ABSTRACT
Intestinal dysbiosis is one of the causes underlying the pathogenesis of inflammatory bowel disease (IBD), encompassing ulcerative colitis (UC) and Crohn’s disease (CD). Besides bacteria, microbiota comprises both prokaryotic and eukaryotic viruses, that together compose the gut virome. Few works have defined the viral composition of stools, while the virome populating intestinal mucosae from early-diagnosed IBD patients has never been studied. Here we show that, by in-depth metagenomic analysis of RNA-Seq data obtained from gut mucosae of young treatment-naïve patients, early-diagnosed for CD and UC, and from healthy subjects (Ctrl), UC patients display significantly higher levels of eukaryotic Hepadnaviridae transcripts by comparison with both Ctrl and CD patients, whereas CD patients show increased abundance of Hepeviridae versus Ctrl. Moreover, we found that UC gut mucosa is characterized by lower levels of Polydnaviridae and Tymoviridae, whereas the mucosa of patients with CD showed a reduced abundance of Virgaviridae. Our findings support the idea that certain eukaryotic viruses might trigger intestinal inflammation and contribute to IBD pathogenesis and pave the way not only for the discovery of novel diagnostic biomarkers but also for the development of anti-viral drugs for the treatment of IBD.
Introduction
The human intestine is progressively colonized after birth by several microbial strains that change during lifespan according to anatomical, dietary and nutritional statuses.Citation1 Alterations in the gut microbiota composition (dysbiosis) are well-recognized contributors to the pathogenesis of gastrointestinal disorders, such as inflammatory bowel disease (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD). In fact, bacteria, fungi, archaea, and viruses, all populating the human intestine, were found to control gut homeostasis, with a continuous pathogen-host interplay that results in both local (mucosal and luminal) and systemic (metabolic and nutritional) effects.1,Citation2
Although bacteria have always gained the greatest attention in gastrointestinal disorders, the viral component of the human gut microbiome, termed the “gut virome”, is understudied.Citation1 The viral community of the microbiota is mainly composed of prokaryotic-infecting viruses (bacteriophages),Citation3,Citation4 thus rendering the gut a dynamic community structure, characterized by continuous “predator-prey interactions” that cause either horizontal gene transfer (viruses to bacteria), or modification in the bacterial composition of the microbiota, impacting on both health and disease.Citation5
Besides bacteriophages, the gut virome hosts also DNA- and RNA-based eukaryotic-targeting viruses able to directly transfer their genetic information to host cells, and demonstrated to be associated with the pathogenesis of both UC and CD.Citation6 Furthermore, eukaryotic viruses have been shown to contribute to intestinal dysbiosis in mice carrying mutations in the IBD-associated Il10 or Atg16L1 genesCitation7,Citation8, suggesting that the gut virome might cooperate with genetic factors, ultimately leading to chronic intestinal inflammation.
Although a consistent number of studies described the viral composition of the intestinal microbiota in human stools, mainly focusing on bacteriophagesCitation4,Citation9–Citation11, the whole eukaryotic virome that colonizes the intestinal mucosa of IBD patients still needs to be defined. A metagenomic analysis performed on a small cohort of UC and CD patients revealed that the inflamed colonic mucosa of these subjects hosts a high level of transcripts belonging to the Herpesviridae family, in comparison with healthy controls.Citation12 However, since they selected late-stage IBD patients undergoing pharmacological treatments, they were not able to assess whether specific eukaryotic viral entities infecting the gut mucosa might have influenced disease onset.
In this work, by exploiting state-of-the-art metagenomics pipelines, we profiled the gut eukaryotic virome in young treatment-naïve patients with early-diagnosed IBD and identified the eukaryotic viral communities that might be involved in IBD onset.
