:1–11.
- Lin C, Raskin L, Stahl DA. Microbial community structure in gastrointestinal tracts of domestic animals: comparetive analyses using rRNA-targeted oligonucleotide probes. FEMS Microbiol Ecol. 1997;22(28):281–294.
- Jarrell KF, Bayley DP, Correia JD, Thomas NA. Recent about Excitement the Archaea The Archaea are valuable for studying basic biological questions and have novel biotechnology applications. Sci York. 2009;49(7):530–541.
- Sirohi SK, Pandey N, Singh B, Puniya AK. Rumen methanogens: A review. Indian J Microbiol. 2010;50(3):253–262.
- Janssen PH, Kirs M. Structure of the Archaeal Community of the Rumen. Appl Environ Microbiol. 2008;74(12):3619–3625.
- Patra A, Park T, Kim M, Yu Z. Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J Anim Sci Biotechnol [Internet]. 2017;8(1):13.
- Leahy SC, Kelly WJ, Ronimus RS, Wedlock N, Altermann E, Attwood GT. Genome sequencing of rumen bacteria and archaea and its application to methane mitigation strategies. Animal. 2013;7(s2):235–243.
- Goopy JP, Donaldson A, Hegarty R, Vercoe PE, Haynes F, Barnett M, Moughan PJ. Low-methane yield sheep have smaller rumens and shorter rumen retention time. Br J Nutr. 2014;111(4):578–585.
- Beauchemin K, McAllister TA, McGinn SM. Dietary mitigation of enteric methane from cattle. CAB Rev Perspect Agric Vet Sci Nutr Nat Resour [Internet]. 2009;4(35):1–18.
- Johnson KA, Johnson DE. Methane emissions from cattle. J Anim Sci. 1995;(8):2483–2492.
- Wallace RJ, Rooke JA, Duthie CA, Hyslop JJ, Ross DW, McKain N, De Souza SM, Snelling TJ, Waterhouse A, Roehe R Archaeal abundance in post-mortem ruminal digesta may help predict methane emissions from beef cattle. Sci Reports [Internet]. 2014;4(2):5892. http://www.ncbi.nlm.nih.gov/pubmed/25081098.
- Denman SE, Tomkins NW, McSweeney CS. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol Ecol. 2007;62(3):313–322.
- Kinley RD, De NR, Vucko MJ, Machado L, Tomkins NW. The red macroalgae Asparagopsis taxiformis is a potent natural antimethanogenic that reduces methane production during in vitro fermentation with rumen fluid. Anim Prod Sci. 2016;56:282–289. DOI:10.1071/AN15576
- Tymensen LD, Beauchemin KA, McAllister TA. Structures of free-living and protozoa-associated methanogen communities in the bovine rumen differ according to comparative analysis of 16S rRNA and mcrA genes. Microbiology [Internet]. 2012;158(7):1808–1817. http://www.ncbi.nlm.nih.gov/pubmed/22539164.
- Jouany J. Rumen microbial metabolism and ruminant digestion. 1991;240.
- Newbold CJ, De La Fuente G, Belanche A, Ramos-Morales E, McEwan NR. The role of ciliate protozoa in the rumen. Front Microbiol. 2015;6(NOV):1–14.
- Finlay BJ, Esteban G, Clarke KJ, Williams AG, Embley TM, Hirt RP. Some rumen ciliates have endosymbiotic methanogens. FEMS Microbiol Lett. 1994;117(2):157–161.
- McSweeney CS, Denman SE, Wright ADG, Yu Z. Application of recent DNA/RNA-based techniques in rumen ecology. Asian-Australasian J Anim Sci. 2007;20(2):283–294.
- Bach A, Calsamiglia S, Stern MD. Nitrogen metabolism in the Rumen. J Dairy Sci [Internet]. 2005;88:E9–21.
- Indikova I, Humphrey TJ, Hilbert F. Survival with a helping hand: campylobacter and microbiota. Front Microbiol. 2015;6(NOV):1–6.
- Kelly WJ, Leahy SC, Li D, Reilly K, Lambie SC, McAllister TA, Valle ER, Attwood GT, Altermann E The complete genome sequence of the rumen methanogen Methanosarcina barkeri CM1. Stand Genomic Sci [Internet]. 2015;10(1):57. http://www.ncbi.nlm.nih.gov/pubmed/26413197.
- Olofsson J, Axelsson-Olsson D, Brudin L, Olsen B, Ellström P. Campylobacter jejuni actively invades the amoeba Acanthamoeba polyphaga and survives within non digestive vacuoles. PLoS One. 2013;8:11.
