ABSTRACT
Probiotics are considered to have multiple beneficial effects on the human gastrointestinal tract, including immunomodulation, pathogen inhibition, and improved host nutrient metabolism. However, extensive characterization of these properties is needed to define suitable clinical applications for probiotic candidates. Lactobacillus johnsonii 456 (LBJ 456) was previously demonstrated to have anti-inflammatory and anti-genotoxic effects in a mouse model. Here, we characterize its resistance to gastric and bile acids as well as its ability to inhibit gut pathogens and adhere to host mucosa. While bile resistance and in vitro host attachment properties of LBJ 456 were comparable to other tested probiotics, LBJ 456 maintained higher viability at lower pH conditions compared to other tested strains. LBJ 456 also altered pathogen adhesion to LS 174T monolayers and demonstrated contact-dependent and independent inhibition of pathogen growth. Genome analyses further revealed possible genetic elements involved in host attachment and pathogen inhibition. Importantly, we show that ingestion of Lactobacillus johnsonii 456 over a one week yogurt course leads to persistent viable bacteria detectable even beyond the period of initial ingestion, unlike many other previously described probiotic species of lactic acid bacteria.
The human gastrointestinal (GI) tract is home to over 500 species of bacteria in a given individual.Citation1 These microbes and their byproducts play as important a function in our bodies as any other organ, and have been subject to co-adaptation with their hosts for at least 500 million years.Citation2 The microbiome has been demonstrated to have an impact on nearly every aspect of human health. Gut microbiota composition is a risk factor for inflammatory bowel diseases (IBDs) such as Crohn’s Disease and ulcerative colitis.Citation3 Resident bacteria play a critical role in the development of healthy immune system function.Citation4 Microbial metabolic processes generate short-chain fatty acids (SCFAs) that provide a primary energy source for the cells of the gut,Citation5,Citation6 as well as vitamins and amino acids necessary for systemic health.Citation7 Microbiome composition affects efficiency of nutrient metabolism, playing a role in obesity risk and even cholesterol levels.Citation8–Citation10 There is even evidence to suggest that the microbiome plays a role in normal CNS function and depression incidence.Citation11 The manipulation of the microbiome by purposefully seeding certain probiotic, or beneficial, strains for their properties may allow us to better control every one of these endpoints – with the right level of understanding. However, these multifactorial effects are often difficult to study in a well-controlled environment.
A probiotic with strong clinically demonstrated effects could be employed in a number of ways to induce a wide variety of health benefits, both as a treatment and as a health maintenance supplement. Probiotic lactobacillus bacteria have been demonstrated to reduce inflammation both in the gut itself and systemically,Citation12 making them a tempting therapy for researchers seeking effective treatments for inflammatory gut conditions, such as ulcerative colitis and Crohn’s disease, although results as of yet have not been particularly strong.Citation13–Citation15 Despite a global market value in the tens of billions of US dollars, growing by over 10% per year,Citation16 there is, as of the end of 2017, no probiotic that is clinically approved by the FDA. The popular probiotic Lactobacillus rhamnosus GG did not yield significant results in a clinical trial against vancomycin-resistant enterococcus (VRE),Citation17 and L. johnsonii NCC 533 failed in a clinical trial against Crohn’s disease.Citation18
Although the use of probiotics to treat chronic states like inflammation and provide subtle benefits to health is an attractive goal, especially due to the millions of people who suffer from some form of inflammatory disease,Citation19 our incomplete understanding of the multifactorial complexity of interactions between the microbiome and immunity makes it difficult to accurately predict which treatments will work and why.Citation4 Instead, we suggest that an interim application, with more clearly definable endpoints, be the primary focus of treatments in the nascent field of clinical probiotics. In particular, the use of probiotic strains to both shorten active and prophylactically prevent instances of pathogenically induced diarrhea should be a top priority – especially given that diarrheal disease led to 1.3 million deaths worldwide in. 2015 Citation20
In this regard, the most clearly demonstrable and valuable attribute of probiotic strains is their capability to reduce the adhesion and subsequent activity of pathogenic strains. Probiotic bacteria can perform this useful service via a number of mechanisms, including indirect competition for nutrients and binding sites in the host,Citation21 and directly through the production of bacteriocins, acids, and other compounds.Citation22–Citation24 In animal models, lactobacillus species have been broadly shown to inhibit gut pathogens. L. johnsonii NCC 533 (formerly referred to as La1) been shown to reduce gastritis induced by H. pylori and infection by the diplomonad G. intestinalis in gerbils, while L. johnsonii FI9785 inhibited C. perfringens colonization in chickens Citation25–Citation27 Multiple probiotic formulations, including Lactobacillus and Bifidobacterium strains, have been shown to reduce the duration of diarrhea and enterocolitis in children.Citation28
Considering the strong evidence for anti-inflammatory and antipathogenic effects, it is clear that both the search for new probiotic strains and the continued testing of existing ones will yield clinically effective treatment methods. For dedicated clinical application, strains will need to be characterized based on their effects on individual disease states. For example, a strain that induces a beneficial cytokine response under certain circumstances might exacerbate pathogen-induced disease in others by interfering with the immune response.Citation29 For this reason, it is imperative that every isolated strain of probiotic bacteria be individually tested and characterized in multiple models. A rationally designed set of experiments demonstrating survival, adhesion, and pathogen inhibition should be carried out with strains that show promising attributes.Citation30
Lactobacillus johnsonii strain 456 (LBJ 456) was discovered by examining bacterial strains overrepresented in the microbiota of a cancer-resistant colony of DNA-repair deficient mice.Citation12 Considering that oral gavage with this strain over the course of 4 weeks was capable of significantly reducing systemic inflammation and genotoxicity in this mammalian model, LBJ 456 represents a strong candidate probiotic strain. As lactobacillus bacteria, this strain is conducive to use not only in traditional manners of application such as supplement pills, but also in active foodstuff delivery methods such as yogurts and kombuchas. In this article, we further demonstrate this strain’s potential for use in humans by characterizing its acid and bile resistance as well as its host adhesion, pathogen inhibition, and colonization properties. We also analyze the LBJ 456 genome to investigate the genetic basis underlying some of these properties.
Results
L. johnsonii 456 shows exceptional resistance to gastric acid and moderate bile acid tolerance
To assess LBJ 456’s viability in the GI tract, we compared its relative tolerance to simulated gastric acid (SGA) and bile against a panel of type strains representing commonly used probiotic species, including the two commercially available strains B. lactis HN019 and L. plantarum 299V (). We also included S. salivarius subsp. thermophilus, which is not a Lactobacillus or Bifidobacterium species but is still considered a “probiotic” by the European Food Safety Administration for its potential assistance in lactose digestion, and traditional role in yogurt preparation.Citation31 We measured the viable bacteria recovered for each strain after incubation for 2 hours in gastric conditions that ranged from pH 3 to pH 1.2 (, Fig. S1). While recoverable CFU from each strain generally decreased with lower pH conditions, the viability of the two L. johnsonii strains LBJ 456 and VPI 7960 was observed to increase beyond the input CFU at pH 3. Moreover, the viability of LBJ 456 in particular was consistently the highest at all pH conditions tested. Importantly, LBJ 456 was also the only strain to show viability at pH 1.2, albeit at a 1000-fold reduction compared to its viability in a control pH 6 incubation.
Table 1. Strains and cell lines used.
Figure 1. L. johnsonii 456 has exceptional resistance to simulated gastric conditions and moderate bile acid tolerance. (a) Survival of Lactobacillus and other probiotic associated strains after 2 hours in SGA. (b) Relative growth capability of test strains after growth in media supplemented with bile acids. Results are expressed as means and SEMs (n = 2). Each experimental pH or bile acid concentration vs. control was run as a separate experiment, and all experiments were repeated at least twice. Specific representation at each pH reading by histogram is included in Supplemental Figures S1 and S2.
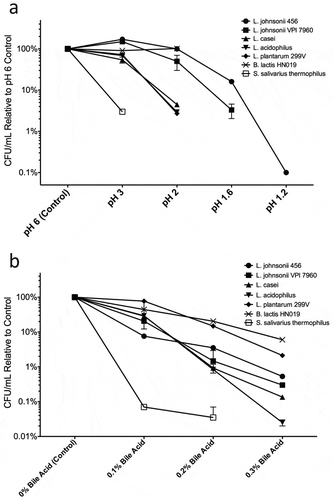
Next, we compared the growth of the probiotic strain panel in media under different physiologically relevant bile acid conditions (, Fig. S2). A relatively bile acid rich environment (0.3%/~ 6mM) impairs the growth of LBJ 456 to a certain extent, but it still reached concentrations of around 4–5 × 106 cells/mL after 24 hours (as opposed to nearly 1 × 109 cells in bile-free media control). Interestingly, bile acid resistance among strains allowed for clear delineation between genera, especially at 0.2 and 0.3%. B. lactis growth was only decreased to about 10% of control at 0.3% bile acid. Lactobacillus species as a whole had moderate resistance, but L. plantarum 299V’s growth was impaired the least of all Lactobacillus strains. S. salivarius was highly sensitive to acid and bile acid exposure and was unable to grow at all in 0.3% bile acid, suggesting that this strain likely does not survive in the human GI tract.