Results
To identify any putative eukaryotic viral entity populating the gut mucosa during early phases of intestinal inflammation, we exploited the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) database (www.ncbi.nlm.nih.gov/geo/) to search for high-throughput sequencing data performed on endoscopic mucosal biopsies from young, treatment-naïve IBD patients at their first diagnosis. Our age-range selection criteria encompassed subjects not older than 20 years. Among all the publicly available datasets, only one RNA-Seq experiment (GSE57945Citation13) performed on total RNA from biopsies derived from a large cohort of age-matched healthy subjects and patients with IBD (RISK study) fulfilled all patient inclusion criteria (patients’ characteristics are listed in ). In details, the RNA-Seq experiment included i) not inflamed ilea from colitic CD patients (cCD), ii) non-involved ilea from patients with UC, iii) ilea from CD patients with macroscopic inflammation (ileal CD [iCD]), and iv) ilea from healthy subjects undergoing colonoscopy surveillance for IBD-unrelated disease (Ctrl). Of note, the comparison between not inflamed mucosae from CD, UC and non-IBD subjects gave us the opportunity to identify those eukaryotic viruses activated independently of the mucosal inflammatory milieuCitation14, that might confer to an individual a virotypeCitation15 susceptible to IBD development. On the other hand, the comparison between not involved and inflamed CD allowed us to define whether a specific eukaryotic viral composition of the gut mucosa is inflammation-site specific.
Figure 1. RNA-Seq analysis reveals an IBD-specific virome signature. A. Table showing the characteristics of patients included in GSE57945 study. B. GSEA functional enrichment of viral response GO categories in the indicated groups. The horizontal line represents statistical significance. C. Bioinformatics pipeline. D. Relative abundance of viral orders in Ctrl, UC, cCD, and iCD. E-F. Principal component analysis (PCA) of viral order (C) and viral family (D) relative abundance in Ctrl, UC, cCD, and iCD. Data are mean± s.e.m.
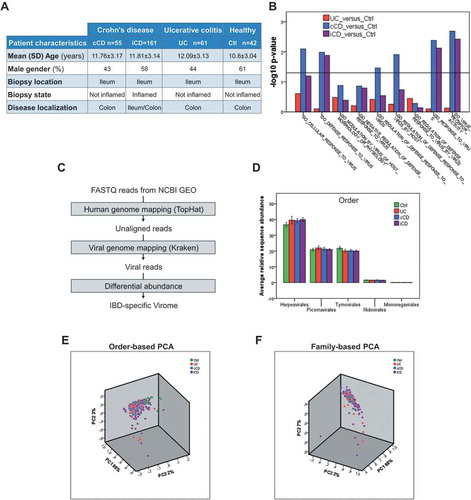
We first questioned whether mucosae from IBD patients were characterized by a viral response-related transcriptional signature in comparison with the Ctrl. By Gene Set Enrichment Analysis (GSEA) we observed a statistically significant over-representation of viral infection-related gene sets in IBD samples (). Specifically, datasets related to “cellular response to virus”, “regulation of defense response to virus” and “virus receptor activity” were found enriched in cCD, iCD, and UC versus Ctrl, confirming that a physiological response against viral infections at transcriptional level occurred at early stages of CD and UC.
Metagenomic analysis revealed a specific eukaryotic virome signature in early-diagnosed IBD
To overcome the limit given by the extensive genetic variation existing in the human gut viral genomesCitation16, we took advantage of KrakenCitation17, an ultrafast metagenomic sequence classification tool based on exact alignments of sequencing reads to several reference genomes simultaneously. Such tool is able to achieve genus-level sensitivity and precision that are very similar to that obtained by the BLAST program,Citation18().
By this analysis, the 0.02–0.03% of the total RNA-Seq FASTQ reads resulted to be viral transcripts (Supplementary Figure 1A). To verify the correct assignment of Kraken-classified reads to the viral genomes, we subsequently performed the same metagenomics analysis against the currently available microbial genome collection, finding a very small residual contamination (8.28 ± 0.4% bacterial over the total viral sequences). Additionally, classified viral reads were submitted to BLASTCitation19 in order to further exclude possible artifacts and to ensure their viral identity. Finally, we manually inspected the average base quality of classified reads; this was higher than 30 in a scale from 0 to 41 (Illumina 1.8+ Phred+ 33, Supplementary Figure 1B), thus confirming the good quality of the data prior to downstream analysis.