- Sahin O, Fitzgerald C, Stroika S, Zhao S, Sippy RJ, Kwan P, Plummer PJ, Han J, Yaeger MJ, Zhang Q. Molecular evidence for zoonotic transmission of an emergent, highly pathogenic Campylobacter jejuni clone in the United States. J Clin Microbiol. 2012;50(3):680–687.
- Dagar SS, Kumar S, Griffith GW, Edwards JE, Callaghan TM, Singh R, Nagpal AK, Puniya AK. A new anaerobic fungus (Oontomyces anksri gen. nov., sp. nov.) from the digestive tract of the Indian camel (Camelus dromedarius). Fungal Biol [Internet]. 2015;119(8):731–737.
- Callaghan TM, Podmirseg SM, Hohlweck D, Edwards JE, Puniya AK, Dagar SS, Griffith GW. Buwchfawromyces eastonii gen. nov., sp. nov.: a new anaerobic fungus (Neocallimastigomycota) isolated from buffalo faeces. MycoKeys [Internet]. 2015;9(34):11–28. http://mycokeys.pensoft.net/articles.php?id=4799.
- Garcia-Vallve S, Romeu A, Palau J. Horizontal gene transfer of glycosyl hydrolases of the rumen fungi. Mol Biol Evol [Internet]. 2000;17(3):352–361.
- Akin DE, Borneman WS. Role of rumen fungi in fiber degradation. J Dairy Sci [Internet]. 1990;114(10):3023–3032.
- Kittelmann S, Naylor GE, Koolaard JP, Janssen PH. A proposed taxonomy of anaerobic fungi (class neocallimastigomycetes) suitable for large-scale sequence-based community structure analysis. PLoS One. 2012;7(5):1–13.
- Fliegerová K, Mrázek J, Hoffmann K, Zábranská J, Voigt K. Diversity of anaerobic fungi within cow manure determined by ITS1 analysis. Folia Microbiol (Praha). 2010;55(4):319–325.
- Klieve AVA, Swain RAB, Nolan JVB. Bacteriophages in the Rumen; types present, population size and implications for the efficiency of feed utilisation. Popul (English Ed. 1996;2(2):92–94.
- Ross EM, Petrovski S, Moate PJ, Hayes BJ. Metagenomics of rumen bacteriophage from thirteen lactating dairy cattle. BMC Microbiol [Internet]. 2013;13(1):242.
- Berg Miller ME, Yeoman CJ, Chia N, Tringe SG, Angly FE, Edwards RA, Flint HJ, Lamed R, Bayer EA, White BA. Phage-bacteria relationships and CRISPR elements revealed by a metagenomic survey of the rumen microbiome. Environ Microbiol. 2012;14(1):207–227.
- Ricard G, McEwan NR, Dutilh BE, Jouany J-PP, Macheboeuf D, Mitsumori M, et al Horizontal gene transfer from Bacteria to rumen Ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics [Internet]. 2006;7:22. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16472398.
- Rufener WH, Nelson W, Wolin MJ. Maintenance of the rumen microbial population in continuous culture. Appl Microbiol. 1962;11(May).
- Fessenden SW. Amino acid supply in lactating dairy cattle. Cornell University; 2016.
- Creevey CJ, Kelly WJ, Henderson G, Leahy SC. Determining the culturability of the rumen bacterial microbiome. Microb Biotechnol [Internet]. 2014;7(5):467–479.
- McCabe MS, Cormican P, Keogh K, O’ Connor A, Eoin OH, Pallandino RA, Kenny DA, Waters SM. Illumina MiSeq phylogenetic amplicon sequencing shows a large reduction of an uncharacterised succinivibrionaceae and an increase of the methanobrevibacter gottschalkii clade in feed restricted cattle. PLoS One [Internet]. 2015;10(7). http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0133234.
- Luton PE, Wayne JM, Sharp RJ, Riley PW. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology. 2016;2002:3521–3530.
- Reuter JA, Spacek D, Snyder MP. High-Throughput Sequencing Technologies. Mol Cell. 2015;58(4):586–597.
- Kunin V, Copeland A, Lapidus A, Mavromatis K, Hugenholtz P. A bioinformatician’s guide to metagenomics. Microbiol Mol Biol Rev [Internet]. 2008;72(4):557–578. Table of Contents. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2593568&tool=pmcentrez&rendertype=abstract.
- Wood DE, Salzberg SL. Kraken : ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46.
- Silva GGZ, Green KT, Dutilh BE, Edwards RA. Sequence analysis SUPER-FOCUS : a tool for agile functional analysis of shotgun metagenomic data. Bioinformatics. 2016;32(October 2015):354–361.
- Beever DE, Doyle PT. Feed conversion efficiency as a key determinant of dairy herd performance :a review. Aust J Exp Agric. 2007;47:645–657.