L. johnsonii 456 adheres most strongly to goblet cell-like monolayer forming line LS 174T
Bacterial adhesion to the intestinal epithelium, as well as the associated mucus secretions, has long been considered an important probiotic criterion.Citation32,Citation33 We evaluated the host attachment capabilities of our probiotic panel by measuring the percentage of CFU-forming cells that could adhere to two monolayer-forming human cancer cell lines, the enterocyte-like Caco-2 line and the goblet cell-like LS 174T line (). All tested probiotics, including LBJ 456, adhered better to the secreted mucin-rich LS 174T cultures than to Caco-2 cultures. The relative adhesion of each strain to the two monolayer types did not directly correlate (r = −0.1429, Spearman’s rank correlation). For example, L. casei showed the lowest rate of adhesion to Caco-2, but was one of the most adherent strains on LS 174T. These data suggest potential specialization of different Lactobacillus strains to better adhere to different gut mucin phenotypes. Adhesion of LBJ 456 to LS 174T was observed to a greater extent than the commercially available probiotic strains L. plantarum 299V and B. lactis HN019. LBJ 456 also adhered to LS 174T an order of magnitude better than L. acidophilus ATCC 4356, which demonstrated strong adhesion in the 5 hour exposure model used by Jung et al.Citation34 Interestingly, S. salivarius adhered relatively well to both cell lines, despite the fact that its survival until that point in the digestive tract would seem unlikely based on acid and bile sensitivity.
L. johnsonii 456 significantly alters pathogen adhesion to LS 174T, but not to caco-2 monolayers
Using the LS 174T and Caco-2 monolayer models described above, we examined the capacity of adherent LBJ 456 to inhibit the attachment of three pathogenic strains of gut bacteria: Enterotoxigenic E. Coli (ETEC), E. faecalis, and S. enterica. (). Pretreatment of Caco-2 monolayers with LBJ 456 did not lead to any significant difference in the level of pathogen adhesion, likely because of the LBJ 456’s limited ability to adhere to this cell type (). However, LBJ 456 pretreatment of LS 174T monolayers led to significant changes in pathogen adhesion (). (ETEC) and S. enterica adhesion were reduced by about 30% (p = 0.0423) and 40% (p = 0.0658), respectively. However, E. faecalis adhesion increased slightly after pretreatment with LBJ 456, indicating that the inhibitory capability of this strain is not universal.
Figure 3. L. johnsonii 456 significantly inhibits pathogenic strain adhesion to goblet cell-like gut epithelial monolayers, but not enterocyte-like monolayers. (a) Adhesion of pathogenic bacteria to a Caco-2 monolayer after 1 hour pre-exposure to LBJ 456. (b) Adhesion of pathogenic bacteria to an LS 174T monolayer after 1 hour pre-exposure to LBJ 456. Data expressed as means and SEM. Relevant statistically significant differences are indicated [* = p < 0.05, ** = p < 0.1 (t test); n = 4]. All experiments were performed twice.
![Figure 3. L. johnsonii 456 significantly inhibits pathogenic strain adhesion to goblet cell-like gut epithelial monolayers, but not enterocyte-like monolayers. (a) Adhesion of pathogenic bacteria to a Caco-2 monolayer after 1 hour pre-exposure to LBJ 456. (b) Adhesion of pathogenic bacteria to an LS 174T monolayer after 1 hour pre-exposure to LBJ 456. Data expressed as means and SEM. Relevant statistically significant differences are indicated [* = p < 0.05, ** = p < 0.1 (t test); n = 4]. All experiments were performed twice.](/cms/asset/b07733c3-1b46-4db9-87de-97c940d9ae30/kgmi_a_1547612_f0003_oc.jpg)
L. johnsonii 456 significantly inhibits pathogen growth in co-culture
We determined whether LBJ 456 can directly inhibit the growth of pathogens by co-culturing it with equal CFU ratios of each pathogenic strain. The growth of all three pathogens was significantly reduced when they were co-cultured with LBJ 456 (). ETEC growth was suppressed from a final concentration of 5 to 2 × 108 cells/mL. Final E. faecalis concentrations were cut by more than half as well. The greatest effect was seen against Salmonella, with a full order of magnitude decrease in viable CFU detected from coincubation. (p < 0.0001 for all comparisons) Lactobacillus was readily capable of growth in media other than its own, although colonies were petite under aerobic growth conditions (Fig. S3).
Filter sterilized supernatant of L. johnsonii 456 significantly inhibits the survival of S. enterica and E. faecalis, but not of ETEC
As shown in , ETEC was unaffected by filtered supernatant (FS) from any Lactobacillus strain except L. plantarum, which completely prevented its survival. LBJ 456 FS significantly inhibited E. faecalis survival by over half (p = 0.0427), while L. plantarum FS killed off over 99% of this strain (p = 0.0051). All tested Lactobacillus strains significantly decreased S. enterica survival, with all tested strains beside L. casei leading to a complete absence of viable CFU after 18–20 hours. Surprisingly, all tested FS led to significantly decreased Bifidobacterium viability as well, even though this strain was introduced as a non-pathogen control. As pH was controlled for, another acellular factor must be responsible for these differences.
Table 2. Inhibition of growth by Lactobacillus filtered supernatant.
Detectable LBJ 456 can persist in the human gut long after initial ingestion
We determined whether LBJ 456 was capable of long-term survival in the human gut. 11 healthy adult individuals completed a 7 day LBJ yogurt trial and supplied fecal samples before yogurt consumption (day 0), immediately after (day 7) and at 30 and 60 days after initiation. Over the course of the study, no adverse side effects or diarrhea symptoms were reported for any of the volunteers. Background levels of live lactobacillus varied significantly between volunteers, from undetectable (6 subjects) to nearly 40 million viable CFU/gram in one subject. After sample collection, the subset of volunteers that tested negative for background Lactobacillus before study initiation were grouped for secondary analysis. First, stool samples were analyzed for the presence of live lactic acid bacteria (LAB) (). In the full group of participants, recovered LAB differed significantly between all time points over the course of the 60 day period (p < 0.001, Friedman test). To ascertain differences between individual time points, post hoc analyses with Wilcoxon signed rank tests were conducted and a Holm-Bonferroni correction was applied. After correction, statistical significance was observed only between baseline and day 7 samples (p = 0.048), indicating that the yogurt course successfully introduced live LAB even under the most stringent conditions of analysis. Individuals with Lactobacillus-negative baseline fecal readings also showed detectable lactobacillus over the 60 day course (p < 0.001, Friedman test). After post hoc analysis with Wilcoxon signed rank tests and Holms-Bonferroni correction, no differences were significant in this group due to the low number of participants. Despite this, there was a clear upward trend in detectable live Lactobacillus counts in both the whole group and the LB-negative background subset that lingered through at least one month after the weeklong course. Viable LAB increased by about an order of magnitude (~ 104.5 to 105.5 CFU/gram feces) at days 7 and 30 in the full group, and remained higher at day 60. Quantitative polymerase chain reaction (qPCR) targeting a LBJ 456-specific gene sequence in fecal DNA confirmed that gut abundance of LBJ 456 was specifically increased by the yogurt trial. (, Fig. S4). Overall, the 16S-normalized abundance of this sequence followed a similar pattern as LAB CFU after ingestion. Background counts increased from LBJ 456-negative at day 0 (< 1 copy/million copies of 16s RNA) to an average of roughly one copy per 50,000 from days 7–30. Although a clear trend towards increased LBJ 456 was observed over time, this difference did not reach significance due to the high variance and low number of participants (p = 0.0547 for Day 0 vs. Day 30, Wilcoxon signed rank test, n = 11).
Figure 5. A 7 day course of LBJ 456 yogurt leads to elevations in both total lactic acid bacteria (LAB) and LBJ 456 specific DNA. (a) Fecal load of viable LAB, as detectable by anaerobic growth on MRS agar, prior to and after 7 day course. Solid line: All volunteers that completed a full course and supplied all four fecal samples (n = 11). Dashed Line: individuals with no detectable LAB at day 0, prior to initiation of yogurt course (n = 6). All individuals with LAB negative backgrounds had detectable LAB at day 7. 5/6 still had detectable levels at 30 days, and half were still detectable at day 60. Lower detection limit of this assay = 4000 CFU/mL. * = adjusted p < 0.05 (Wilcoxon signed rank test with Holm-Bonferroni correction) for Day 0–7 (all volunteers). (b) qPCR of detectable DNA sequences in fecal samples, expressed as ratio of LBJ 456 specific DNA sequence to 16S gene as a universal bacterial ribosomal marker sequence. Ratio of LBJ 456 specific DNA to 16S DNA sequence is approximately 1:1 in cultured LBJ 456, and undetectable in LBJ VPI 7960 control (control data not shown). Data expressed as means and SEM.
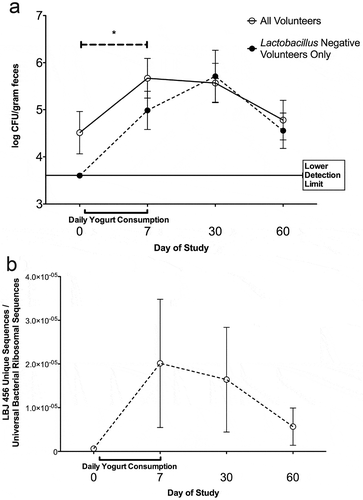
According to recent estimates, about 3.8 × 1013 bacteria make their home in the average human, with most of those in the gut.Citation35 Of these, approximately 1011 cells are shed with every gram of feces. Based on our qPCR estimates, at least two million LBJ 456 genomes would be represented in the feces of individuals recently inoculated with the strain, assuming the average cell has one detectable copy of the 16sRNA gene (a conservative estimate, considering that many species encode multiple copies).Citation36 This number is relatively consistent with a viable CFU estimate on the order of 105.5 CFU/gram, especially considering cells rendered nonviable during fecal storage at freezing temperature.