We next assessed the presence of the classified viral transcripts in endoscopic mucosal biopsies derived from patients with IBD who were recruited independently of disease stage, pharmacological treatments or age (Supplementary Table 1). In these samples viral amplicons were detected by RT-PCR, using multiple sets of primers (Supplementary Figure 1C, 1D, 1E and Supplementary Table 2). Not all mucosal samples presented viral amplicons (Supplementary Figure 1E), confirming the heterogeneity of biopsies in terms of viral composition. To further strengthen the reliability of our metagenomic analysis, RT-PCR-detected viral amplicons were Sanger-sequenced and successfully aligned to the respective viral genomes, in the expected region (Supplementary Figure 2A), conclusively indicating that the computational methodology identified real viral sequences.Citation17,Citation20–Citation22
Being the relative composition of viral communities more important than the absolute number of viral readsCitation18,Citation23, we then analyzed the fraction of specific viral reads on the total viral reads (hereafter referred as relative sequence abundances), rather than the absolute Reads Per Kilobase of transcript, per Million mapped reads (RPKM). By excluding prokaryotic-infecting viruses and viral entities with relative abundances lower than 1%, we found the Herpesvirales order to be the main component of both IBD- and Ctrl-gut viromes (~ 40% of total orders detected), followed by Picornavirales and Tymovirales (~ 40% of total orders detected). However, no statistically significant variation was observed ().
Moreover, Principal Component Analysis (PCA) performed on the relative sequence abundances of viral orders failed to distinguish Ctrl- from IBD-derived samples (). Similar results were obtained when PCA was performed on viral families’ relative abundances (), thus highlighting the extreme heterogeneity among the gut viromes of these patients. Nevertheless, at family level, we observed Partitiviridae to be the most abundant in all the experimental groups, followed by Herpesviridae, accounting for ~ 25% and ~ 12% of total families detected, respectively ( and Supplementary Table 3). Of note, the Hepadnaviridae family resulted to be highly enriched in UC patients by comparison not only with Ctrl, in which this viral family was not detected at all, but also with cCD and iCD (-), pointing out these transcripts as a peculiar feature of the gut virome populating the mucosa in the early stages of UC pathogenesis.
Figure 2. Early-diagnosed IBD patients showed increased abundance of specific eukaryotic viruses. A. Viral family relative abundance in Ctrl, UC, cCD, and iCD. B-E. Dot-plot (B) and MA-plots (C-E) showing Hepadnaviridae differential abundance for the indicated comparisons. F. Dot-plot showing HBx transcript differential enrichment in UC vs Ctrl, iCD, and cCD. G-K. Dot-plots for Hepeviridae (G) and Baculoviridae (J) and MA-plots (H, I, K) showing the differential abundance of the indicated comparisons. Red dots in MA-plot represent statistically enriched viral families (P ≤ 0.05). Data are represented as mean ± s.e.m. *P < 0.05; ***P < 0.005.
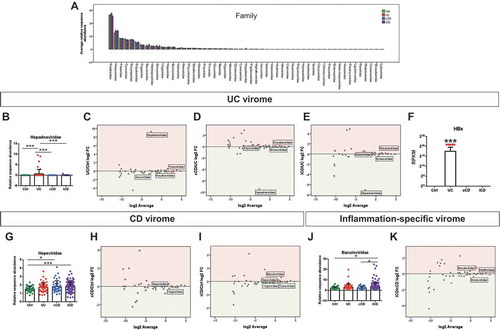
The most investigated and characterized member of the Hepadnaviridae family is Hepatitis B virus (HBV). HBV genome encodes for a small protein named HBx, whose function has yet to be fully elucidated.Citation24 Interestingly, the not-inflamed mucosa of UC patients showed a higher level of HBx transcripts by comparison with the other groups ( and Supplementary Figure 2B), thus suggesting this protein to be a conceivable factor that may influence the mucosal immune response in UC.Citation24
A different signature was found in CD-specific eukaryotic viromes. In fact, both iCD and cCD samples showed higher levels of Hepeviridae by comparison with Ctrl (-), indicating this family to characterize the intestinal mucosa of early-diagnosed CD patients. We also observed that the eukaryotic virome signature in CD patients was inflammation-specific; in fact, Baculoviridae were significantly enriched in actively inflamed iCD in comparison with unaffected ilea from cCD ( and ).
Since the intestinal microbiota is characterized by a dynamic equilibrium among its residentsCitation25, we asked whether the over-representation of defined families in the human gut virome might be counterbalanced by the reduction of others.
