ABSTRACT
SifA is a bi-functional Type III Secretion System (T3SS) effector protein that plays an important role in Salmonella virulence. The N-terminal domain of SifA binds SifA-Kinesin-Interacting-Protein (SKIP), and via an interaction with kinesin, forms tubular membrane extensions called Sif filaments (Sifs) that emanate from the Salmonella Containing Vacuole (SCV). The C-terminal domain of SifA harbors a WxxxE motif that functions to mimic active host cell GTPases. Taken together, SifA functions in inducing endosomal tubulation in order to maintain the integrity of the SCV and promote bacterial dissemination. Since SifA performs multiple, unrelated functions, the objective of this study was to determine how each functional domain of SifA becomes processed. Our work demonstrates that a linker region containing a caspase-3 cleavage motif separates the two functional domains of SifA. To test the hypothesis that processing of SifA by caspase-3 at this particular site is required for function and proper localization of the effector protein domains, we developed two tracking methods to analyze the intracellular localization of SifA. We first adapted a fluorescent tag called phiLOV that allowed for type-III secretion system (T3SS) mediated delivery of SifA and observation of its intracellular colocalization with caspase-3. Additionally, we created a dual-tagging strategy that permitted tracking of each of the SifA functional domains following caspase-3 cleavage to different subcellular locations. The results of this study reveal that caspase-3 cleavage of SifA is required for the proper localization of functional domains and bacterial dissemination. Considering the importance of these events in Salmonella pathogenesis, we conclude that caspase-3 cleavage of effector proteins is a more broadly applicable effector processing mechanism utilized by Salmonella to invade and persist during infection.
Introduction
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a Gram-negative, facultative, intracellular anaerobe that causes gastroenteritis. In the United States, S. Typhimurium is responsible for nearly 25% of all food-borne infections and continues to be a major public health and economic burden.Citation1S. Typhimurium infection in humans is typically acquired by ingestion of contaminated food or water leading to acute gastroenteritis, and can also lead to severe complications or death in persons at risk.Citation2 Salmonella species, like other Gram-negative pathogens, have a sophisticated virulence mechanism called a type III secretion system (T3SS), which is responsible for the delivery of a series of bacterial effectors into host cells aimed at reprogramming eukaryotic cell functions.Citation3–Citation5 S. enterica use two distinct type III T3SSs encoded on Salmonella pathogenicity islands (SPIs)-1 and-2 to inject their arsenal of effectors. In general, the SPI-1-encoded T3SS is primarily required for the invasion of non-phagocytic cells, where expression is induced by the intestinal microenvironment, enabling Salmonella to cross the epithelial gut barrier and promote intestinal inflammation. Subsequently, the SPI-2-encoded T3SS mediates intracellular bacterial replication and is necessary for the establishment of systemic disease.
The type III secreted effectors (T3SEs) constitute a large and diverse group of virulence proteins that mimic eukaryotic proteins in structure and function. In fact, over 30 different effector proteins are delivered into host cells by S. Typhimurium through both SPI-1 and SPI-2 T3SSs.Citation6,Citation7 A prominent feature shared by bacterial effectors is their modular architecture, which is often comprised of well-defined regions that confer a subversive function. Strikingly, the distinct modules within an effector often mediate very different, unrelated functions, strongly suggesting that they evolved independently of each other and subsequently combined to form a chimeric protein.Citation8,Citation9 Chimerization is a common theme shared by many effectors,Citation10 and this forms the basis of a provocative hypothesis termed ‘terminal reassortment’, proposed by Guttman and colleagues to explain the diversity of bacterial effectors.Citation9 The terminal reassorment tenet is substantiated by their finding that 32% of all T3SE families contain chimeric effectors, far greater than any other analyzed protein family. Additionally, other studies suggest that terminal reassortment is important for the evolution of these virulence proteins.Citation8,Citation9 In keeping with this theory, we previously observed that many T3SEs harbor a functional caspase-3 cleavage motif (DxxD) uniquely positioned at the junction separating the two distinct functional domains.Citation11 This finding revealed that T3SEs had evolved to use the host defense system in a manner pivotal to the pathogenicity of the organism.Citation11 More specifically, S. Typhimurium appears to have evolved a mechanism to deliver effector proteins in a precursor form to the host cell where they are processed into independent functionally active domains.
Although much attention has been given to identifying the functions of T3SEs, mechanisms underlying host mediated effector processing (e.g., cleavage) are far less appreciated. To further our understanding in this regard, we had previously examined at two SPI-1 effector proteins, SipA and SopA, and found that caspase-3 processing was necessary for the function of these T3SEs.Citation11 In this report, we chose to take a closer look at SPI-2, which mediates the translocation across the vacuolar membrane of a set of bacterial effector proteins that support collectively the intra-vacuolar replication. One such SPI-2 effector protein SifACitation12 plays a significant role in Salmonella virulence by maintaining the integrity of the Salmonella-containing vacuole (SCV),Citation13 and hence promoting the formation of tubular membranous structures connected to SCVs that are named Salmonella-induced tubules (also referred to as Sif filaments).Citation14 Unique to this effector protein, the N-terminal domain of SifA interacts with the pleckstrin homology domain of the host kinesin-binding protein SKIP, and the C-terminal domain harbors a WxxxE motif that mimics the active the form of Rho-GTPases. Cooperatively these functional domains, along with another SPI-2 T3SE SseJ, promote host membrane tubulation.Citation14
The resolution of the crystal structure and domain function analysis of SifA has shown that the protein is divided into two distinct major domainsCitation14–Citation16 separated by a potential caspase-3 cleavage site,Citation11 suggesting the two domains of SifA might act independently of each other upon cleavage. Herein we characterize the structural features of the caspase-3 cleavage motif in SifA, and describe a new method of tracking T3SEs to show that cleavage of SifA by caspase-3 is indeed critical for proper localization of its functional domains, and is essential for bacterial dissemination. Understanding how T3SEs are functionally controlled through caspase-3 processing, will shed new light concerning the co-evolutionary interplay between S. Typhimurium and its host.
Materials and methods
Bacterial strains, plasmids, and growth conditions
Wild type S. Typhimurium SL1344, and isogenic mutants thereof (), were used throughout this study. All cloning was carried out in Escherichia coli strain BL21 DE3 (Invitrogen). All bacterial strains were constructed and cultured in LB as previously described,Citation17 unless otherwise specified. Primers used for construction of bacterial plasmids are listed in .
Table 1. Bacterial strains and plasmid constructs.
Table 2. Primers used for construction of bacterial plasmids (5ʹ→ 3ʹ).
-GST-SifA
SifA was PCR-amplified from wild-type (WT) S. Typhimurium (SL1344) using primers SP5 and SP6. The PCR product was then digested, purified, and ligated to the GST-containing vector, pGEX-6p-1 (GE Healthcare), as described previously.Citation14
-GST-SifAcsm
SifAcsm was synthetically generated and cloned into pUCIDT-AMP by Integrated DNA Technologies (IDT) using the same cloning sites as GST-SifA. sifAcsm was then cloned into pGEX-6p-1 and sequences were verified using the same methods used for GST-SifA.