Potential genetic basis of L. johnsonii 456 persistence and inhibition properties revealed by comparative genomics
To better understand the genomic basis underlying the probiotic properties of LBJ 456, we sequenced its genome and compared it to 23 publically available L. johnsonii genomes. The 24 L. johnsonii genomes share 1.2 Mb of conserved genomic sequence. Alignment of 101,088 SNPs in this core genome revealed two groups of highly similar L. johnsonii genomes, one consisting of 117a, 117c, 117d, 117k, 117q, and 117x, and the other consisting of UMNLJ21 and UMNLJ22 (). The isolates within these groups differ by less than 300 SNPs and may represent isolates of the same strain. Considering 117a and UMNLJ22 as representative isolates for these groups, we found that the L. johnsonii strains differ by an average of 29,943 core genome SNPs. In particular, LBJ 456 differs more from VPI 7960 (33,051 core SNPs), an isolate from human blood, than W1 (20,369 core SNPs), another isolate from mouse gut,Citation37 consistent with previous observations that genetic similarity among L. johnsonii strains is highest among strains from the same host organism.Citation38
Figure 6. L. johnsonii 456 is genetically distinct from other described L. johnsonii strains. (a) Phylogenetic tree of 24 L. johnsonii isolates based on 101,088 SNPs in core genomic regions. Distances represent percentage of SNP differences out of total compared SNPs. (b) Frequency of non-core regions among the 24 L. johnsonii strains. Yellow cells indicate presence of a non-core region (along columns) in a strain (along rows). Dendrograms based on complete linkage hierarchical clustering of strains or regions based on Euclideans distances. Non-core region lengths are not represented here.
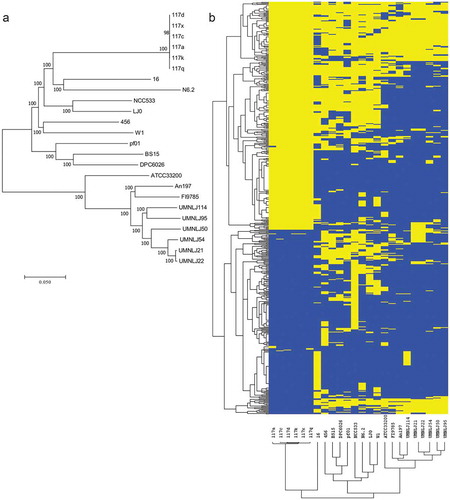
We then identified 550 genomic regions that are not shared among all 24 L. johnsonii strains. These non-core regions total 1,067,226 bp and include regions unique to each of LBJ 456, NCC 533, 117c, 117d, 117q, and 16 (). Within the LBJ 456 genome, we identified 11 non-core regions totaling 41,781 bp. As shown in , these regions encode proteins mostly involved in replication (e.g. FtsK/SpoIIIE family protein, plasmid replication protein), and antiviral defense (restriction-modification enzymes), suggesting that cell proliferation strategies and phage exposure may be important processes that determine the compatibility and viability of LBJ 456, and L. johnsonii strains in general, within particular hosts.
Table 3. Genes within noncore regions unique to L. johnsonii 456.
Moreover, we observed two mucus binding proteins (MBPs) encoded in the non-core regions unique to LBJ 456. A more comprehensive comparison of MBPs across the 24 L. johnsonii strains revealed between 3 and 18 MBPs (average 7) in each genome, with LBJ 456 containing the most. These MBPs represent a repertoire of 34 unique homologs based on clustering by 70% identity. 13 of these homologs are encoded only by single strains, including two that are uniquely encoded by LBJ 456 (PROKKA_00690 and PROKKA_00875).
We also identified two bacteriocin-encoding loci in the LBJ 456 genome (Figure S6, Table S2). The first locus contains three putative type II bacteriocins (PROKKA_00793, PROKKA_00800, and PROKKA_00806) and two other genes (PROKKA_00798 and PROKKA_00799) predicted to encode proteins involved in bacteriocin processing and secretion. The second locus contains a single predicted bacteriocin gene, PROKKA_00732, surrounded by genes with no clear bacteriocin processing or secretion function. However, as two transposase-related proteins, PROKKA_00728 and PROKKA_00729, were also detected in this locus, it is possible that this putative bacteriocin may be part of a mobile genetic element.
Discussion
Immediately after ingestion, bacteria face the twin challenges of low pH and protease activity in the gastric environment. Resistance to gastric acid is one of most important selection criteria for any potential probiotic. Strong acid tolerance is not universal amongst LAB, even at the species level, and must be tested on a strain-by-strain basis.Citation39 Survival at pH 3 is generally considered to be the absolute minimum necessary for a probiotic strain to remain viable in vivo, although pH 2 is a much more commonly encountered level of acidity.Citation40 In a fasting stomach, pH can drop as far as 1.2–1.5Citation41 The results of our viability assays are consistent with literature values for well-researched strains, including the high tolerance of B. lactis HN019 to pH 2, but relatively poor tolerance of L. acidophilus 4356 to the same level of acidity.Citation42,Citation43
LAB isolated from both probiotic foods and human samples have shown large decreases in viability when exposed to pH values between 1.5 and 2, though some strains with higher resistance to acid have been documented.Citation44–Citation46 For example, Aiba et al identify an L. johnsonii and an L. gasseri strain that maintain over 10% viability in growth media acidified to pH. 1Citation47 However, it is still unclear how well resistance to simulated gastric conditions translates to tolerance of the gastric environment in vivo. The SGA used in our study simulates a scenario in which probiotic strains are introduced, with minimal protective adjuvants, into a gastric environment consisting of low pH, few nutrients, and proteolytic activity from added pepsin enzyme, as would be expected in an empty stomach. Based on the detectable viability maintained by LBJ 456 at all tested pH levels in SGA, we predict robust survival of this probiotic even under the most restrictive gastric conditions, although the mechanism of this strong resistance has yet to be determined. As the presence of milk and metabolizable sugar have both been found to afford some protection from low pH over buffered saline of the same acidity, the growth of LBJ 456 should be further enhanced when delivered in most conventional formats, such as dairy products.Citation48,Citation49
Bile acids in the small intestine inhibit growth via their detergent effects on bacterial cell membranes. Some probiotic species can hydrolyze these bile acids directly.Citation50,Citation51 They are very rarely lethal for LAB at the lower end of physiological concentrations, which can vary from up to 10mM in the upper ileum to 2mM in the lower ileum after a meal.Citation52,Citation53 We observed moderately retarded growth rates in all LAB strains exposed to physiologically relevant bile acid concentrations. Literature values generally agree with our observations, including our assessment of L. acidophilus 4356 being particularly sensitive, and B. lactis being relatively bile acid tolerant.Citation42,Citation43 Although LBJ 456 is only moderately bile tolerant relative to other strains, it remains viable and capable of slower growth at bile acid concentrations between 0.1 and 0.3% (~ 2–6 mM).
Even in light of the strain’s in vitro survival through simulated GI tract barriers, we find that LBJ 456 demonstrates exceptional persistence in the human gut. A weeklong course of a small, daily amount of live culture led to elevated fecal abundance through at least a month, as determined by both culture-based and qPCR analyses. It is important to note that tests included here, especially detection of live CFU, revealed heavy variation in lactic acid bacterial load between individuals, and even in the same individual over time. As previously mentioned, background levels varied from under 4000 CFU/gram feces to over 40 million. This heavy interpersonal variability is consistent with that found in other studies. Tannock et al. found CFU counts ranging from 100 to 4 billion per gram, and Goossens et al found between 2500 and 80 million CFU/gram in human fecal samples.Citation54,Citation55 Dietary intake of lactic acid bacteria likely explains a great deal of this variation, as countless strains and varieties of Lactobacillus are naturally found in common fruits and vegetables.Citation56 Despite this near constant incidental intake, probiotic genera like Lactobacillus and Bifidobacterium, are generally transient and do not colonize the human gut for long periods.Citation57 Most clinically tested strains, including L. plantarum 299V, L. rhamnosus GG, and L. casei Shirota, are not recoverable in host feces after more than a week or so post ingestion.Citation55,Citation58,Citation59 Of Lactobacillus species investigated so far, L. reuteri ATCC 55730 seems to have one of the longest reported persistence records, with live bacteria detectable by biopsy (though not in the feces) up to four weeks after the cessation of a monthlong course of ingestion.Citation60 This persistence has not been fully reproduced consistently, though, with shorter courses of inoculation or by standard fecal assay, and other authors have concluded that it does not effectively establish long term colonization.Citation61 Despite the fact that the elevated gut counts observed beyond the week of inoculation were not significant (likely due to the small pilot nature of the trial) LBJ 456’s long duration of detectability warrants further investigation, particularly of the factors that promote survival and host attachment. Future clinical studies in humans (particularly those that would measure a specific effect on other bacteria or levels of inflammation) should include the use of a placebo control group and multiple benchmark samples to control for natural variation within an individual.