- Carberry CA, Waters SM, Waters SM, Kenny DA, Creevey CJ. Rumen methanogenic genotypes differ in abundance according to host residual feed intake phenotype and diet type. Appl Environ Microbiol [Internet]. 2014;80(2):586–594.
- Nkrumah JD, Okine EK, Mathison GW, Schmid K, Li C, Basarab JA, Price MA, Wang Z, Moore SS. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J Anim Sci. 2006;84:145–153.
- Maulfair D, Heinrichs J, Ishler V. Feed efficiency for lactating dairy cows and its relationship to income over feed costs. 2011;DAS 11-183:1–6. College of Agricultural Sciences.
- Heinrichs J, Suarez J, Jones C. Feed efficiency in dairy heifers [Internet]. 2011. http://extension.psu.edu/animals/dairy/nutrition/heifers/heifer-feeding-and-management/feed-efficiency-in-dairy-heifers/extension_publication_file.
- Guan L, Li F, Bulumulla A, Zhou M. The role of rumen microbiome on feed efficiency of grazing cattle. Dep Agric Food Nutr Sci Univ Alberta [Internet]. 2014;137–147. http://dairy.ifas.ufl.edu/rns/2017/Guan.pdf.
- Li F, Guan LL. Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle. Appl Environ Microbiol. 2017;83(9):1–16.
- Engelking LR. Chapter 37 – gluconeogenesis. In: Textbook of Veterinary Physiological Chemistry. 2015. p. 225–230. ISBN: 9780123919106
- Lovett DK, Shalloo L, Dillon P, Mara FPOÕ. A systems approach to quantify greenhouse gas fluxes from pastoral dairy production as affected by management regime. Agric Syst. 2006;88:156–179.
- Hurley AM, López-Villalobos N, McParland S, Kennedy E, Lewis E, O’Donovan M, Burke JL, Berry DP. Inter-relationships among alternative definitions of feed efficiency in grazing lactating dairy cows. J Dairy Sci [Internet]. 2016;99(1):468–479. http://www.sciencedirect.com/science/article/pii/S0022030215008255.
- Martinez-Fernandez G, Denman SE, Yang C, Cheung J, Mitsumori M, McSweeney CS. Methane inhibition alters the microbial community, hydrogen flow, and fermentation response in the rumen of cattle. Front Microbiol [Internet]. 2016;7(July):1–14.
- Liu C, Li XH, Chen YX, Cheng ZH, Duan QH, Meng QH, Tao XP, Shang B, Dong HM. Age-related response of rumen microbiota to mineral salt and effects of their interactions on enteric methane emissions in cattle. Microb Ecol. 2017;(3):590–601.
- Niu D, Zuo S, Jiang D, Tian P, Zheng M, Xu C. Treatment using white rot fungi changed the chemical composition of wheat straw and enhanced digestion by rumen microbiota in vitro. Anim Feed Sci Technol [Internet]. 2018;237(July 2017):46–54.
- Vyas D, Alemu AW, McGinn SM, Duval SM, Kindermann M, Beauchemin KA. The combined effects of supplementing monensin and 3-nitrooxypropanol on methane emissions, growth rate, and feed conversion efficiency in beef cattle fed high forage and high grain diets. J Anim Sci. 2018;96(7):2923–2938.
- Bielecka M. Probiotics in Food. FAO/WHO [Internet]. 2006;413–426. http://www.crcnetbase.com/doi/abs/10.1201/9781420009613.ch16.
- Deng KD, Xiao Y, Ma T, Tu Y, Diao QY, Chen YH, Jiang JJ. Ruminal fermentation, nutrient metabolism, and methane emissions of sheep in response to dietary supplementation with Bacillus licheniformis. Anim Feed Sci Technol. 2018;241(November 2017):38–44.
- Chee-Sanford JC, Krapac IJ, Yannarell AC, Mackie RI. Environmental Impacts of Antibiotic Use in the Animal Production Industry. Ecol Anim Heal Ecosyst Heal Sustain Agric. 2012;2:228–368.
- Thanner S, Drissner D, Walsh F. Antimicrobial resistance in Agriculture. MBio. 2016;7(2):67–75.
- Auffret MD, Dewhurst RJ, Duthie C-A, Rooke JA, John Wallace R, Freeman TC, Stewart R, Watson M, Roehe R. The rumen microbiome as a reservoir of antimicrobial resistance and pathogenicity genes is directly affected by diet in beef cattle. Microbiome [Internet]. 2017;5(1):159. https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-017-0378-z.
- Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature [Internet]. 2018;555(7698):623–628.
- Carberry CA, Kenny DA, Kelly AK, Waters SM. Quantitative analysis of ruminal methanogenic microbial populations in beef cattle divergent in phenotypic residual feed intake (RFI) offered contrasting diets. J Anim Sci Biotechnol [Internet]. 2014;5(1):41.