-∆SifA
The ∆SifA mutant was constructed from SL1344 by deletion mutagenesis using a chloramphenicol cassette as described by Datsenko and Wanner.Citation18 Briefly, a chloramphenicol resistance cassette was PCR amplified from the pKD3 plasmid using primers SP1 and SP2. The pKD3 PCR product was then concentrated and transformed into electrocompetent SL1344 expressing the pKD46 plasmid. The sifA deletion was sequence verified using SP3 and SP4 primers.
-∆SifA/pGST-SifA and ∆SifA/pGST-SifAcsm
The ∆SifA/pGST-SifA and ∆SifA/pGST-SifAcsm complemented strains were made using the ∆SifA mutant. pGST-SifA and pGST-SifAcsm plasmid DNA were transformed into the electrocompetent ∆SifA strain of Salmonella. Transformants were sequence verified using SP3 and SP4 primers.
-∆SifA/SifA-phiLOV
SifA-phiLOV was made using pUC57-SipA-phiLOV. sifA was PCR amplified from SL1344 using SP7 and SP8 primers. These primers added a SacI and XhoI site on the 5ʹ and 3ʹ ends of sifA, respectively. Since sipA was cloned into pUC57-phiLOV using the same cloning sites, the pUC57-SipA-phiLOV plasmid was digested with SacI and XhoI (NEB) to remove sipA, and then gel purified.Citation19 The sifA PCR product was similarly digested and purified and ligated to pUC57-phiLOV before being transformed in the same manner as pGST-SifA. Plasmid DNA was then isolated from transformants that had ampicillin resistance and transformed into electrocompetent ∆SifA.
-Single/dual tagged SifA constructs for lentiviral transduction
V5-SifA-HA, V5-SifA, SifA-HA, and V5-SifAcsm-HA constructs were generated and cloned into pUC57 with AgeI and EcoRI cloning sites by Genscript. Both V5 and HA are small ectopic tags that can be visualized using fluorescent antibodies. Plasmids were then digested with AgeI and EcoRI, and cloned into pLVX-TetOne-Puromycin and pLVX-TetOne-Blasticidin using the same restriction sites.
SifA purification
A 10 mL overnight culture of Escherichia coli BL21 expressing pGST-SifA in LB/Ampicillin media was back diluted 1:100 into a 1 L LB/Ampicillin. The culture was grown at 37°C for 3.5 hr and then induced with 1 mM IPTG at 22°C for 3 hr. The bacteria were resuspended in GST Lysis Buffer (25 mM Tris pH 8, 150 mM NaCl, 3 mM DTT, and 1 mM PMSF) and then sonicated for 30 sec intervals 4 times. The lysate was then clarified at 14,000 rpm for 1 hr at 4 degrees Celsius. The clarified lysate was then run through glutathione sepharose beads in a column (GE Healthcare), washed with 1 X phosphate buffered solution (PBS), and then eluted using reduced glutathione (GE Healthcare).
Caspase-3 cleavage assay
Purified GST-SifA and GST-SifAcsm were incubated with 10U and 20U of active recombinant human caspase-3 (BioVision) for 1 hr at 37°C. The resulting products were then Western blotted using Anti-GST antibody (GE Healthcare). Densitometry analysis was performed using the free online software FIJI.
Mouse dissemination and colonization experiments (both oral and tail vein infection)
For intestinal colonization, mice were treated with 40 uL of 100 mg/mL streptomycin 24 hrs prior to infection, as described previously.Citation20 Mice were then infected with 1 × 10Citation7 colony forming units (CFUs) of each bacterial strain by oral gavage. At 48 hr post-infection (hpi) the liver and proximal colon were harvested for dissemination analysis. Dissemination analysis was completed by homogenizing the tissues and serial dilution plating. For tail vein injection, mice were injected with 1 × 10Citation6 bacteria and dissemination analysis was again carried out but at 24 hpi. Statistical analyses were completed using an unpaired Student’s t test.
Generation of caspase-3 KO HeLa cells
The caspase-3 KO HeLa cells were generated according to the Caspase-3 CRISPR/Cas9 construct manufacturer’s instructions (Santa Cruz Biotechnology). Briefly, 1µg of the Caspase-3 CRISPR/Cas9 plasmid and 1µg of the HDR plasmid (contains puromycin resistance marker for selection) were incubated with Plasmid Transfection Medium (Santa Cruz Biotechnology). Ten µL of the UltraCruz Transfection Reagent (Santa Cruz Biotechnology) was incubated with Plasmid Transfection Medium. The solution containing the plasmids and the solution containing the UltraCruz Transfection Reagent were then combined and incubated at room temperature for 20 min. After incubation, the combined solution was added to HeLa cells in fresh H1 media. H1 media was replaced 24 hr later, and selection with H1 media containing 2 µg/mL puromycin was started 72 hr post-transfection. Cells were sub-cultured for 2 weeks before use in experiments.
Lentiviral transduction of WT and caspase-3 KO HeLa cells
Six µg of each construct was cloned into pLVX-TetOne-Puro (for WT HeLa cells) or pLVX-TetOne-Blasticidin (for Caspase-3 KO HeLa cells), 4µg of a packaging plasmid psPAX2 and 2ug of an envelope plasmid pMD2.G were added to Opti-MEM media (Difco). Thirty-six µL of TransIT-293 transfection reagent was added to the Opti-MEM mixture and incubated at room temperature for 30 min. The transfection mixture was then added dropwise to individual 10 cm dishes of HEK293T cells. HEK293T cells were plated at 6–8 x 10Citation6 in 8 mLs of D10 media (made using 500 mL DMEM, 10% FBS, 5 mL Pen/Strep, 10 mL L- glutamine, and 50 µL plasmocin, Invivogen).
After 48 hr, the supernatant containing the virus from each plate was collected and added dropwise to WT HeLa cells (constructs in pLVX-TetOne-Puro) or caspase-3 KO HeLa cells (constructs in pLVX-TetOne-Blasticidin) that were >60% confluent. Both WT and caspase-3 KO HeLa cells were incubated with virus overnight, and then selected using puromycin or blasticidin, respectively.
In vitro infection experiments
A 3 mL overnight LB culture of SL1344 or LB/Ampicillin cultures of ∆SifA/pSifA-phiLOV were back-diluted 1:5 into 10 mL LB broth or LB/Ampicillin/1 mM IPTG, respectively, and grown at 37°C for 1hr. Fifty µL of this culture was then added to 1mL H1 media, and 200 µL per well was used for infection of WT and caspase-3 KO HeLa cells. Following a 1 hr infection, the cells were incubated with H1 media containing 50 µg/mL gentamicin for 1 hr, before incubation with H1 media containing 10 µg/mL gentamicin until 1, 2, 4, 8, and/or 10 hpi. Cells were fixed and permeabilized in 1% bovine serum albumin (BSA)/0.2% saponin then stained using Image-iT LIVE Caspase-3 Detection Kit (ThermoFisher) for SifA-phiLOV/Caspase-3 colocalization experiements. For dual-tagged SifA constructs, cells were stained using anti-V5 (Abcam) and anti-HA primary antibodies (Santa Cruz), and goat anti-rabbit Alexa Fluor 488 and goat anti-mouse Alexa Fluor 568 secondary antibodies (Invitrogen). Cells were then visualized using a 63X objective lens on a confocal microscope Leica SP5 confocal microscope. Colocalization data was determined using the Coloc 2 tool in the free online software FIJI. The Mander’s coefficient was the selected output, which is a pixel intensity spatial correlation analysis.