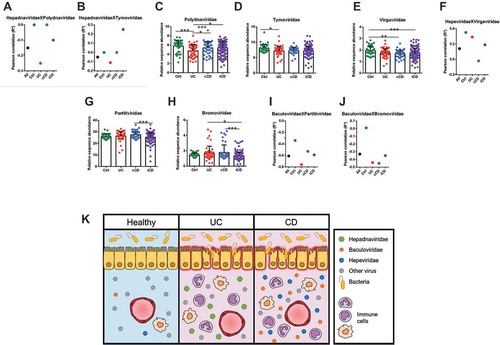
Pearson correlation analysis revealed that higher levels of Hepadnaviridae in UC patients correlated with low levels of Polydnaviridae and Tymoviridae ( and ); consistently, both Polydnaviridae and Tymoviridae were significantly decreased in UC patients by comparison with Ctrl ( and ). Similarly, higher levels of Hepeviridae in cCD and iCD was compensated by the reduced abundance of the Virgaviridae family, versus Ctrl (), with a significant negative correlation in cCD (). Finally, the enrichment of Baculoviridae in iCD was associated with lower levels of both Partitiviridae and Bromoviridae by comparison with cCD ( and ). Also in this case we found a negative correlation between Baculoviridae and Partitiviridae and Bromoviridae (-).
Figure 3. Defined viral entities prevail over others in the IBD-specific eukaryotic gut virome. Dot plots displaying statistical significant Pearson RCitation2 correlation coefficients for the following comparisons: Hepadnaviridae vs Polydnaviridae (A), Hepadnaviridae vs Tymoviridae (B), Hepeviridae vs Virgaviridae (F), Baculoviridae vs Partitiviridae (I), Baculoviridae vs Bromoviridae (J), and histogram showing relative sequence abundance for the Polydnaviridae (C), Tymoviridae (D), Virgaviridae (E), Partitiviridae (H), and Bromoviridae (I) viral families. Data are represented as mean ± s.e.m. *P < 0.05; ** P < 0.01; ***P < 0.005.
Overall, our metagenomic analysis revealed that this cohort of young, treatment-naive early- diagnosed patients displayed a peculiar eukaryotic gut virome signature that characterizes mucosae at early stages of IBD pathogenesis.
Discussion
The human virome includes human, plantCitation26 and insectCitation27 viruses, ancient virus-derived elements inserted in our chromosomes, and bacteriophages infecting bacteria that reside within our organism.Citation3 Since systemic viruses are frequently inherited from parents very early in life, either by vertical or horizontal transmission soon after birth, the virome may be seen as a significant part of the individual genetic identity.Citation28
Viruses may have both beneficial and detrimental effects on human health, depending on their interactions with other viruses and bacteria within the host.Citation29 Of note, in the gut of patients with CD, the specific expansion of bacteriophages Caudovirales has been associated with decreased bacterial diversity, supporting the concept that an unbalanced virome composition may contribute to bacterial dysbiosis and intestinal inflammation.Citation11 On the other hand, the concept that viruses might be protective has been already introduced by pioneering studies demonstrating that lymphocytic choriomeningitis virus (LCMV) prevents diabetes in miceCitation30; this finding was reinforced by several other epidemiological observations in animal models.Citation15,Citation31 In this regard, some works in mice reported that viral entities, such as Herpesvirus, might enhance resistance to Listeria monocytogenes and Yersinia pestisCitation32, or activate NK cells and gain resistance to tumor graftsCitation33, as well as increased susceptibility to autoantigen-driven experimental allergic encephalomyelitis.Citation34
Eukaryotic viruses residing in the gut may infect host cells without symptoms; however, even if asymptomatic, a virus-carrying host may hold a persistent immune response with a “continuum” of inflammatory mediators that might increase host susceptibility to disease.Citation15 The scientific community now agrees with the concept that viruses are sensed by host’s specific molecules thus eliciting a specific transcriptional response not only in infected cellsCitation15 but also in bystander cells that may be induced to release inflammatory cytokines, eventually influencing systemic immunity.Citation35,Citation36
So far, the occurrence of gut viral infections has been evaluated only in cohorts of IBD patients with long-lasting chronic intestinal inflammation. However, in these studies, either immunosuppressant or disease-induced stressing conditions may have led to the activation of latent viral infectionsCitation37 such as those from Hepatitis B and C (HBV and HCV)Citation38, CMVCitation39 and Epstein-Bar virus (EBV).Citation40 For example, a recent metagenomic analysis on colonic mucosae revealed Herpesviridae as the most enriched viral community in IBD patients.Citation12 Nonetheless, since in these works most of the enrolled patients were under pharmacological treatments and not early diagnosed, the identification of any putative viral entity responsible for the onset of IBD might not have been accurate.