Host mucins provide a common binding site for both beneficial and pathogenic bacterial strains.Citation62 We observed a broad array of MBPs encoded by L. johnsonii strains, some of which appeared to be unique to LBJ 456. Different combinations of these MBPs may confer unique binding properties and contribute to host specificities previously observed among L. johnsonii subtypes.Citation38 The secreted mucin Muc2 constitutes the main mucus glycoprotein in the mouse small intestine, from which LBJ 456 was derived.Citation63 Its ortholog, MUC2, is the main secreted mucin in the human gut as well, comprising most of the upper, gel-like layer of mucus.Citation64 The mucin profile of the enterocyte-like cell line Caco-2 is almost purely limited to the expression of membrane-bound mucins like MUC1. The goblet cell-like line LS 174T, however, has much higher expression of secretory mucins, including MUC.2Citation65 As the secreted mucus layer is the major site of microbe-host interaction, increased adhesion of LBJ 456 to LS 174T could explain this strain’s persistence in vivo, especially if MUC2 or another secreted mucin is assumed to be a putative binding target.Citation66
Although adhesion to mucins and other mucosal proteins is difficult to study in the human intestine proper, cell monolayer assays correlate reasonably well with in vivo persistence data and provide a method for the investigation of both relative adhesion and adhesion inhibition between microbes.Citation67,Citation68 The adhesion of LAB to monolayer models such as Caco-2 and LS 174T is highly variable between strains. Many probiotic Lactobacillus species barely adhere to enterocyte-like Caco-2 cells at all, while others adhere reasonably well.Citation69 Interestingly, while none of the strains tested here were especially adherent to Caco-2 monolayers, LBJ 456’s adhesion was particularly low, roughly 1/20th that of its type strain LBJ VPI 7960. This intraspecies difference may reflect adaptations to the different natural hosts or environments from which the substrains were derived. Todoriki et al report that the likewise murine-derived L. johnsonii strain JCM 8792 exhibits very low adhesion to Caco-2 cells relative to such strains as the chicken derived L. reuteri JCM 1081,Citation70 strengthening our suggestion that Lactobacillus of murine origin may be less specialized to adhere to secreted mucin-poor culture.
The pathogenic activity of many diarrheagenic bacteria, such as ETEC and Salmonella, is dependent upon adhesion to the gut mucosa and can therefore also be modeled with monolayers in vitro.Citation71,Citation72 Salmonella enterica typhimurium and ETEC strain H10407 are both prototypical diarrhea inducers that require close adhesion to the host cell in order to cause disease.Citation73–Citation75 E. faecalis, although normally considered a commensal, can also become an opportunistic pathogen and diarrhea inducer in immunocompromised individuals, with multidrug resistant strains causing particularly stubborn nosocomial infections.Citation76 A number of studies have demonstrated that adherent Lactobacillus species can inhibit subsequent pathogen adhesion. Todoriki showed that L. crispatus JCM 8779 itself reduced E. faecalis adhesion by 99% in a Caco-2 model, as well as Salmonella and ETEC adhesion by 28 and 47% respectively. Filtered supernatant from L. crispatus inhibited E. faecalis growth, but not Salmonella or ETEC.Citation70 Maragkoudakis et al showed that Caco-2 adherent strains could reduce E. coli and Salmonella adhesion by 10–50%, although they noted no inhibition from supernatant-localized factors.Citation51 L. johnsonii 456’s capacity to inhibit pathogen adhesion appears to depend on its own ability to adhere to the monolayer in question. Pre-treatment with one hour of LBJ 456 led to no significant change in pathogen adhesion on Caco-2 cells. On LS 174T monolayers, both ETEC and Salmonella adhesion were cut by about 33 and 40%, respectively. Unexpectedly, E. faecalis adhesion increased slightly with LBJ 456 incubation, suggesting that LBJ 456 would not necessarily displace other commensals from its milieu, but may potentially promote the attachment of certain gut flora, perhaps through direct binding to the cell surface or via substrates secreted, induced, or modified by it.
In co-culture conditions, LBJ 456 drastically reduced the growth of all three pathogenic strains, particularly S. enterica. This inhibition appears to occur through different mechanisms. FS from LBJ 456 grown to the beginning of stationary phase was capable of inhibiting S. enterica and E. faecalis growth but not ETEC. As L. johnsonii VP 7960 FS produced a very similar pattern of inhibition, a common L. johnsonii factor may be responsible. Interestingly, L. plantarum 299V FS inhibited all three pathogenic strains. L. plantarum strains are known to produce a class of two-peptide, class IIB bacteriocins called plantaricins.Citation77,Citation78 The 299V genome is specifically known to encode a number of predicted bacteriocins including plantaricin components (BT929_RS02485, BT929_RS02490, BT929_RS02515, BT929_RS02535, BT929_RS02545, and BT929_RS02550). Most plantaricins are effective against other Gram-positive bacteria, like Listeria and Enteroccoccus, so this particular class of peptide antimicrobial likely does not explain the strong inhibition of ETEC. However, FS inhibition of E. faecalis growth could be due to a bacteriocin. L. johnsonii strains have been shown to produce a bacteriocin, lactacin F, that inhibits the growth of both E. faecalis and other LAB through membrane-disrupting pore formation.Citation79 Based on our genomic analysis, it is likely that LBJ 456 does in fact produce functional bacteriocins, some of which may be responsible for this cell-independent inhibitory effect.
At least two proposed mechanisms exist regarding probiotic-mucin interaction and pathogen binding inhibition. Through competitive adhesion, Lactobacillus or other beneficial species could compete directly for mucin binding sites with pathogens, preventing them from having a chance to interact with host cells.Citation80 Alternatively, it has been suggested that Lactobacillus binding to host mucins can lead to the secretion of even more mucins, essentially flushing pathogens from the lumen.Citation81,Citation82 This increased mucin production could potentially counteract the mucin degradation induced by pathogens like ETEC.Citation83 Regardless of the precise mechanism, secreted mucins are critical for gut homeostasis. Lower levels of Muc2 expression are associated with increased inflammation, colitis, and even rates of colon cancer in mice.Citation84,Citation85 These conditions have all been associated with gut pathogen infection.Citation86,Citation87
Our laboratory previously demonstrated LBJ 456’s anti-inflammatory properties in a mouse model.Citation12 However, the specific mechanisms of this effect remain unclear. Lactobacillus species have been suggested to induce regulatory T cells and modulate host inflammatory factors.Citation88,Citation89 It is possible that the adhesion inhibition shown by LBJ 456 in our study may act in concert with direct host immunomodulation to reduce pathogen-associated inflammation, as has been observed with other probiotics.Citation90
Probiotic bacteria represent a potential method for both prevention and treatment of diarrheal diseases.Citation91,Citation92 Diarrheal infections are a major complication in hospital patients. Gao et al showed that prophylactic administration of a blend of two Lactobacillus species cut antibiotic and C. dificile associated diarrhea by over half in a clinical environment.Citation93 Intervention is even more important in children. Diarrheal diseases, including those induced by ETEC and salmonella infection, are responsible for an eighth of childhood deaths below the age of 5 worldwide.Citation20 Probiotics are effective here, too; vigilant and repeated L. rhamnosus GG and B. lactis BB-12 supplementation have been demonstrated to reduce the duration of acute diarrhea in a number of studies.Citation94–Citation97 Unfortunately, the majority of this burden occurs in developing nations with lower rates of regular access to healthcare. Large outbreaks of diarrheal disease are also common after disasters, like floods, that disrupt stable access to clean water and services.Citation98 An inexpensive probiotic supplement with a relatively wide “useful prophylactic duration” could be of great use in situations where repeated supplementation is difficult.
L. johnsonii 456 represents a promising probiotic lactobacillus strain
Unique attributes include exceptional acid resistance and well-documented, inoculation-inducible anti-inflammatory effect in mice. The strain is capable of inhibiting the growth and adhesion of multiple types of pathogens in vitro. Importantly, the human pilot study described here suggests that L. johnsonii 456 may be persistent in the human gut for longer than many other documented strains of probiotic bacteria. Although larger scale clinical studies are needed, the combination of attributes demonstrated here suggest future use as part of an antidiarrheal regimen, or even in the treatment of gut inflammation.
Materials and methods
Bacterial strains and growth culture conditions used
Bacterial strains used are detailed in . Lactobacillus johnsonii 456 was isolated from wildtype mice with restricted gut microflora, housed under specific pathogen free (SPF) conditions at UCLA, by Yamamoto et al.Citation12 The samples used in this study were derived from frozen stock stored by the Schiestl laboratory. Lactobacillus plantarum 299V (NCBI Refseq genome accession NZ LEAV00000000.1) was isolated from Goodbelly Probiotic Juice Drink (NextFoods; Boulder, CO). Bifidobacterium lactis HN019 was isolated from Tropicana Essentials Probiotic Juice (Tropicana Products; Chicago, IL). Lactobacillus johnsonii VPI 7960, Lactobacillus casei 03, Lactobacillus acidophilus ATCC 4356, Streptococcus salivarius subsp. thermophilus NCDO 573, Escherichia coli H10407, Enterococcus faecalis NCTC775, and Salmonella enterica subsp. enterica serovar typhimurium were obtained from the American Type Culture Collection (ATCC; Manassas, VA).
All Lactobacillus species were cultured in MRS (De Man, Rogosa, and Sharpe) broth (Sigma-Aldrich; St. Louis, MO) for 18–20 hours at 37° C under microaerophilic conditions with sealed test tubes. Colony-forming units (CFU) of Lactobacillus were enumerated after 48 hours of growth at 37° C on MRS agar (Sigma) incubated in chambers with anaerobic sachets (Sigma). L. johnsonii 456 colonies are distinguishable as smooth bordered, white colonies (Supp. Fig S5) B. lactis was cultured and enumerated similarly, except that MRS broth and agar were supplemented with 0.5g/L Cysteine-HCl. S. salivarius was cultured in tryptic soy (TS) broth (Sigma) for 18–20 hours at 37° C with no special anaerobic considerations (aerobically), and enumerated on TS agar plates after 48 hours aerobically at 37° C. ETEC and S. enterica were cultured in TS broth for 18–20 hours at 37° C aerobically, and enumerated on TS agar plates after 24 hours at 37° C aerobically. E. faecalis was cultured in Brain-Heart (BH) broth (Sigma) for 18–20 hours at 37° C aerobically, and enumerated on BH agar plates after 24 hours at 37° C aerobically.