Results
Structural features of the caspase-3 motif in S. Typhimurium T3SE SifA
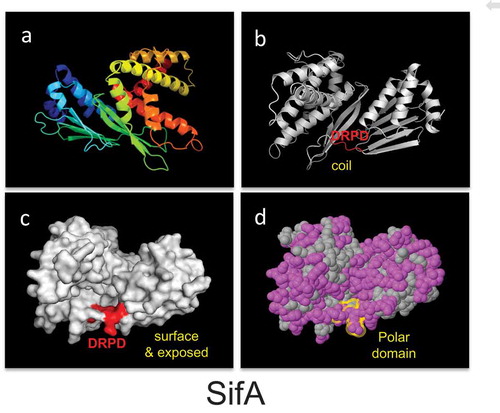
Given that the resolution of the crystal structure and domain function analysis of SifA has shown that the protein is divided into two distinct major domainsCitation14–Citation16 separated by a potential caspase-3 cleavage site,Citation11 we investigated the structural features of the caspase-3 cleavage motifs by in silico modeling of the crystallized caspase-3 substrate.Citation14,Citation16 Protein sequences were retrieved from the NCBI protein database, screened for caspase-3 motifs, and then analyzed in the web server Phyre2 for the identification of structural homologues. We found that the caspase-3 motif is surrounded by hydrophilic residues, exposed at the surface, and localized in coiled regions of proteins (). Additionally, we studied the frequency of the DxxD site by analyzing the residue downstream of the caspase-3 cutting site (DxxD-x) in 33 known caspase-3 substrates.Citation21 Of these residues, serine, glycine, and asparagine (all hydrophilic/polar residues) are most frequently located downstream of the caspase-3 cleavage site; serine was found 13 times, glycine 10 times, and asparagine 4 times and is consistent with our observation of the CASP3 motif being surrounded by hydrophilic residues.
SifA harbors a functionally active caspase-3 cleavage site
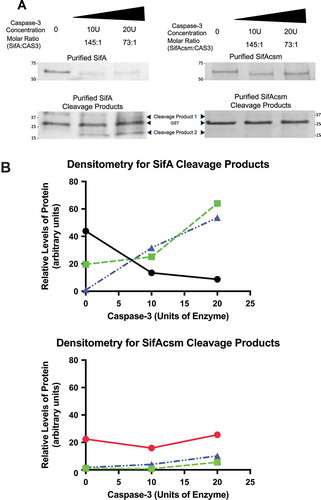
SifA is a SPI-2 S. Typhimurium effector protein necessary for intracellular survival. Based on the structural data as shown in , as well as the observation that each of the SifA domains function independently,Citation14,Citation16 we next sought to determine whether the caspase-3 motif of SifA (DRPD) is functionally active. To do this we PCR amplified SifA from SL1344 and cloned it into pGEX-6p-1 (GE Healthcare), which adds an N-terminal GST tag to SifA that has been previously used to purify SifA protein using a glutathione sepharose matrix.Citation14 We found that that exogenous addition of purified fractions of SifA (5µg) with commercially obtained active caspase-3 enzyme (10U and 20U) (Biovision) resulted in the cleavage of this effector protein at the expected site as evidenced by a decrease in the amount of SifA protein and concomitant increase in degradation products (,b)). Additionally, a single amino acid substitution in the caspase-3 recognition site is known to render substrates insensitive to caspase-3. Thus, we altered the caspase-3 site (DRPD) by one amino acid to DRPA (refer to methods). The plasmid bearing the mutant clone was sequence confirmed and named pSifAcsm (for SifA caspase site mutant). Such mutation of the caspase-3 site by one amino acid to alanine rendered SifA insensitive to caspase-3 cleavage (,).
Figure 2. SifA harbors a functionally active caspase-3 cleavage site. (a) Purified fractions of SifA and SifAcsm (5µg) were incubated with 10 units or 20 units of human recombinant caspase-3 (Biovision) for 1 hr at 37 ºC. Each sample was then Western blotted using an Anti-GST primary antibody (GE Healthcare) at 1:20,000 and Donkey anti-Goat IgG HRP secondary antibody (Santa Cruz) at 1:5,000. The cleavage products for both SifA and SifAcsm were visualized using coomassie blue staining (BioRad). SifA protein, but not SifAcsm protein, was cleaved by caspase-3 in a concentration dependent manner. (b) Densitometry of Western Blot in (a), SifA (Black), SifAcsm (Red), Cleavage Product 1 (Green), and Cleavage Product 2 (Blue). SifA, but not SifAcsm, displays a caspase-3 concentration dependent decrease, which coincides with an increase in both Cleavage Product 1 and Cleavage Product 2.
SifA colocalizes with caspase-3 during infection
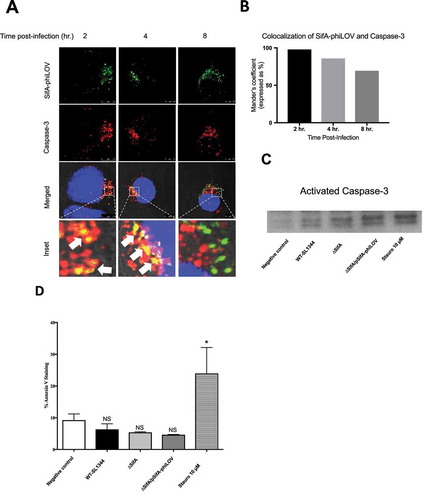
To more deeply understand how the processing of SifA occurs through caspase-3 cleavage, we used the small fluorescent phiLOV tag to determine the extent to which we can observe caspase-3 and SifA colocalizing during an in vitro infection. This method does not interfere with secretion kinetics of the effector through the T3SS and can be used to examine de novo expression of T3SEs in vitro and in vivo.Citation19,Citation22,Citation23 As shown in ), following infection of HeLa cells with Salmonella, SifA-phiLOV colocalizes with caspase-3 beginning at 2 hpi (seen by yellow), with colocalization continuing through at 4 hpi After 4 hr, colocalization was found to decrease (,). Our prior studies found that increased caspase-3 activation occurs in SL1344-infected intestinal epithelial cells over the first 5 hr of infection, with the effector protein SipA playing a central roleCitation11; thus such co-localization of SifA with caspase-3 is consistent with the timing of these events. Moreover, consistent with our previous findingsCitation11, we show that early after Salmonella infection of epithelial cells this pathogen activates caspase-3, without inducing apoptosis as determined by a Western blot for activated caspase-3 ()) and Annexin V staining ()); as shown, infection with Salmonella expressing SifA-phiLOV results in an increase in activated caspase-3 expression (via the function of SipA), but no significant difference in Annexin V positive cells in comparison to uninfected HeLa cells.