Here we report for the first time a comprehensive metagenomic analysis on gut mucosae from a large cohort of treatment-naïve young patients with IBD at their first diagnosis. We included data from not inflamed mucosae to understand whether a host-specific virotype might occur and lie beneath IBD pathogenesis, independently of the inflammatory milieu. In agreement with the study performed by Wang and colleagues on long-lasting IBD patients, also in our cohort Herpesviridae family was highly abundant in all the analyzed eukaryotic gut viromes, although not differentially enriched among the groups.
By contrast, we found Hepadnaviridae to be highly abundant in UC patients, along with the small protein HBx. Despite the huge efforts in characterizing the life-cycle of Hepadnaviridae, and in particular of HBV, the function of HBx has yet to be elucidated. This has been proposed to indirectly impact on host’s transcriptional activity by interacting with nuclear transcription factors or by activating specific signal transduction pathways.Citation24 Moreover, many cell-signaling cascades, including JAK/STAT and Ras-Raf-MAPK pathways, seemed to be affected by HBx. Thus, we may argue that HBx, or similar proteins, might influence host’s immune response, ultimately leading to gut chronic inflammation in some subjects predisposed to UC, although this concept definitely needs further investigations.
Analogous events may occur in those CD patients that displayed a high abundance of Hepeviridae, the viral family encompassing Hepatitis E virus (HEV). HEV anti-viral response is very complex and intricate and leads to the release of multiple cytokines and antiviral molecules.Citation41 Thus, similarly to HBx, also Hepeviridae-derived proteins may have an impact on host immunity, eventually triggering intestinal inflammation.
Conversely, other viral families, such as Polydnaviridae and Tymoviridae in UC, and Virgaviridae in CD, that we observed to be less enriched in IBD patients and to negatively correlate with the presence of other viruses, might be somehow considered protective in the human host.Citation15 This is interesting, because Polydnaviridae, Tymoviridae, and Virgaviridae are viruses that typically infects plants and insects and may have reached the gut through the diet.Citation3 The trans-kingdom interactionCitation15 between viruses and hosts, such as plant and insect viruses that colonize human tissues, has already been reported in the past for Tobacco Mosaic Virus (TMV), against which antibodies were found in human sera.Citation42 Unlike animal viruses, plant viruses cannot replicate in humans or other animals because of the lack of specific receptors. Nevertheless, they still can induce the host immune response, as shown for the cowpea mosaic virus in mice.Citation43,Citation44
Altogether, our metagenomic analysis introduces a quite new concept that sheds new light on IBD aetiogenesis as likely being related to a peculiar mucosal eukaryotic virome composition where some viral entities clearly prevail over others (). These virome signatures could be acquired early in life possibly because of environmental factors (i.e., diet), and ultimately be responsible for higher host susceptibility to IBD.
Notably, the metagenomic analysis we performed for accomplishing this study exploited pre-existing RNA-Seq data, generated with the final goal to characterize the entire human transcriptomic profile of mucosal biopsies derived from pediatric IBD patients. This means that the sequencing coverage used in the original workCitation13 did not properly cover the non-human extra genomes present in the preparation, as the authors’ intentions were not indeed to look for exogenous viral sequences. We believe that future works aimed to finely dissect this aspect will need to consider this important technical limitation and increase sequencing power accordingly.
Conclusively, further studies adequately planned to elucidate the molecular mechanisms through which these viruses may lead to gut chronic inflammation are urgently needed, as this may help to develop novel biomarkers for the diagnosis and drugs for the treatment of IBD.