Acid resistance during simulated gastric transit
SGA was prepared by dissolving 3.3ppm pepsin (Sigma) and 0.2% NaCl w/v in 0.1% peptone water (Becton Dickinson; Franklin Lakes, NJ). The pH of this solution was then brought to 1.2 with the addition of 11.65M hydrochloric acid to recapitulate concentrated gastric fluid in an otherwise empty human stomach. This solution was diluted using additional 0.1% peptone water to pHs of 1.6, 2.0, and 3.0. The probiotic strains (L. johnsonii 456, L. johnsonii VPI 7960, L. casei, L. acidophilus, L. plantarum, B. lactis, and S. salivarius) were grown to a concentration of roughly 1 × 108 CFU/mL by the methods described above. 106 mid-log phase cells were inoculated into 10mL SGA or 0.1% peptone water control (pH 6) and incubated for 2 hours at 37° C to simulate gastric transit. After incubation, samples were diluted in 0.1% peptone water and plated on agar for enumeration.
Bile acid tolerance
Bile acid tolerance was evaluated using a modified version of the method of Gilliland and Walker.Citation99 Each probiotic strain was evaluated based on addition of bile salts to their standard growth conditions. Freshly inoculated culture media was vortexed heavily and then split evenly into either a fresh vial or one containing ox gall extract (Sigma) to 0.1, 0.2, or 0.3% of final solution by weight (2.12, 4.24, and 6.36 mM based on rough Sigma ox gall extract bile acid salt composition: 10% glycocholic acid, 15% glycodeoxycholic acid, 30% taurocholic acid, 55% cholic acid) Culture media was then incubated anaerobically for 18–20 hours at 37° C, and samples were plated and enumerated.
Human gut monolayer culture conditions
Caco-2 and LS 174T cell lines were initially obtained from ATCC. Cells were cultured in Eagle’s Minimal Essential Media with 4mM glutamine (Caisson Labs; Smithfield, UT) supplemented with the following: 20% fetal Bovine Serum by volume (Corning Cellgro; Manassas, VA), non-essential amino acids (from 100x, Corning), sodium pyruvate (from 100x, Lonza; Walkersville, MD), and 100u/mL PEN-STREP (penicillin-streptomycin mixture, from 100x, Corning). Cells were grown at up to 50% confluence, trypsinized, and subcultured at a 1:4 ratio roughly every 3 days. Conditions were maintained at 37° C in a 5% CO2 atmosphere.
Caco-2 monolayers were prepared with small modifications to the method described by Natoli et al.Citation100 Approximately 3 × 105 Caco-2 cells/cm2 were seeded into a 12 or 24 well plate (BD). Cells were maintained in these plates while growing to confluence under the same controlled conditions as previous. The nascent monolayers were rinsed with warmed PBS and given fresh EMEM media containing all previous additives except antibiotics three times a week. After 15 days of culture, the Caco-2 monolayer was considered to be “mature” for adhesion experimental purposes.
LS174 T monolayers were prepared similarly to Caco-2 cells, with a few modifications to the conditions used by by Jung et al.Citation34 After approaching 50% confluence in growth culture, cells were trypsinized and resuspended in fresh EMEM without antibiotics, then seeded into a 12 or 24 well plate at approximately 3 × 105 cells/cm2. After 3 days, the LS174T monolayer was inspected and rinsed in warmed PBS. Small areas of the growing monolayer that became detached from the substrate were carefully removed during this rinse step, then the media was replaced. Fresh media without antibiotics was then added after PBS rinsing 3 times a week. After roughly 10 days, the LS 174T monolayer was fully confluent and considered “mature” for adhesion experimental purposes.
Monolayer adhesion assay
Bacterial adhesion to monolayers was assayed with small modifications to previously described methods.Citation69,Citation70 Overnight cultures of probiotic associated test strains (L. johnsonii 456, L. johnsonii VPI 7960, L. casei, L. acidophilus, L. plantarum, B. lactis, and S. salivarius) were pelleted via centrifuge (10 minutes, 3K RPM) (Allegra 6R, Beckmann-Coulter), rinsed, and resuspended in antibiotic-free EMEM and an equal amount of their bacterial culture media to a concentration of 2–5 × 108 cells/mL. Mature Caco-2 or LS 174T monolayers were rinsed twice with warmed PBS. A 1 mL volume of resuspended bacterial sample was then applied to each well of the tissue culture plate. Plates were incubated at 37° C in a 5% CO2 atmosphere for 2 hours with gentle intermittent rocking. After incubation, supernatant was removed and monolayers were rinsed 3 times with warmed PBS. The monolayers were then covered in 1 mL fresh PBS per well and vigorously agitasted with micropipette until disrupted and fully resuspended. 10-fold serial dilutions were then plated on strain specific agar media to enumerate adherent cells.
Pathogen adhesion inhibition assay
Pathogen adhesion was determined by the method of Todoriki et al.Citation70 Overnight cultures of 3 potentially pathogenic strains (ETEC, E. faecalis, and S. enterica) and Lactobacillus johnsonii 456 were pelleted via centrifuge, rinsed, and resuspended in antibiotic-free EMEM to a concentration of 2 × 108 cells/mL, as determined by microscope count. Samples of this media were taken aside, diluted, and plated on specific agar media to control precisely for viable CFU plated. Mature Caco-2 or LS 174T monolayers were rinsed twice with warmed PBS. 0.5 mL of L. johnsonii in EMEM was added to each well. Monolayers were then incubated at 37° C in a 5% CO2 atmosphere for an hour with gentle intermittent rocking. After initial incubation, wells were rinsed twice with warmed PBS to remove nonadherent LBJ 456. 0.5 mL of pathogen suspensions were then applied to experimental and control (no LBJ pretreatment) wells. Plates were returned to the incubator for an additional hour. After incubation, supernatant was removed and monolayers were rinsed 3 more times. The monolayers were then covered in 1 mL fresh PBS per well and vigorously agitated with micropipette until disrupted and fully resuspended. 10-fold serial dilutions were then plated on specific agar media to enumerate colonies from adherent bacteria of the test strain.
Co-culture inhibition assay
Cultures of the 3 potentially pathogenic strains (ETEC, E. faecalis, and S. enterica) and LBJ 456 were grown overnight as described earlier to concentrations of 2–10 × 108 cells/mL, by the method of Hsieh et al.Citation101 Cells were rinsed and pelleted via centrifuge, then 1 × 108 cells of each pathogenic strain were co-inoculated with 1 × 108 cells of LBJ 456 into TS (ETEC and S. enterica) or BH (E. faecalis). 1 × 108 cells of each test strain were also resuspended alone in their respective media as a control. Co-cultures were incubated for 18–20 hours at 37° C, then plated for enumeration. After 24 hours at 37° C, colonies of the pathogenic strain were enumerated, and easy to distinguish morphologically from the inhibited growth of Lactobacillus colonies under aerobic conditions. LBJ 456 controls were enumerated after 48 hours at 37° C on MRS agar.
Supernatant inhibition assay
A slight modification on the method used by Sgouras et al was used to determine whether an acellular factor could inhibit pathogen growth.Citation102 Overnight cultures of Lactobacillus strains L. johnsonii 456, L. johnsonii VPI 7960, L. casei, and L. plantarum were cultured in MRS broth for 18–20 hours at 37° C under microaerophilic conditions. The test strains B. lactis, ETEC, E. faecalis, and S. enterica were also inoculated into their respective growth media, and cultured 18–20 hours at 37° C. Spent supernatant from the Lactobacillus cultures was vacuum filtered via a 0.45 micron filter (Nalge Nunc; Rochester NY) to exclude cellular components. B. lactis, ETEC, E. faecalis, and S. enterica overnight growth cultures were rinsed and pelleted via centrifuge, then 1 × 108 CFU of each test strain were inoculated into 5 mL of their fresh respective growth media, and either 5 mL of filter sterilized supernatant (FS) from one of the Lactobacillus cultures or fresh MRS adjusted to pH 4 with lactic acid as a control. Cultures were grown for 18–20 hours at 37° C, then serially diluted and plated on each strain’s respective growth agar for enumeration. All plates were incubated at 37° C under their respective growth conditions described above. B. lactis was grown for 48 hours under anaerobic conditions while ETEC, S. enterica, and E. faecalis were incubated for 24 hours aerobically.
Human gut survival trial and enumeration of viable lactic acid bacteria
Yogurt containing a starter culture of LBJ 456 was generated using commercially available whole fat milk. The yogurt was kept fermenting at room temperature until fully solidified, then refrigerated at 4° C. 11 mixed gender individuals in good health (no inflammatory gut conditions or known disease states) received a 7 day course of this yogurt, and consumed 100mL, or roughly 1 × 1010 CFU, every morning over the course of the trial week. A baseline fecal sample was taken prior to first yogurt consumption, and then at 7, 30, and 60 days after study initiation. Fecal samples were stored at −20° C . 11 individuals supplied fecal samples for each time point, and the others were excluded. To determine viable LAB load at each timepoint, 0.1g of fecal matter was thawed and serially diluted in PBS, then plated on MRS agar to select for LAB. Plates were enumerated after 48 hours at 37° C. Volunteers were asked to refrain from consuming other probiotics for the duration of the study, but otherwise maintain a normal diet. Researchers were blinded to the identity of volunteers. Study design was reviewed and volunteer consent was obtained by MicroBio Pharma, Inc., and determined to be ethically and clinically sound based on previous demonstrations of Lactobacillus johnsonii safety in the literature.