- Fouts DE, Szpakowski S, Purushe J, Torralba M, Waterman RC, MacNeil MD, Alexander LJ, Nelson KE. Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS One. 2012;7(11) :e48289. doi:10.1371/journal.pone.0048289.
- Morgavi DP, Rathahao-Paris E, Popova M, Boccard J, Nielsen KF, Boudra H. Rumen microbial communities influence metabolic phenotypes in lambs. Front Microbiol. 2015;6(OCT):1–13.
- Droge J, Mchardy AC. Taxonomic binning of metagenome samples generated by next-generation sequencing technologies. Brief Bioinform. 2012;13(6):646–655.
- Hess M. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science (80-) [Internet]. 2011;463(6016):463–467.
- Svartström O, Alneberg J, Terrapon N, Lombard V, De Bruijn I, Malmsten J, Dalin AM, El Muller E, Shah P, Wilmes P, et al. Ninety-nine de novo assembled genomes from the moose (Alces alces) rumen microbiome provide new insights into microbial plant biomass degradation. ISME J. 2017;11(11):2538–2551.
- Stewart RD, Auffret MD, Warr A, Wiser AH, Press MO, Langford KW, Liachko I, Snelling TJ, Dewhurst RJ, Walker AW, et al. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat Commun [Internet]. 2018;9(1):1–11.
- Cao MD, Nguyen SH, Ganesamoorthy D, Elliott AG, Cooper MA, Coin LJM. Scaffolding and completing genome assemblies in real-time with nanopore sequencing. Nat Commun [Internet]. 2017;8:1–10.
- Miller JR, Zhou P, Mudge J, Gurtowski J, Lee H, Ramaraj T, Walenz BP, Liu J, Stupar RM, Denny R, Song L. Hybrid assembly with long and short reads improves discovery of gene family expansions. BMC Genomics. 2017;18(1):1–12. doi:10.1186/s12864-016-3396-5.
- Hino T, Asanuma N. Suppression of ruminal methanogenesis by decreasing the substrates available to methanogenic bacteria. Nutr Abstr Rev. 2003;1R–8R. (Series B: Livestock Feeds and Feeding).
- Lopes J, De Matos L, Harper M, Giallongo F, Oh J, Gruen D, Ono S, Kindermann M, Duval S, Hristov AN. Effect of 3-nitrooxypropanol on methane and hydrogen emissions, methane isotopic signature, and ruminal fermentation in dairy cows. J Dairy Sci. 2016;99:5335–5344. DOI:10.3168/jds.2015-10832
- Hristov AN, Oh J, Giallongo F, Frederick TW, Harper MT, Weeks HL, Branco AF, Moate PJ, Deighton MH, Williams SR, et al An inhibitor persistently decreased enteric methane emission from dairy cows with no negative effect on milk production. Proc Natl Acad Sci U S A. 2015;112(34):10663–10668.
- Reid M, O’Donovan M, Elliott CT, Bailey JS, Watson CJ, Stj L, Corrigan B, Fenelon MA, Lewis E. The effect of dietary crude protein and phosphorus on grass-fed dairy cow production, nutrient status, and milk heat stability. J Dairy Sci [Internet]. 2015;98(1):517–531. http://www.ncbi.nlm.nih.gov/pubmed/25465549.
- Tadele Y. Use of different non protein nitrogen sources in ruminant nutrition: a review. Adv Life Sci Technol. 2015;29:100–106.
- Murphy SC, Martin NH, Barbano DM, Wiedmann M. Influence of raw milk quality on processed dairy products: how do raw milk quality test results relate to product quality and yield. J Dairy Sci. 2016;99:10128–10149. ISBN 1478611200
- Campbell JR, Marshall RT. Dairy production and processing: the science of milk and milk products. 2016;105–109.
- Castillo AR, Kebreab E, Beever DE, France J A review of efficiency of nitrogen utilisation in lactating dairy cows and its relationship with environmental pollution. J Anim Feed Sci [Internet]. 2000;9(1):1–32. http://www.scopus.com/inward/record.url?eid=2-s2.0-0034552301&partnerID=40&md5=fb9f0a85c7eaf652d6cca82cd991fecc.
- Ipharraguerre IR, Clark JH. Impacts of the source and amount of crude protein on the intestinal supply of nitrogen fractions and performance of dairy cows. J Dairy Sci. 2005;88(Supplement):E22–37.
- Hall MB, Huntington GB. Nutrient synchrony: sound in theory, elusive in practice. J Anim Sci. 2008;86(14 Suppl):287–292.
- Broderick GA Effects of varying dietary protein and energy levels on the production of lactating dairy cows. J Dairy Sci [Internet]. 2003;86(4):1370–1381. http://www.ncbi.nlm.nih.gov/pubmed/12741562.