Material and methods
Rna-Seq transcriptome and metagenomic analyses
Patient cohort
The National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) database (www.ncbi.nlm.nih.gov/geo/) was interrogated to search for high-throughput sequencing data performed on endoscopic biopsies collected from IBD patients and healthy controls satisfying the following criteria: (1) all subjects had to be not older than 20 years; (2) IBD patients enrolled had to be at their first diagnosis (with clinical symptoms for less than 5 years) and treatment-naïve. One RNA-Seq experiment (GEO accession number GSE57945Citation13) displaying these patients’ characteristics was performed on total RNA isolated from ileal biopsies obtained during diagnostic colonoscopy of early-diagnosed CD, UC, and non-IBD (Ctrl) patients, with an age comprised between 2 up to 17 years and belonging to the RISK study, an ongoing, prospective observational inflammatory bowel disease (IBD) inception cohort sponsored by the Crohn’s and Colitis Foundation of America (CCFA) (www.crohnscolitisfoundation.org). Confirmation of active colitis/ileitis was done by histology prior to diagnosis and treatment, in a standardized fashion. Information about diagnosis and biopsy collection is available in Haberman et al., 2014Citation13 and at http://www.crohnscolitisfoundation.org/science-and-professionals/research/current-research-studies/pediatric-risk-successes.html?referrer=https://www.google.it/#.
Transcriptomics and metagenomics
FASTQ reads were retrieved from the NCBI GEO repository, quality filtered, and adaptor trimmed, with NCBI fastq-dump. For transcriptome analysis, reads were aligned to the hg38 reference human genome with TopHat for splice junction specific mapping.Citation45 Gene expression read counts were measured with featureCounts.Citation46 Functional enrichment analysis was performed with GSEA.Citation47 For metagenomics analysis, the reads that failed to be aligned to the human genome with TopHat were subsequently mapped to the complete collection of all available virus genomes (https://www.ncbi.nlm.nih.gov/genome/) with KrakenCitation17 for exact alignment of k-mers and accurate viral read classification. Relative abundances were calculated with Pavian.Citation23 Viral read calls were confirmed by manually aligning Kraken classified reads to the respective viral genomes with TopHat and visualizing the resulting BAM alignments with the Integrative Genomics Viewer (IGV).Citation48 Prior to statistical analysis, classified reads were double checked with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to confirm quality filtering and adaptor trimming, and then submitted to BLASTCitation19 in order to exclude possible artifacts resulting from the in silico analysis. Analyzed data and FASTQ read quality check can be found at https://doi.org/10.17605/OSF.IO/EKD3C.
Statistical analysis
All statistical analysis and plotting were performed with either GraphPad Prism 7 or the IBM SPSS statistical packages. Differential relative viral read abundances were calculated by one-way ANOVA, with Bonferroni post-hoc test and FDR correction for multiple comparisons (see Supplementary Table 3). Pearson correlation coefficients were used for multivariate analyses with linear regression assumptions.
Collection of human biopsies for viral amplicon detection after in silico analysis
Colonic biopsies were collected from patients with UC and CD undergoing endoscopy. IBD patients were diagnosed by physicians based on clinical, endoscopic and histological criteria. Endoscopic activities were assessed using the endoscopic Mayo scoreCitation49, and the Riley histological scoring systemCitation50 as described in Ungaro et al.., 2017Citation51 Healthy colonic biopsies were obtained from patients undergoing routine check-up for non-IBD related diseases. For patients’ characteristics see the Supplementary Table 1. Samples were not randomized, and allocation and evaluation of clinical disease parameters were not blinded. The study was approved by the Humanitas Research Hospital ethics committee. All subjects provided written informed consent.
Reverse transcription polymerase chain reaction (RT-PCR) and sanger sequencing
Total RNA was extracted from human biopsies using the PureZOL® RNA isolation reagent (catalog #7326880; Bio-Rad) according to manufacturer’s instructions. RNA retro-transcription was performed with High Capacity cDNA Reverse Transcription Kits (catalog #4368814; Applied Biosystems). RT-PCR reaction was performed with GoTaq® DNA Polymerase (catalog #M300; Promega Corporation) according to the manufacturer’s instructions. For melting curve analysis, RT-PCRs was performed with SYBR® Green Real-Time PCR Master Mix (catalog #4309155; Thermo Fisher Scientific) according to the manufacturer’s instructions and analyzed on ViiA7 Real-Time PCR System (Applied Biosystems). PCR products were loaded on 2% agarose gel and then visualized at ChemiDoc™ MP Imaging System (Bio-Rad Laboratories), using Quantity One software. GAPDH expression was used as internal control. Primer pairs were specifically designed for viral nucleotide sequences obtained by metagenomic analysis, tested for the viral identity by BLAST and are listed in the Supplementary Table 2.