LBJ 456 DNA detection in human fecal samples by rt-qpcr
Fecal bacterial DNA was purified using a Zymo QuickDNA Fecal/Soil Microbe Miniprep Kit (Zymo; Irvine, CA) according to manufacturer instructions. DNA content was verified using a Nanodrop (Thermo Fischer; Canoga Park, CA). Real-Time quantitative Polymerase Chain Reaction (RT-qPCR) was performed on the CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using PerfeCTa SYBR Green SuperMix Low ROX reagent (Quantabio; Beverly, MA). DNA was diluted to a working solution of 10 ng/uL and 1 μl was used per replicate of each sample. Samples were analyzed in technical triplicates. Presence of LBJ in each sample was normalized to 16S levels. Primer sequences used are listed in Table S1. Optimal qPCR temperature of 55° C was determined by temperature gradient. Specificity of primers was determined by melt curve analysis and comparison of strains (Supp. Sig. S4).
Genome sequencing, assembly and annotation
Whole genome sequencing of Lactobacillus johnsonii 456 was performed by Genewiz (South Plainfield, NJ). In brief, 6 × 109 CFU of LBJ 456 bacteria were shipped to Genewiz. Cells were lysed and total DNA was purified. Sequencing was performed using PacBio Sequel to an average coverage of 100X per sample. Read assembly was performed using CanuCitation103 Coding and tRNA genes were annotated using Prodigal 2.6Citation104 and Aragorn 1.2Citation105, respectively, via the PROKKA pipeline.Citation106 Genome uploaded to NCBI database under accession code QGQW00000000.
Core genome comparison
We compared the genome of Lactobacillus johnsonii 456 (LBJ 456) with 23 other L. johnsonii genomes available from NCBI on September 7, 2017. We first calculated the core genomic regions shared by all 24 L. johnsonii strains, as described by Tomida et al.Citation107 Briefly, Nucmer was used to identify homologous regions between the genome of NCC 533 and each of the other 23 genomes. The set of core genomic regions was determined to be the regions homologous to NCC 533 that were present in the genomes of the 23 other strains. Single nucleotide polymorphisms (SNPs) were identified within core regions using Nucmer and were used to construct a phylogenetic tree in MEGA 7 using the Neighbor-Joining method on p-distances.Citation108 Bootstrapping was performed using 500 replicates.
Non-core genome comparison
Non-core genomic regions among the L. johnsonii genomes were also identified as described by Tomida et al.Citation107 Briefly, a pan-genome across all 24 L. johnsonii strains was constructed by first using Nucmer to compare the NCC 533 genome with one of the 23 other genomes. Regions in this genome without homology to NCC 533 were concatenated to the NCC 533 genome sequence. This concatenated sequence was then iteratively compared using the same method to each of the remaining genomes to construct the pan-genome. Finally, the pan-genome was compared to each of the 24 genomes individually to identify non-core regions ≥ 500 bp that were absent in at least one of the genomes.
Genome analysis
To determine whether the 24 L. johnsonii isolates, and LBJ 456 in particular, may possess distinct host-binding properties, we identified putative MBPs within the isolate genomes. A set of reference MBPs was compiled from amino acid sequences matching the search term “((((mucus[Title] OR mucin[Title])) AND binding[Title])) OR ((mucus-binding[Title] OR mucin-binding[Title]))”, downloaded from the NCBI protein database as of September 9, 2017. A non-redundant set of reference MBPs was obtained by clustering sequences with ≥ 97% identity using CD-HIT,Citation109 and all proteins from all 24 L. johnsonii strains were aligned to this reference set using BLASTP. Putative L. johnsonii MBPs were identified as sequences showing at least 60% identity to at least one reference MBP. L. johnsonii MBPs were further clustered by 70% identity using CD-HIT. Bacteriocin-encoding loci in the LBJ 456 genome were detected usingBAGEL.4Citation110
Statistical analyses
Statistical analyses were carried out using Graphpad Prism 5 software and Microsoft Excel. T tests were used to assess significance of differences in bacterial survival and adhesion assays. Changes within fecal CFU were carried out using the non-parametric Friedman test and Wilcoxon signed rank post-hoc analysis; P values were adjusted with a Bonferroni correction.
Significance
Bacterially derived diarrheal disease is a major contributor to worldwide deaths for children under the age of five. On the other end of the spectrum, chronic inflammation contributes to cancer, which claims more lives in developed countries than any other illness beside cardiovascular disease. Probiotic bacteria offer a method of intervention that could reduce fatalities from both of these seemingly disparate diseases. It is imperative that new strains of probiotic bacteria be characterized while we simultaneously develop our understanding of their mechanisms of action. Lactobacillus johnsonii 456 is associated with reduced inflammation and genotoxicity in vertebrate models, pathogen inhibition in vitro, and long persistence in human trials, making it a powerful option for populations without consistent access to resources.
Author contributions
R.H.S. designed research and prepared manuscript, M.J.D. and J.L. designed research, performed research, analyzed data, and prepared manuscript; J.C. and N.I.R.M. performed research, analyzed data, and prepared manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download MS Word (16.1 KB)Supplemental Material
Download PDF (4 MB)Acknowledgments
We thank Dr. Patrick Allard (University of California, Los Angeles) for generous facilities and supply sharing, MicroBio Pharma, Inc. (Los Angeles) for organizing the volunteer trial and providing us with fecal samples, and the lab of Dr. Huiying Li (University of California, Los Angeles) for equipment sharing. Jocelyn Castellanos was supported by an NIEHS Student to Scientist grant (5R25ES025505-02).
Conflict of Interest Statement
R.H.S. has a financial interest in MicroBio Pharma, Inc, which might benefit from the commercialization of the results of this research.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- Ciorba MA. A gastroenterologist’s guide to probiotics. Clin Gastroenterol Hepatol. 2012;10(9):960–968.
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13(4):260–270. doi: 10.1038/nrg3182.
- Sheehan D, Moran C, Shanahan F. The microbiota in inflammatory bowel disease. J Gastroenterol. 2015;50(5):495–507. doi: 10.1007/s00535-015-1064-1.
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490.
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer R-J. The role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27(2):104–119. doi: 10.1111/j.1365-2036.2007.03562.x.
- LeBlanc JG, Chain F, Martín R, Bermúdez-Humarán LG, Courau S, Langella P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Fact. 2017;16(1):79. doi: 10.1186/s12934-017-0691-z.
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449(7164):804. doi: 10.1038/nature06244.
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540.
- Jones ML, Martoni CJ, Ganopolsky JG, Labbé A, Prakash S. The human microbiome and bile acid metabolism: dysbiosis, dysmetabolism, disease and intervention. Expert Opin Biol Ther. 2014;14(4):467–482. doi: 10.1517/14712598.2014.880420.
- Hartstra AV, Bouter KEC, Bäckhed F, Nieuwdorp M. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care. 2015;38(1):159–165. doi: 10.2337/dc14-0769.
- Foster JA, Neufeld K-AM. Gut–brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36(5):305–312. doi: 10.1016/j.tins.2013.01.005.
- Yamamoto ML, Maier I, Dang AT, Berry D, Liu J, Ruegger PM, Yang J-I, Soto PA, Presley LL, Reliene R, et al. Intestinal bacteria modify lymphoma incidence and latency by affecting systemic inflammatory state, oxidative stress, and leukocyte genotoxicity. Cancer Res. 2013;73(14):4222–4232. doi: 10.1158/0008-5472.CAN-13-0022.
- Zocco M, DAL VERME LZ, Cremonini F, Piscaglia AC, Nista EC, Candelli M, Novi M, Rigante D, Cazzato IA, Ojetti V, et al. Efficacy of Lactobacillus GG in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther. 2006;23(11):1567–1574. doi: 10.1111/apt.2006.23.issue-11.
- Prantera C, Scribano ML, Falasco G, Andreoli A, Luzi C. Ineffectiveness of probiotics in preventing recurrence after curative resection for Crohn’s disease: a randomised controlled trial with Lactobacillus GG. Gut. 2002;51(3):405–409.
- Ghouri YA, David MR, Erik FR, Joseph TK, Katherine AJ, Andrew WD. Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clin Exp Gastroenterol. 2014;7:473.
- Di Cerbo A, Palmieri B. The market of probiotics. Pak J Pharm Sci. 2015;28(6):2199–2206.
- Doron S, Hibberd PL, Goldin B, Thorpe C, McDermott L, Snydman DR. Effect of Lactobacillus rhamnosus GG administration on vancomycin-resistant enterococcus colonization in adults with comorbidities. Antimicrob Agents Chemother. 2015;59(8):4593–4599. doi: 10.1128/AAC.00300-15.
- Van Gossum A, Dewit O, Louis E, de Hertogh G, Baert F, Fontaine F, DeVos M, Enslen M, Paintin M, Franchimont D. Multicenter randomized‐controlled clinical trial of probiotics (Lactobacillus johnsonii, LA1) on early endoscopic recurrence of Crohn’s disease after ileo‐caecal resection. Inflamm Bowel Dis. 2007;13(2):135–142. doi: 10.1002/ibd.20063.
- Loftus EV Jr. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004;126(6):1504–1517.
- Kotloff KL. The burden and etiology of diarrheal illness in developing countries. Pediatr Clin. 2017;64(4):799–814. doi: 10.1016/j.pcl.2017.03.006.
- Servin AL, Coconnier M-H. Adhesion of probiotic strains to the intestinal mucosa and interaction with pathogens. Best Pract Res Clin Gastroenterol. 2003;17(5):741–754. doi: 10.1016/S1521-6918(03)00052-0.
- Barefoot SF, Klaenhammer TR. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl Environ Microbiol. 1983;45(6):1808–1815.
- Ocaña VS, Nader-Macías ME. Production of antimicrobial substances by lactic acid bacteria II: screening bacteriocin-producing strains with probiotic purposes and characterization of a Lactobacillus bacteriocin. In: Public Health Microbiology: Methods and Protocols. New York: Humana Press; 2004. p. 347–353.