Figure 3. SifA colocalizes with caspase-3 during infection. (a) HeLa cells were infected with ∆ SifA/pSifA-phiLOV, fixed at 2, 4, and 8 hrs post-infection, stained with Image-iT LIVE Caspase-3 Detection Kit (ThermoFisher) and visualized using fluorescent confocal microscopy. Caspase-3 (Red) and SifA-phiLOV (Green) can be seen colocalizing at 2 hrs post-infection (Yellow; bottom, left panel), with colocalization decreasing through 8 hrs. post-infection (bottom, bottom middle, and bottom right panels). (b) Colocalization of SifA-phiLOV and caspase-3 quantified using FIJI Software. (c) Western Blot for activated-Caspase-3. HeLa cells were infected with indicated strains, lysed, and analyzed for activated caspase-3 expression. Protein concentration was determined using a Bradford Assay to ensure equal amounts of protein were loaded into each well (d) Annexin V Staining Assay. HeLa cells were infected with indicated strains, trypsinized, and stained with Annexin V. Flow cytometric analysis was then completed on a MACSQuant Analyzer to determine the number of Annexin positive cells. In comparison to the negative control (uninfected), HeLa cells only showed a significant increase in Annexin V positive cells with the use of 10µM of staurosporine. HeLa cells infected with WT-SL1344, ∆SifA, and ∆SifA/pSifA-phiLOV all do not exhibit a significant change in Annexin V positive cells in comparison to negative control (uninfected). P Values (statistics calculated using unpaired Student’s t test): NS, not significant; *P < 0.05. Experiments were performed at least three times using cells of different passage.
Caspase-3 cleavage of SifA is necessary for dissemination
SifA plays a key role in maintaining the integrity of the Salmonella-containing vacuole (SCV)Citation13, through the formation of tubular membranous structures connected to SCVs named Salmonella-induced tubules. Because the formation of such tubules is an essential virulence requirement for intracellular replication in macrophages, and hence contributes to the ability of Salmonella to successfully disseminate to extraintestinal organs (i.e., liver and spleen), we next sought to determine whether caspase-3 cleavage of SifA is necessary for Salmonella dissemination. Using the murine model of Salmonella-induced enteritis, as described in the Methods, we found that consistent with prior reports,Citation24 deletion of the sifA gene from S. Typhimurium results in a 1 log decrease in dissemination of bacteria to the liver ()).
Figure 4. Caspase-3 cleavage of SifA is necessary for dissemination. (a) Mice were infected by oral gavage and livers were harvested 48 hrs. post-infection. Bacterial burdens were determined by homogenizing the livers and serial dilution plating. ∆SifA and ∆SifA/pSifAcsm exhibit over 1 log decrease in bacterial burden in the liver. This decrease is restored upon complementation with pSifA. Results are averages of 6 mice per group ± SD. B.) Mice were infected by oral gavage and proximal colon samples were taken 48 hr post-infection. Bacterial burdens were determined similarly to (a). Results are averages of 6 mice per group +/- SD. ∆SifA, ∆SifA/pSifA, and ∆SifA/pSifAcsm do not exhibit any significant defects in colonization of the proximal colon in comparison to WT-SL1344. C.) Mice were infected via the tail vein, and livers were harvested 24 hr post-infection. Bacterial burden was determined similar to (a). In comparison to WT-SL1344, ∆SifA, ∆SifA/pSifA, and ∆SifA/pSifAcsm all show similar levels of bacteria in the liver. P Values (statistics calculated using unpaired Student’s t test): NS, not significant; *P < 0.05. Data shown are representative of at least three independent experiments.
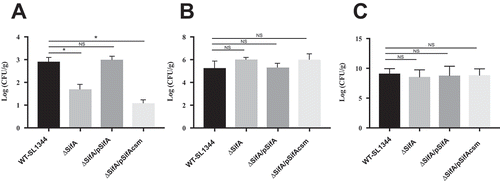
Next, we complimented this SifA deletion strain of Salmonella with two plasmids; one expressing SifA and the other SifAcsm. We reasoned that if cleavage of SifA at the caspase-3 cleavage site was a critical event required for dissemination, we would expect to see the SifAcsm phenocopy the SifA deletion strain. As shown in ), we found that when the caspase-3 cleavage site is mutated, we indeed observed a similar 1 log decrease in dissemination to the liver. However, when the SifA deletion strain is complemented with a plasmid containing SifA, there was a rescue of the phenotype to near wild-type levels of dissemination to the liver.
While all strains colonized the large intestine to similar levels ()), we needed to rule out the possibility that the decrease in dissemination observed for the SifAcsm mutant strain resulted from a defect in the ability to disseminate via the blood to the liver. We therefore delivered the Salmonella strains systemically through tail vein injection, and found that both the ∆SifA strain as well as the SifAcsm strain, showed similar levels of bacterial load in the liver as compared to the wild type and SifA-complemented strain ()). Given that the SifAcsm strain does not demonstrate an impairment in either the ability to colonize the intestine or reach the liver when delivered systemically suggests that the defect observed results from a failure to disseminate from the intestine.
Subcellular localization of SifA functional domains is dependent upon caspase-3 cleavage
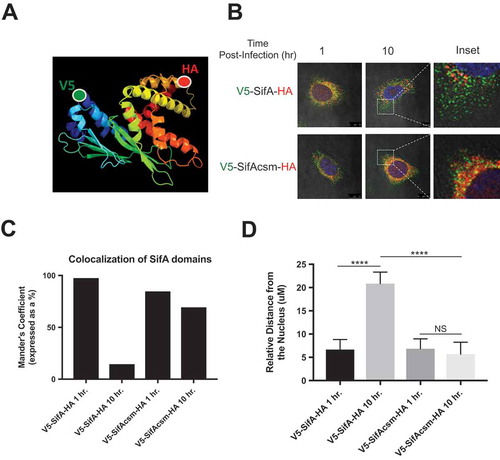
Although we have been able to link SifA function to cleavage at the caspase-3 cleavage site, we next sought to determine whether the each of the SifA functional domains could be sub-cellularly tracked following caspase-3 cleavage. SifA is ideal for addressing this question given that the N-terminal domain of SifA interacts with the pleckstrin homology domain of the host kinesin-binding protein SKIP, and the C-terminal domain harbors a WxxxE motif that mimics the active the form of Rho-GTPases. If caspase-3 cleavage is necessary for SifA domain sub-cellular localization, then each of the functional domains would remain in the perinuclear region of the cell, where the SCV resides, until after its interaction with caspase-3. Then following caspase-3 cleavage, the individual functional domains would move to distinct subcellular locations to perform their functions. To this end, we designed dual-tagged V5-SifA-HA and dual-tagged V5-SifAcsm-HA ()), allowing both red (V5) and green (HA) probes to be displayed on a single effector. In addition to the full-length protein, we also designed the individual domains (N and C terminal domain) with a single tag; – V5-SifA (green) and SifA-HA (red).