RT-PCR products obtained with Expand High Fidelity PCR System (Roche®) were loaded on 1% agarose gel and run at 130 V in TAE1X buffer. For each viral amplicon, the band of the expected molecular weight (Supplementary Table 4) was purified with Wizard® SV Gel and PCR Clean-Up System (Promega Corporation) and underwent Sanger Sequencing at Hunimed Genomic Unit. Primers used for Sanger sequencing are listed in the Supplementary Table 4. Sanger sequencing results are available at https://doi.org/10.17605/OSF.IO/EKD3C.
Author Contributions
Conceptualization, F.U., L.M., and S.DA.; Methodology, F.U., L.M. and V.R.; Formal Analysis, F.U., and L.M.; Investigation, F.U., L.M.; Resources F.F. and S.D.; Writing – Original Draft, F.U., L.M., and S.DA.; Writing – Review & Editing, F.U., L.M., F.F. S.DA., L.P., V.R. and S.D.; Visualization, F.U., and L.M.; Supervision, S.DA. and S.D.; Project Administration, F.U., and S.DA.; Funding Acquisition, F.U.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download MS Power Point (1.6 MB)Supplementary materials
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Scarpellini E, Ianiro G, Attili F, Bassanelli C, De Santis A, Gasbarrini A. The human gut microbiota and virome: potential therapeutic implications. Dig Liver Dis. 2015;47:1007–1012.
- Dave M, Higgins PD, Middha S, Rioux KP. The human gut microbiome: current knowledge, challenges, and future directions. Transl Res. 2012;160:246–257.
- Focà A, Liberto MC, Quirino A, Marascio N, Zicca E, Pavia G. Gut inflammation and immunity: what is the role of the human gut virome? Mediators Inflamm. 2015;2015:1–7.
- Carding SR, Davis N, Hoyles L. Review article: the human intestinal virome in health and disease. Aliment Pharmacol Ther. 2017;46:800–815.
- Cadwell K. Expanding the role of the virome: commensalism in the gut. J Virol. 2015;89:1951–1953.
- Lopes S, Andrade P, Conde S, Liberal R, Dias CC, Fernandes S, Pinheiro J, Simões JS, Carneiro F, Magro F, et al Looking into enteric virome in patients with IBD. Inflamm Bowel Dis. 2017;23:1278–1284.
- Cadwell K, Patel KK, Maloney NS, Liu T-C, Ng ACY, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145.
- Basic M, Keubler LM, Buettner M, Achard M, Breves G, Schröder B, Smoczek A, Jörns A, Wedekind D, Zschemisch NH, et al Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm Bowel Dis. 2014;20:431–443.
- Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008. p. 4.
- Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proc Natl Acad Sci. 2013;110:12450–12455.
- Jm N, Sa H, Mt B, Droit L, Cy L, Bc K, Kambal A, Cl M, Zhao G, Fleshner P, et al Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460.
- Wang W, Jovel J, Halloran B, Wine E, Patterson J, Ford G, OʼKeefe S, Meng B, Song D, Zhang Y, et al. Metagenomic analysis of microbiome in colon tissue from subjects with inflammatory bowel diseases reveals interplay of viruses and bacteria. Inflamm Bowel Dis. 2015. 1.
- Haberman Y, Tickle TL, Dexheimer PJ, Kim M-O, Tang D, Karns R, Baldassano RN, Noe JD, Rosh J, Markowitz J, et al Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–3633.
- Traylen CM, Patel HR, Fondaw W, Mahatme S, Williams JF, Walker LR, Dyson OF, Arce S, Akula SM. Virus reactivation: a panoramic view in human infections. Future Virol. 2011;6:451–463.
- Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157:142–150.
- Minot S, Grunberg S, Wu GD, Lewis JD, Bushman FD. Hypervariable loci in the human gut virome. Proc Natl Acad Sci. 2012;109:3962–3966.
- Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46.
- Waller AS, Yamada T, Kristensen DM, Kultima JR, Sunagawa S, Koonin EV, Bork P. Classification and quantification of bacteriophage taxa in human gut metagenomes. ISME J. 2014;8:1391–1402.
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693.
- Brady A, Salzberg S. PhymmBL expanded: confidence scores, custom databases, parallelization and more. Nat Methods. 2011;8:367.
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386.
- Brady A, Salzberg SL. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat Methods. 2009;6:673–676.