- Midolo P, Lambert JR, Hull R, Luo F, Grayson ML. In vitro inhibition of helicobacter pylori NCTC 11637 by organic acids and lactic acid bacteria. J Appl Bacteriol. 1995;79(4):475–479.
- Isobe H, Nishiyama A, Takano T, Higuchi W, Nakagawa S, Taneike I, Fukushima Y, Yamamoto T. Reduction of overall helicobacter pylori colonization levels in the stomach of Mongolian gerbil by Lactobacillus johnsonii La1 (LC1) and its in vitro activities against H. pylori motility and adherence. Biosci Biotechnol Biochem. 2012;76(4):850–852. doi: 10.1271/bbb.110921.
- Humen MA, De Antoni GL, Benyacoub J, Costas ME, Cardozo MI, Kozubsky L, Saudan K-Y, Boenzli-Bruand A, Blum S, Schiffrin EJ, et al. Lactobacillus johnsonii La1 antagonizes Giardia intestinalis in vivo. Infect Immun. 2005;73(2):1265–1269. doi: 10.1128/IAI.73.2.1265-1269.2005.
- La Ragione R, Narbad A, Gasson MJ, Woodward MJ. In vivo characterization of Lactobacillus johnsonii FI9785 for use as a defined competitive exclusion agent against bacterial pathogens in poultry. Lett Appl Microbiol. 2004;38(3):197–205.
- Reid G, Jass J, Sebulsky MT, McCormick JK. Potential uses of probiotics in clinical practice. Clin Microbiol Rev. 2003;16(4):658–672.
- Mileti E, Matteoli G, Iliev ID, Rescigno M, Fritz JH. Comparison of the immunomodulatory properties of three probiotic strains of Lactobacilli using complex culture systems: prediction for in vivo efficacy. PLoS One. 2009;4(9):e7056. doi: 10.1371/journal.pone.0007056.
- Klaenhammer TR. Probiotic bacteria: today and tomorrow. J Nutr. 2000;130(2):415S–416S. doi: 10.1093/jn/130.2.415S.
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. Expert consensus document: the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol. 2014;11(8):506–514. doi: 10.1038/nrgastro.2014.66.
- Brassart D, Brassart D, Neeser JR, Michetti P, Servin AL. The selection of dairy bacterial strains with probiotic properties based on their adhesion to human intestinal epithelial cells. In: Proceedings Lactic; Adria Normandie, Caen, France; 1994. p. 94.
- Salminen S, Laine M, Vonwright A, Vuopio-Varkila J, Korhonen T, Mattila-Sandholm T. Development of selection criteria for probiotic strains to assess their potential in functional foods: a Nordic and European approach. Biosci Microflora. 1996;15(2):61–67. doi: 10.12938/bifidus1996.15.61.
- Jung T-H, Park JH, Jeon W-M, Han K-S. Butyrate modulates bacterial adherence on LS174T human colorectal cells by stimulating mucin secretion and MAPK signaling pathway. Nutr Res Pract. 2015;9(4):343–349. doi: 10.4162/nrp.2015.9.4.343.
- Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14(8):e1002533. doi: 10.1371/journal.pbio.1002533.
- Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One. 2013;8(2):e57923. doi: 10.1371/journal.pone.0057923.
- Wu X, Zhao C, Guo Z, Hao Y, Li J, Shi H, Sun Y. Genome sequence of Lactobacillus johnsonii strain W1, isolated from mice. Genome Announc. 2016;4(3):e00561–16. doi: 10.1128/genomeA.00561-16.
- Buhnik-Rosenblau K, Matsko-Efimov V, Jung M, Shin H, Danin-Poleg Y, Kashi Y. Indication for co-evolution of Lactobacillus johnsonii with its hosts. BMC Microbiol. 2012;12(1):149. doi: 10.1186/1471-2180-12-149.
- Kailasapathy K, Chin J. Survival and therapeutic potential of probiotic organisms with reference to Lactobacillus acidophilus and bifidobacterium spp. Immunol Cell Biol. 2000;78(1):80–88. doi: 10.1046/j.1440-1711.2000.00886.x.
- Charteris W, Kelly PM, Morelli L, Collins JK. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J Appl Microbiol. 1998;84(5):759–768.
- Mudie DM, Murray K, Hoad CL, Pritchard SE, Garnett MC, Amidon GL, Gowland PA, Spiller RC, Amidon GE, Marciani L. Quantification of gastrointestinal liquid volumes and distribution following a 240 mL dose of water in the fasted state. Mol Pharm. 2014;11(9):3039–3047. doi: 10.1021/mp500210c.
- Prasad J, Gill H, Smart J, Gopal PK. Selection and characterisation of Lactobacillus and bifidobacterium strains for use as probiotics. Int Dairy J. 1998;8(12):993–1002. doi: 10.1016/S0958-6946(99)00024-2.
- Liong M, Shah N. Acid and bile tolerance and cholesterol removal ability of lactobacilli strains. J Dairy Sci. 2005;88(1):55–66. doi: 10.3168/jds.S0022-0302(05)72662-X.
- Hassanzadazar H, Ehsani A, Mardani K, Hesari J. Investigation of antibacterial, acid and bile tolerance properties of lactobacilli isolated from Koozeh cheese. In: Veterinary research forum. Urmia, Iran: Faculty of Veterinary Medicine, Urmia University; 2012. p. 181.
- Sahadeva RPK, Leong SF, Chua KH, Tan CH, Chan HY, Tong EV, Wong SYW, Chan HK. Survival of commercial probiotic strains to pH and bile. Int Food Res J. 2011;18(4);1515–1522.
- Pan X, Chen F, Wu T, Tang H, Zhao Z. The acid, bile tolerance and antimicrobial property of Lactobacillus acidophilus NIT. Food Control. 2009;20(6):598–602. doi: 10.1016/j.foodcont.2008.08.019.
- Aiba Y, Nakano Y, Koga Y, Takahashi K, Komatsu Y. A highly acid‐resistant novel strain of Lactobacillus johnsonii No. 1088 has antibacterial activity, including that against helicobacter pylori, and inhibits gastrin‐mediated acid production in mice. Microbiologyopen. 2015;4(3):465–474.
- Corcoran B, Stanton C, Fitzgerald GF, Ross RP. Survival of probiotic lactobacilli in acidic environments is enhanced in the presence of metabolizable sugars. Appl Environ Microbiol. 2005;71(6):3060–3067. doi: 10.1128/AEM.71.6.3060-3067.2005.
- Conway P, Gorbach S, Goldin B. Survival of lactic acid bacteria in the human stomach and adhesion to intestinal cells. J Dairy Sci. 1987;70(1):1–12. doi: 10.3168/jds.S0022-0302(87)79974-3.
- Begley M, Hill C, Gahan CG. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol. 2006;72(3):1729–1738. doi: 10.1128/AEM.72.3.1729-1738.2006.
- Maragkoudakis PA, Zoumpopoulou G, Miaris C, Kalantzopoulos G, Pot B, Tsakalidou E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int Dairy J. 2006;16(3):189–199. doi: 10.1016/j.idairyj.2005.02.009.
- Di Ciaula A, Garruti G, Lunardi Baccetto R, Molina-Molina E, Bonfrate L, Wang DQ, Portincasa P. Bile acid physiology. Ann Hepatol. 2017:16(1):6-7.
- Northfield T, McColl I. Postprandial concentrations of free and conjugated bile acids down the length of the normal human small intestine. Gut. 1973;14(7):513–518.
- Tannock G, Munro K, Harmsen HJ, Welling GW, Smart J, Gopal PK. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosusDR20. Appl Environ Microbiol. 2000;66(6):2578–2588.
- Goossens D, Jonkers D, Russel M, Stobberingh E, Van Den Bogaard A, StockbrUgger R. The effect of Lactobacillus plantarum 299v on the bacterial composition and metabolic activity in faeces of healthy volunteers: a placebo‐controlled study on the onset and duration of effects. Aliment Pharmacol Ther. 2003;18(5):495–505. doi: 10.1046/j.1365-2036.2003.01708.x.
- Vitali B, Minervini G, Rizzello CG, Spisni E, Maccaferri S, Brigidi P, Gobbetti M, Di Cagno R. Novel probiotic candidates for humans isolated from raw fruits and vegetables. Food Microbiol. 2012;31(1):116–125. doi: 10.1016/j.fm.2011.12.027.
- Bezkorovainy A. Probiotics: determinants of survival and growth in the gut. Am J Clin Nutr. 2001;73(2):399s–405s. doi: 10.1093/ajcn/73.2.399s.
- Yuki N, Watanabe K, Mike A, Tagami Y, Tanaka R, Ohwaki M, Morotomi M. Survival of a probiotic, Lactobacillus casei strain Shirota, in the gastrointestinal tract: selective isolation from faeces and identification using monoclonal antibodies. Int J Food Microbiol. 1999;48(1):51–57.
- Alander M, Satokari R, Korpela R, Saxelin M, Vilpponen-Salmela T, Mattila-Sandholm T, von Wright A. Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosusGG, after oral consumption. Appl Environ Microbiol. 1999;65(1):351–354.
- Valeur N, Engel P, Carbajal N, Connolly E, Ladefoged K. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl Environ Microbiol. 2004;70(2):1176–1181.
- Smith TJ, Anderson D, Margolis LM, Sikes A, Young AJ. Persistence of Lactobacillus reuteri DSM17938 in the human intestinal tract: response to consecutive and alternate-day supplementation. J Am Coll Nutr. 2011;30(4):259–264.