Figure 5. Subcellular localization of SifA functional domains is dependent upon caspase-3 cleavage. (a) Schematic depicting the dual-tagging strategy for SifA. The V5 tag (green) is used on the N-terminus and the HA tag (red) is used on the C-terminus. (b) Wild-type HeLa cells containing either V5-SifA-HA or V5-SifAcsm-HA were infected with WT-SL1344. The cells were fixed and stained (refer to methods) at 1 and 10 hr post-infection. The N-terminus of SifA begins to branch out following interaction with caspase-3 at 2 hr post-infection (follow localization of green V5 tag from 1 – 10 hr post-infection), and the C-terminus of SifA remains in the perinuclear region for all time points following infection (red). When the caspase-3 cleavage site is mutated (V5-SifAcsm-HA), both domains (red and green) are observed in the perinuclear region at all time points following infection. Experiments were performed at least three times using cells of different passage. (c) Colocalization of the SifA N-terminal and C-terminal domains following caspase-3 cleavage quantified using FIJI Software. (d) Relative distance (measured using a micron scale) of the distance travelled by the N-terminal domain in both V5-SifA-HA and V5-SifAcsm-HA at 1 and 10 hrs. post-infection. P Values (statistics calculated using unpaired Student’s t test): NS, not significant; **** P < 0.0001.
The benefit of using these constructs is that they permit us to directly investigate the involvement of the caspase-3 motif in SifA domain localization. The V5-SifA-HA and V5-SifAcsm-HA constructs can therefore be used to determine where each domain localizes during infection, and if the domain localization is dependent on caspase-3 cleavage. Additionally, the V5-SifA and SifA-HA individual domain constructs can be used in combination with caspase-3 KO HeLa cells to determine if we restore domain localization when we use individual domains that resemble their post-caspase-3 cleavage form. Two of these constructs (V5-SifA-HA and V5-SifAcsm-HA) were transduced into wild-type Hela cells using lentiviral transduction. The plasmids used also had an inducible promoter, which allowed us to control expression of all four constructs.
In order to stimulate caspase-3 activation typical of Salmonella infection, we infected the HeLa cells transduced with all four constructs with wild-type Salmonella, and then looked for the redistribution of each of the functional domains at 1 and 10 hpi using immunofluorescent confocal microscopy. Maximum Sif filament formation has been observed previously between 8 and 10 hpi (Citation25,Citation26; supplementary Figure 1). Since we found that SifA colocalizes with caspase-3 at 2 hpi (), we included this earlier point (1 hr) to determine the extent to which subcellular localizations change following the interaction of the SifA with caspase-3.
At 1 hpi (prior to caspase-3 activation), we observe both SifA domains located in the perinuclear region for both V5-SifA-HA and V5-SifAcsm-HA (), top left panel, )). However, following interaction with caspase-3 at 2 hpi, the N-terminal domain of V5-SifA-HA begins to branch out from the perinuclear region towards the extremities of the cell and continues to spread throughout the cell at 10 hpi. The role of caspase-3 processing in the domain localization of V5-SifA-HA domains is further supported by the profound decrease in colocalization of the SifA domains at 10 hpi (,). Additionally, we observed a significant increase in the relative distance of the N-terminal domain from the nucleus for V5-SifA-HA at 10 hpi relative to 1 hpi ()). We also observed this distance to be markedly larger when the caspase-3 site was intact, as evidenced by the significant difference at 10 hpi between V5-SifA-HA and V5-SifAcsm-HA ()). This result is consistent with current literature indicating that the N-terminal domain of SifA binds to a protein called SKIP, and then forms Sif filaments via an interaction with kinesin.Citation14 Such Sif filaments are tubular membrane extensions that emanate from the SCV, which is located in the perinuclear region, towards the outer part of the cell. Considering the localization of the N-terminal domain for V5-SifA-HA, our results are in line with current understanding of SifA’s role in Sif filament formation.
Since the C-terminal domain of SifA mimics host cell GTPases, and these host cell GTPases function in a variety of roles, including cellular division and actin cytoskeletal rearrangements, it is difficult to predict where the C-terminal domain of SifA would localize during Salmonella infection.Citation27–Citation29 Furthermore, the C-terminal domain of SifA also contains a CaaX motif. The CaaX motif is prenylated by the geranylgeranyl transferase PGGT-1, which is a process that has been shown to facilitate attachment to cellular membranes.Citation30 On the balance of these reports, it would be feasible to observe the C-terminal domain in the perinuclear region at the SCV membrane, the outer cell membrane, or both. However, as shown in ) and ), the C terminal of SifA appears to remain primarily in the perinuclear region even after interaction with caspase-3 at the 2 hr time point, indicating the C-terminal domain of SifA likely remains at the SCV membrane through the 10 hpi time point.
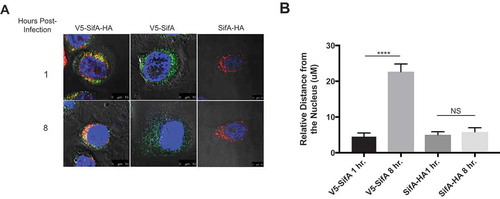
Figure 6. Activated individual SifA domains restore SifA domain localization in caspase-3 KO Cells. Caspase-3 KO HeLa cells containing either V5-SifA-HA, V5-SifA or SifA-HA were infected with WT-SL1344. The cells were fixed and stained (refer to methods) at 1 and 8 hr post-infection. Without caspase-3, both domains of V5-SifA-HA remain in the perinuclear region at all time points following infection (top and bottom left panels), suggesting caspase-3 cleavage is important for domain localization (compare to top panels in ). When using V5-SifA, which resembles the post-caspase-3 cleavage form of the SifA N-terminal domain, we restore domain localization (top and bottom middle panels). Experiments were performed at least three times using cells of different passage. (b) Relative distance (measured using a micron scale) of the distance travelled by the N-terminal and C-terminal in both V5-SifA and SifA-HA at 1 and 8 hrs. post-infection. P Values (statistics calculated using unpaired Student’s t test): NS, not significant; **** P < 0.0001.
We next determined the extent to which the caspase-3 site in SifA plays a role in the localization events we observed with V5-SifA-HA. To do this we used the V5-SifAcsm-HA construct and found that both of the functional domains of this strain remained in the perinuclear region for all time points following infection ()). Additionally, we did not observe a similar decrease in V5-SifAcsm-HA domain colocalization as observed with V5-SifA-HA ()). This result suggests that processing of SifA at the caspase-3 site is essential for proper localization of each of the SifA domains, and further infers that the C-terminal domain appears to perform its “GTPase mimicry” function in the perinuclear region after it’s interaction with caspase-3. Nonetheless, further studies are required to discern whether the C-terminal domain of SifA is indeed functionally active at this location.