- Breitwieser FP, Salzberg SL. Pavian: interactive analysis of metagenomics data for microbiomics and pathogen identification. bioRxiv. 2016. 2014–7.
- Nguyen DH, Ludgate L, Hepatitis HJ. B virus–cell interactions and pathogenesis. J Cell Physiol. 2008;216:289–294.
- Rodriguez-Valera F, A-B M-C, Rodriguez-Brito B, Pašić L, Thingstad TF, Rohwer F, Mira A. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836.
- Balique F, Lecoq H, Raoult D, Colson P. Can plant viruses cross the kingdom border and be pathogenic to humans? Viruses. 2015;7:2074–2098.
- Delwart EA. Roadmap to the Human Virome. PLoS Pathog. 2013;9:e1003146.
- Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50.
- Zou S, Caler L, Colombini-Hatch S, Glynn S, Srinivas P. Research on the human virome: where are we and what is next. Microbiome. 2016;4:32.
- Oldstone M. Prevention of type I diabetes in nonobese diabetic mice by virus infection. Science (80-). 1988;239:500–502.
- Roossinck MJ. The good viruses: viral mutualistic symbioses. Nat Rev Microbiol. 2011;9:99–108.
- Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin HW. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329.
- White DW, Keppel CR, Schneider SE, Reese TA, Coder J, Payton JE, Ley TJ, Virgin HW, Fehniger TA. Latent herpesvirus infection arms NK cells. Blood. 2010;115:4377–4383.
- Peacock JW, Elsawa SF, Petty CC, Hickey WF, Bost KL. Exacerbation of experimental autoimmune encephalomyelitis in rodents infected with murine gammaherpesvirus-68. Eur J Immunol. 2003;33:1849–1858.
- Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534.
- Canny SP, Goel G, Reese TA, Zhang X, Xavier R, Virgin HW. Latent gammaherpesvirus 68 infection induces distinct transcriptional changes in different organs. J Virol. 2014;88:730–8.38.
- Mazzola G, Fs M, Adamoli L, Renna S, Cascio A, Orlando A. Diagnostic and vaccine strategies to prevent infections in patients with inflammatory bowel disease. J Infect. 2017;74:433–441.
- Degasperi E, Caprioli F, El Sherif O, Back D, Colombo M, Aghemo A. Challenges in treating patients with inflammatory bowel disease and concurrent viral hepatitis infection. Expert Rev Gastroenterol Hepatol. 2016;10:1373–1383.
- Sager K, Alam S, Bond A, Chinnappan L, Probert CS. Review article: cytomegalovirus and inflammatory bowel disease. Aliment Pharmacol Ther. 2015;41:725–733.
- Rizzo AG, Orlando A, Gallo E, Bisanti A, Sferrazza S, Montalbano LM, Macaluso FS, Cottone M. Is Epstein-Barr virus infection associated with the pathogenesis of microscopic colitis? J Clin Virol. 2017;97:1–3.
- Kang S, Myoung J. Host innate immunity against hepatitis e virus and viral evasion mechanisms. J Microbiol Biotechnol. 2017;27:1727–1735.
- Liu R, Vaishnav RA, Roberts AM, Friedland RP. humans have antibodies against a plant virus: evidence from tobacco mosaic virus. PLoS One. 2013;8:e60621.
- Olszewska W, Steward MW. The molecular basis of the antigenic cross-reactivity between measles and cowpea mosaic viruses. Virology. 2003;310:183–189.
- Rae CS, Wei Khor I, Wang Q, Destito G, Gonzalez MJ, Singh P, Thomas DM, Estrada MN, Powell E, Finn MG, et al Systemic trafficking of plant virus nanoparticles in mice via the oral route. Virology. 2005;343:224–235.
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36.
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930.
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550.
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26.
- Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 1987;317:1625–1629. doi:10.1056/NEJM198712243172603
- Riley SA, Mani V, Mj G, Dutt S, Me H. Microscopic activity in ulcerative colitis: what does it mean? Gut. 1991;32:174–178.
- Ungaro F, Tacconi C, Massimino L, Corsetto PA, Correale C, Fonteyne P, Piontini A, Garzarelli V, Calcaterra F, Della Bella S, et al. MFSD2A promotes endothelial generation of inflammation-resolving lipid mediators and reduces colitis in mice. Gastroenterology. 2017. doi:10.1053/j.gastro.2017.07.048.