Activated individual SifA domains restore SifA domain localization in caspase-3 KO cells
To further confirm the role of caspase-3 in the localization of the SifA functional domains, we generated caspase-3 KO HeLa cells using CRISPR/Cas9 (refer to Methods). Additionally, we designed the V5-SifA and SifA-HA constructs containing individual domains (without the caspase-3 site) to assess whether we could restore localization each SifA functional domain by using a form of each domain that resembles their post-caspase-3 cleavage structure. V5-SifA-HA, V5-SifA and SifA-HA were transduced into caspase-3 KO HeLa cells in the exact same manner as they were transduced into wild-type HeLa cells. As shown in ), both domains of V5-SifA-HA remain in the perinuclear region in the caspase-3 KO cells for all time points following infection, phenocopying what we see in V5-SifAcsm-HA in wild-type HeLa cells. When we use the post-caspase-3 cleavage form of each functional domain (V5-SifA and SifA-HA), we restore the domain localization we observed for V5-SifA-HA in wild-type HeLa cells, further implying that caspase-3 plays a pivotal role in the functional domain localization of SifA ()). Although SifA-HA resembles the post-caspase-3 cleavage form of the SifA C-terminal domain, the relative distance from the nucleus does not change following infection as it does for V5-SifA ()). These results agree with what is observed in for V5-SifA-HA in wild-type HeLa cells, indicating the perinuclear region of the HeLa cell is where the C-terminal domain of SifA is inferred to perform its function.
Discussion
In silico modeling of the SifA caspase-3 motif at the structural level revealed that this motif is surrounded by hydrophilic residues, exposed at the surface, and localized in coiled regions of proteins. This finding is consistent with two other crystallized structural homolog proteins of Salmonella effectors (SopA and SptP) that we examined (Supplementary Figures 2, and 3). This information coupled to the known structural data of SifA,Citation14 suggested that cleavage of the caspase-3 motif in this effector divides this protein into an interacting partner for the host protein SKIP (N-terminal) and a member of the WxxxE family of proteins with GTPase mimicry function (C-terminal). We further report that cleavage of SifA by caspase-3 is indeed critical for proper localization of its functional domains, and is essential for bacterial dissemination from the intestine. Such data are consistent with prior evidence that each of these domains can function independently,Citation14 and is also in step with recent findings that SifA can be split into two functional domains.Citation16 Our demonstration that caspase-3 cleavage of SifA is required for bacterial dissemination, adds to the emerging concept that caspase-3 sites are present and functionally active in T3SE. Such processing has been described for Salmonella invasion (SipA),Citation31 ubiquitination (SopA),Citation31 and now in this report for SCV maintenance and bacterial dissemination (SifA), implying that caspase-3 cleavage may have a broad impact on the bacteria’s ability to both invade and persist during infection.
Although we have previously shown that the Salmonella T3SE, SipA, induces increased caspase-3 activity upon S. Typhimurium infection, such activation remains insufficient to induce the signatures of apoptotic cell death (Citation31, and this report). Thus, our data suggest that SipA is not only critical in inducing its own proteolytic cleavage but also that of other effectors such as SifA via its role in caspase-3 activation. This idea is consistent with our prior observations that Salmonella is significantly less virulent in caspase-3 knockout (CASP3−/-) mice and Salmonella invasiveness in bone-marrow-derived macrophages from CASP3−/- mice is profoundly reduced. Citation31 Moreover, in Salmonella strains lacking SipA, it has been previously described that SifA and SCVs are incorrectly positioned in the cellCitation32 and in a manner shown herein, since the function of SifA depends on activation by caspase-3.
Method development for dual-tagging effectors permitted the tracking of effector proteins that also reflects the timeline of effector protein activation and localization during Salmonella infection. Unlike other strategies, this method does not rely on the T3SS secretion system for delivery of effector proteins, avoiding any complications associated with effector protein secretion kinetics or lack of secretion signals. The benefit to using this method is that it allows for the simultaneous tracking of domain localization and activation. One potential limitation to this method, however, is that it may be difficult to determine activation of T3SE domains if the localization does not change upon processing. For example, the subcellular localization of the N-terminal domain of SifA changes upon activation, whereas the C-terminal domain remains in the perinuclear region ( and ). Although we speculate the C-terminal domain is active, additional experimentation will be required to determine if this is indeed the case. Nevertheless, dual-tagging of SifA demonstrated for the first time that the individual domains of SifA have different subcellular localizations. Additionally, the results from our dual tagging method, in conjunction with the findings ofCitation32 suggest that the C-terminal domain alone may be acting to maintain the SCV in the perinuclear region, though further testing of this hypothesis is required to determine if this is indeed the case. Moreover, as Reinicke and colleagues demonstrated in,Citation30 the process termed prenylation occurs at the CAAX motif in the C-terminal domain of SifA, which is a process important for membrane association. Taking into account the localization of the C-terminal domain determined in this study and the perinuclear localization of the SCV determined in Citation32, the prenylation of the C-terminal domain may be required for interaction with the SCV membrane in the perinuclear region, although further studies would be required to determine if this is true.
Our prior studies revealed that S. Typhimurium T3SEs evolved the ability to use host enzymes, such as caspase-3 to activate themselves, displaying a high degree of co-evolution.Citation31 We speculate that the cleavage motifs of T3SEs have evolved over time and have targeted their cleavage and activation via different host enzymes depending on the host and cell type infected.Citation33 SipA, the first T3SE delivered into intestinal epithelial cells during infection is an excellent example of the evolution of a T3SE in response to selective pressures at the host-pathogen interface. With type III secretion requiring the delivery of a number of effectors rapidly over a short span of time, the formation of a chimeric effector with a lone signal sequence and requiring just a single chaperone, greatly reduces the workload at the T3SS interface.
The relationship between bacterial effectors and the activation of caspase-3 is an area of increasing interest. While the outcome of such rearrangements results in independently functional domains coming together as one effector, this also creates a unique challenge in separation of the domains upon host cell delivery to ensure their correct location and function. It is now understood that caspase-3 is constantly present in the cell at levels below those required to induce apoptosisCitation19, suggesting that caspase-3 processing of effectors likely occurs throughout the cell rather than being dependent on contact merely at the cell surface or within the cytosol. Moreover, the interaction between effector proteins and caspase-3 may play a role in preventing caspase-3 from performing other important functions during infection, as has been described during Yersinia infection.Citation34
Caspase-3 activation during bacterial infection is also likely a common by-product of the co-evolution of bacterial-host interactions, perhaps precipitated by the stress placed on host cells during invasion.Citation35–Citation39 In addition to non-specific or indirect activation of caspase-3, bacterial effectors are also able to promote caspase-3 activation through subtle changes within cellular pathways or even through direct interaction with the enzyme. The outcome for the pathogenic intruder, such as S. Typhimurium is often an increase in infectivity rather than a clearing of the infection as expected by the conventional understanding of the protective role of apoptosis.
The study herein therefore supports the concept that that caspase-3 cleavage of T3SS secreted effectors represents a common mechanism by which effector functions are regulated in host cells. Understanding of this biological phenomenon as well as advancing methods to interrogate T3SE function inside host cells will allow us to dissect specific aspects of Salmonella-host interactions that have yet to be documented, and in general could have broad impact on the field of bacterial pathogenesis.
The structural features of the caspase-3 motif found in the Salmonella effector protein SifA (PDB ID 3CXB). Protein structures are displayed as cartoon (Panels a and b), surface (Panel c), and spheres (Panel c). Panel (a) shows protein structures colored with the spectrum color set of PyMOL to display the different protein domains, α-helixes, β-sheets, and coil regions. In Panels (b) and (c), the location of the caspase-3 motif is colored in red, whereas in Panel (d) in yellow. In Panel (d), polar residues are colored in pink and hydrophobic in grey. DRPD, DVHD, and DGQD are putative caspase-3 motifs. PDB; Protein data bank. SifA: Confidence 100%; Identity 79%; Coverage 90%.
Author contributions
Study conception and design: B.A.M., D.M.W., S.P.
Acquisition of data: S.P., A.C.
Analysis and interpretation of data: B.A.M., D.M.W, S.P., A.C.,
Drafting of manuscript: B.A.M., D.M.W., S.P.
Conflict of interest statement
The authors declare no conflict of interest.
Acknowledgments
We would like thank William M. McDougall and Jill M. Perreira for technical assistance.
Additional information
Funding
References
- Sandler RS, Everhart JE, Donowitz M, Adams E, Cronin K, Goodman C, Gemmen E, Shah S, Avdic A, Rubin R. The burden of selected digestive diseases in the United States. Gastroenterology. 2002;122(5):1500–1511. Epub 2002/05/02.PubMed PMID: 11984534.
- Graham SM, Molyneux EM, Walsh AL, Cheesbrough JS, Molyneux ME, Hart CA. Nontyphoidal Salmonella infections of children in tropical Africa. Pediatr Infect Dis J. 2000;19(12):1189–1196. Epub 2001/ 01/06.PubMed PMID: 11144383.
- Galan JE. Molecular and cellular bases of Salmonella entry into host cells. Curr Top Microbiol Immunol. 1996;209:43–60. Epub 1996/01/01.PubMed PMID: 8742245.
- Galan JE. Molecular genetic bases of Salmonella entry into host cells. Mol Microbiol. 1996;20(2):263–271. Epub 1996/04/01.PubMed PMID: 8733226.
- Stebbins CE, Galan JE. Priming virulence factors for delivery into the host. Nat Rev Mol Cell Biol. 2003;4(9):738–743. Epub 2003/09/25, PubMed PMID: 14506477. doi:10.1038/nrm1201.
- Kenny B, Valdivia R. Host-microbe interactions: bacteria. Curr Opin Microbiol. 2009;12(1):1–3. Epub 2009/ 01/29, PubMed PMID: 19174324. doi:10.1016/j.mib.2009.01.002.
- Hansen-Wester I, Stecher B, Hensel M. Type III secretion of Salmonella enterica serovar Typhimurium translocated effectors and SseFG. Infect Immun. 2002;70(3):1403–1409. Epub 2002/02/21.PubMed PMID: 11854226; PMCID: 127782.
- Fookes M, Schroeder GN, Langridge GC, Blondel CJ, Mammina C, Connor TR, Seth-Smith H, Vernikos GS, Robinson KS, Sanders M, et al. Salmonella bongori provides insights into the evolution of the Salmonellae. PLoS Pathogens. 2011;7(8):e1002191. Epub 2011/08/31, PubMed PMID: 21876672; PMCID: 3158058. doi:10.1371/journal.ppat.1002191.
- Stavrinides J, Ma W, Guttman DS. Terminal reassortment drives the quantum evolution of type III effectors in bacterial pathogens. PLoS Pathogens. 2006;2(10):e104. Epub 2006/10/17, PubMed PMID: 17040127; PMCID: 1599762. doi:10.1371/journal.ppat.0020104.
- Kaniga K, Uralil J, Bliska JB, Galan JE. A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol Microbiol. 1996;21(3):633–641. Epub 1996/08/01.PubMed PMID: 8866485.
- Srikanth CV, Wall DM, Maldonado-Contreras A, Shi H, Zhou D, Demma Z, Mumy KL, McCormick BA. Salmonella pathogenesis and processing of secreted effectors by caspase-3. Science. 2010;330(6002):390–393. Epub 2010/ 10/16, PubMed PMID: 20947770; PMCID: PMC4085780. doi:10.1126/science.1194598.
- Stein MA, Leung KY, Zwick M, Garcia-del Portillo F, Finlay BB. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol Microbiol. 1996;20(1):151–164. Epub 1996/04/01.PubMed PMID: 8861213.
- Beuzon CR, Meresse S, Unsworth KE, Ruiz-Albert J, Garvis S, Waterman SR, Ryder TA, Boucrot E, Holden DW. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. Embo J. 2000;19(13):3235–3249. Epub 2000/07/06, PubMed PMID: 10880437; PMCID: PMC313946. doi:10.1093/emboj/19.13.3235.
- Ohlson MB, Huang Z, Alto NM, Blanc MP, Dixon JE, Chai J, Miller SI. Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell Host Microbe. 2008;4(5):434–446. Epub 2008/11/11, PubMed PMID: 18996344; PMCID: 2658612. doi:10.1016/j.chom.2008.08.012.
- Diacovich L, Dumont A, Lafitte D, Soprano E, Guilhon AA, Bignon C, Gorvel JP, Bourne Y, Meresse S. Interaction between the SifA virulence factor and its host target SKIP is essential for Salmonella pathogenesis. J Biol Chem. 2009;284(48):33151–33160. Epub 2009/10/06, PubMed PMID: 19801640; PMCID: PMC2785157. doi:10.1074/jbc.M109.034975.
- Zhao W, Moest T, Zhao Y, Guilhon AA, Buffat C, Gorvel JP, Meresse S. The Salmonella effector protein SifA plays a dual role in virulence. Sci Rep. 2015;5:12979. Epub 2015/08/14, PubMed PMID: 26268777; PMCID: PMC4534788. doi:10.1038/srep12979.
- Lee CA, Silva M, Siber AM, Kelly AJ, Galyov E, McCormick BA. A secreted Salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proc Natl Acad Sci U S A. 2000;97(22):12283–12288. Epub 2000/10/26, PubMed PMID: 11050248; PMCID: 17333. doi:10.1073/pnas.97.22.12283.
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645. PubMed PMID: 10829079. doi:10.1073/pnas.120163297.
- McIntosh A, Meikle LM, Ormsby MJ, McCormick BA, Christie JM, Brewer JM, Roberts M, Wall DM. SipA activation of caspase-3 is a decisive mediator of host cell survival at early stages of Salmonella enterica Serovar Typhimurium infection. Infect Immun. 2017;85(9). Epub 2017/06/21, PubMed PMID: 28630067. doi:10.1128/IAI.00393-17.
- Wall DM, Nadeau WJ, Pazos MA, Shi HN, Galyov EE, McCormick BA. Identification of the Salmonella enterica serotype typhimurium SipA domain responsible for inducing neutrophil recruitment across the intestinal epithelium. Cell Microbiol. 2007;9(9):2299–2313. PubMed PMID: 17697195. doi:10.1111/j.1462-5822.2007.00960.x.
- Chan SL, Griffin WS, Mattson MP. Evidence for caspase-mediated cleavage of AMPA receptor subunits in neuronal apoptosis and Alzheimer’s disease. J Neurosci Res. 1999;57(3):315–323. Epub 1999/07/21.PubMed PMID: 10412022. doi:10.1002/(SICI)1097-4547(19990801)57:3<315::AID-JNR3>3.0.CO;2-#.
- Gawthorne JA, Audry L, McQuitty C, Dean P, Christie JM, Enninga J, Roe AJ. Visualizing the translocation and localization of bacterial type III effector proteins by using a genetically encoded reporter system. Appl Environ Microbiol. 2016;82(9):2700–2708. Epub 2016/02/28, PubMed PMID: 26921426; PMCID: PMC4836418. doi:10.1128/AEM.03418-15.
- Christie JM, Hitomi K, Arvai AS, Hartfield KA, Mettlen M, Pratt AJ, Tainer JA, Getzoff ED. Structural tuning of the fluorescent protein iLOV for improved photostability. J Biol Chem. 2012;287(26):22295–22304. Epub 2012/05/11, PubMed PMID: 22573334; PMCID: PMC3381190. doi:10.1074/jbc.M111.318881.
- Freeman JA, Ohl ME, Miller SI. The Salmonella enterica serovar Salmonella translocated effectors SseJ and SifB are targeted to the Salmonella containing vacuole. Infect Immun. 2003;71(1):418–427. PubMed PMID: 12496192; PMCID: PMC143161.
- Drecktrah D, Levine-Wilkinson S, Dam T, Winfree S, Knodler LA, Schroer TA, Steele-Mortimer O. Dynamic behavior of Salmonella-induced membrane tubules in epithelial cells. Traffic. 2008;9(12):2117–2129. Epub 2008/09/13, PubMed PMID: 18785994; PMCID: PMC2682622. doi:10.1111/j.1600-0854.2008.00830.x.
- Knodler LA, Steele-Mortimer O. The Salmonella effector PipB2 affects late endosome/lysosome distribution to mediate Sif extension. Mol Biol Cell. 2005;16(9):4108–4123. Epub 2005/07/01, PubMed PMID: 15987736; PMCID: PMC1196323. doi:10.1091/mbc.E05-04-0367.
- Alto NM, Dixon JE. Analysis of Rho-GTPase mimicry by a family of bacterial type III effector proteins. Methods Enzymol. 2008;439:131–143. Epub 2008/04/01, PubMed PMID: 18374161. doi:10.1016/S0076-6879(07)00410-7.
- Suzuki N, Buechner M, Nishiwaki K, Hall DH, Nakanishi H, Takai Y, Hisamoto N, Matsumoto K. A putative GDP-GTP exchange factor is required for development of the excretory cell in Caenorhabditis elegans. EMBO Rep. 2001;2(6):530–535. Epub 2001/06/21, PubMed PMID: 11415987; PMCID: PMC1083904. doi:10.1093/embo-reports/kve110.
- Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, et al. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124(1):133–145. Epub 2006/01/18, PubMed PMID: 16413487. doi:10.1016/j.cell.2005.10.031.
- Reinicke AT, Hutchinson JL, Magee AI, Mastroeni P, Trowsdale J, Kelly AP. A Salmonella typhimurium effector protein SifA is modified by host cell prenylation and S-acylation machinery. J Biol Chem. 2005;280(15):14620–14627. Epub 2005/02/16, PubMed PMID: 15710609. doi:10.1074/jbc.M500076200.
- Srikanth CV, Wall DM, Maldonado-Contreras A, Shi HN, Zhou D, Demma Z, Mumy KL, McCormick BA. Salmonella pathogenesis and processing of secreted effectors by caspase-3. Science. 2010;330(6002):390–393. Epub 2010/ 10/16, PubMed PMID: 20947770. doi:10.1126/science.1194598.
- Brawn LC, Hayward RD, Koronakis V. Salmonella SPI1 effector SipA persists after entry and cooperates with a SPI2 effector to regulate phagosome maturation and intracellular replication. Cell Host Microbe. 2007;1(1):63–75. Epub 2007/11/17, PubMed PMID: 18005682; PMCID: PMC1885946. doi:10.1016/j.chom.2007.02.001.
- Hansen-Wester I, Stecher B, Hensel M. Analyses of the evolutionary distribution of Salmonella translocated effectors. Infect Immun. 2002;70(3):1619–1622. Epub 2002/02/21.PubMed PMID: 11854253; PMCID: 127817.
- Ye Z, Gorman AA, Uittenbogaard AM, Myers-Morales T, Kaplan AM, Cohen DA, Straley SC. Caspase-3 mediates the pathogenic effect of Yersinia pestis YopM in liver of C57BL/6 mice and contributes to YopM’s function in spleen. PLoS One. 2014;9(11):e110956. Epub 2014/11/06, PubMed PMID: 25372388; PMCID: PMC4220956. doi:10.1371/journal.pone.0110956.
- Wall DM, McCormick BA. Bacterial secreted effectors and caspase-3 interactions. Cell Microbiol. 2014;16(12):1746–1756. Epub 2014/09/30, PubMed PMID: 25262664; PMCID: PMC4257569. doi:10.1111/cmi.12368.
- Heine K, Pust S, Enzenmuller S, Barth H. ADP-ribosylation of actin by the Clostridium botulinum C2 toxin in mammalian cells results in delayed caspase-dependent apoptotic cell death. Infect Immun. 2008;76(10):4600–4608. Epub 2008/08/20, PubMed PMID: 18710868; PMCID: PMC2546856. doi:10.1128/IAI.00651-08.
- Ionin B, Hammamieh R, Shupp JW, Das R, Pontzer CH, Jett M. Staphylococcal enterotoxin B causes differential expression of Rnd3 and RhoA in renal proximal tubule epithelial cells while inducing actin stress fiber assembly and apoptosis. Microb Pathog. 2008;45(5–6):303–309. Epub 2008/ 08/30, PubMed PMID: 18721871. doi:10.1016/j.micpath.2008.07.002.
- Lee BC, Choi SH, Kim TS. Vibrio vulnificus RTX toxin plays an important role in the apoptotic death of human intestinal epithelial cells exposed to Vibrio vulnificus. Microbes Infect. 2008;10(14–15):1504–1513. Epub 2008/ 10/14, PubMed PMID: 18849006. doi:10.1016/j.micinf.2008.09.006.
- Cheung GY, Kelly SM, Jess TJ, Prior S, Price NC, Parton R, Coote JG. Functional and structural studies on different forms of the adenylate cyclase toxin of Bordetella pertussis. Microb Pathog. 2009;46(1):36–42. Epub 2008/ 11/11, PubMed PMID: 18992319. doi:10.1016/j.micpath.2008.10.005.