- Juge N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 2012;20(1):30–39. doi: 10.1016/j.tim.2011.10.001.
- Van Klinken BJ-W, Einerhand AWC, Duits LA, Makkink MK, Tytgat KMAJ, Renes IB, Verburg M, Büller HA, Dekker J. Gastrointestinal expression and partial cDNA cloning of murine Muc2. Am J Physiol Gastrointest Liver Physiol. 1999;276(1):G115–G124. doi: 10.1152/ajpgi.1999.276.1.G115.
- Tytgat KM, Büller HA, Opdam FJ, Kim YS, Einerhand AW, Dekker J. Biosynthesis of human colonic mucin: muc2 is the prominent secretory mucin. Gastroenterology. 1994;107(5):1352–1363.
- van Klinken BJ-W, Oussoren E, Weenink JJ, Strous GJ, Büller HA, Dekker J, Einerhand AW. The human intestinal cell lines Caco-2 and LS174T as models to study cell-type specific mucin expression. Glycoconj J. 1996;13(5):757–768.
- Johansson ME, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host–microbial interactions. Proc Natl Acad Sci. 2011;108(Supplement 1):4659–4665. doi: 10.1073/pnas.1006451107.
- Ouwehand AC, Salminen S. In vitro adhesion assays for probiotics and their in vivo relevance: a review. Microb Ecol Health Dis. 2003;15(4):175–184. doi: 10.1080/08910600310019886.
- Crociani J, Grill JP, Huppert M, Ballongue J. Adhesion of different bifidobacteria strains to human enterocyte‐like Caco‐2 cells and comparison with in vivo study. Lett Appl Microbiol. 1995;21(3):146–148.
- Chauviere G, Coconnier, MH, Kernéis SOPHIE, Fourniat J, Servin AL. Adhesion of human Lactobacillus acidophilus strain LB to human enterocyte-like Caco-2 cells. Microbiology. 1992;138(8):1689–1696.
- Todoriki K, Mukai T, Sato S, Toba T. Inhibition of adhesion of food‐borne pathogens to Caco‐2 cells by Lactobacillus strains. J Appl Microbiol. 2001;91(1):154–159.
- Bernet M-F, Brassart D, Neeser JR, Servin AL. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut. 1994;35(4):483–489.
- Luo Q, Kumar P, Vickers TJ, Sheikh A, Lewis WG, Rasko DA, Sistrunk J, Fleckenstein JM. Enterotoxigenic escherichia coli secretes a highly conserved mucin-degrading metalloprotease to effectively engage intestinal epithelial cells. Infect Immun. 2014;82(2):509–521. doi: 10.1128/IAI.01106-13.
- Crossman LC, Chaudhuri RR, Beatson SA, Wells TJ, Desvaux M, Cunningham AF, Petty NK, Mahon V, Brinkley C, Hobman JL, et al. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic escherichia coli strain H10407. J Bacteriol. 2010;192(21):5822–5831. doi: 10.1128/JB.00710-10.
- Taxt A, Aasland R, Sommerfelt H, Nataro J, Puntervoll P. Heat-stable enterotoxin of enterotoxigenic escherichia coli as a vaccine target. Infect Immun. 2010;78(5):1824–1831. doi: 10.1128/IAI.01397-09.
- Gagnon M, Zihler Berner A, Chervet N, Chassard C, Lacroix C. Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate salmonella adhesion and invasion. J Microbiol Methods. 2013;94(3):274–279. doi: 10.1016/j.mimet.2013.06.027.
- Moellering RC Jr. Emergence of enterococcus as a significant pathogen. Clin Infect Dis. 1992:1173–1176. doi: 10.1093/clinids/14.6.1173.
- Diep DB, Straume D, Kjos M, Torres C, Nes IF. An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides. 2009;30(8):1562–1574. doi: 10.1016/j.peptides.2009.05.014.
- Da Silva Sabo S, Vitolo M, González JMD, Oliveira RPDS. Overview of Lactobacillus plantarum as a promising bacteriocin producer among lactic acid bacteria. Food Res Int. 2014;64:527–536. doi: 10.1016/j.foodres.2014.07.041.
- Abee T, Klaenhammer T, Letellier L. Kinetic studies of the action of lactacin F, a bacteriocin produced by Lactobacillus johnsonii that forms poration complexes in the cytoplasmic membrane. Appl Environ Microbiol. 1994;60(3):1006–1013.
- Ouwehand AC, Tuomola EM, Tölkkö S, Salminen S. Assessment of adhesion properties of novel probiotic strains to human intestinal mucus. Int J Food Microbiol. 2001;64(1):119–126.
- Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am J Clin Nutr. 2001;73(6):1131S–1141S. doi: 10.1093/ajcn/73.6.1131S.
- Mack D, Ahrne S, Hyde L, Wei S, Hollingsworth MA. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut. 2003;52(6):827–833.
- Kumar P, Luo Q, Vickers TJ, Sheikh A, Lewis WG, Fleckenstein JM. EatA, an immunogenic protective antigen of enterotoxigenic Escherichia coli, degrades intestinal mucin. Infect Immun. 2014;82(2):500–508. doi: 10.1128/IAI.01078-13.
- Van der Sluis M, De Koning BAE, De Bruijn ACJM, Velcich A, Meijerink JPP, Van Goudoever JB, Büller HA, Dekker J, Van Seuningen I, Renes IB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131(1):117–129. doi: 10.1053/j.gastro.2006.04.020.
- Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, Kucherlapati R, Lipkin M, Yang K, Augenlicht L. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295(5560):1726–1729. doi: 10.1126/science.1069094.
- Kelly CP, Pothoulakis C, LaMont JT. Clostridium difficile colitis. N Engl J Med. 1994;330(4):257–262. doi: 10.1056/NEJM199401273300406.
- Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661. doi: 10.1038/nrmicro3344.
- van Baarlen P, Troost F, van der Meer C, Hooiveld G, Boekschoten M, Brummer RJM, Kleerebezem M. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc Natl Acad Sci. 2011;108(Supplement 1):4562–4569. doi: 10.1073/pnas.1000079107.
- Smits HH, Engering A, van der Kleij D, de Jong EC, Schipper K, van Capel TMM, Zaat BAJ, Yazdanbakhsh M, Wierenga EA, van Kooyk Y, et al. Selective probiotic bacteria induce IL-10–producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell–specific intercellular adhesion molecule 3–grabbing nonintegrin. J Allergy Clin Immunol. 2005;115(6):1260–1267. doi: 10.1016/j.jaci.2005.03.036.
- Roselli M, Finamore A, Britti MS, Mengheri E. Probiotic bacteria Bifidobacterium animalis MB5 and Lactobacillus rhamnosus GG protect intestinal Caco-2 cells from the inflammation-associated response induced by enterotoxigenic escherichia coli K88. Br J Nutr. 2006;95(6):1177–1184.
- Saavedra J. Probiotics and infectious diarrhea. Am J Gastroenterol. 2000;95(1):S16–S18.
- Huang JS, Bousvaros A, Lee JW, Diaz A, Davidson EJ. Efficacy of probiotic use in acute diarrhea in children: a meta-analysis. Dig Dis Sci. 2002;47(11):2625–2634.
- Gao XW, Mubasher M, Fang CY, Reifer C, Miller LE. Dose–response efficacy of a proprietary probiotic formula of Lactobacillus acidophilus CL1285 and Lactobacillus casei LBC80R for antibiotic-associated diarrhea and clostridium difficile-associated diarrhea prophylaxis in adult patients. Am J Gastroenterol. 2010;105(7):1636. doi: 10.1038/ajg.2010.11.
- Guandalini S, Pensabene L, Zikri MA, Dias JA, Casali LG, Hoekstra H, Kolacek S, Massar K, Micetic-Turk D, Papadopoulou A, et al. Lactobacillus GG administered in oral rehydration solution to children with acute diarrhea: a multicenter European trial. J Pediatr Gastroenterol Nutr. 2000;30(1):54–60.
- Isolauri E, Juntunen M, Rautanen T, Sillanaukee P, Koivula T. A human Lactobacillus strain (Lactobacillus casei sp strain GG) promotes recovery from acute diarrhea in children. Pediatrics. 1991;88(1):90–97.
- Saavedra JM, Bauman NA, Perman JA, Yolken RH, Saavedra JM, Bauman NA, Oung I. Feeding of bifidobacterium bifidum and streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. The Lancet. 1994;344(8929):1046–1049. doi: 10.1016/S0140-6736(94)91708-6.
- Szajewska H, Skórka A, Ruszczyński M, Gieruszczak-Białek D. Meta‐analysis: lactobacillus GG for treating acute diarrhoea in children. Aliment Pharmacol Ther. 2007;25(8):871–881. doi: 10.1111/j.1365-2036.2007.03282.x.
- Watson JT, Gayer M, Connolly MA. Epidemics after natural disasters. Emerging Infect Dis. 2007;13(1):1. doi: 10.3201/eid1301.060779.
- Gilliland S, Walker D. Factors to consider when selecting a culture of Lactobacillus acidophilus as a dietary adjunct to produce a hypocholesterolemic effect in humans. J Dairy Sci. 1990;73(4):905–911. doi: 10.3168/jds.S0022-0302(90)78747-4.
- Natoli M, Leoni BD, D’Agnano I, Zucco F, Felsani A. Good Caco-2 cell culture practices. Toxicol In Vitro. 2012;26(8):1243–1246. doi: 10.1016/j.tiv.2012.03.009.
- Hsieh PS, Tsai Y-C, Chen Y-C, Teh S-F, Ou C-M, King VA-E. Eradication of helicobacter pylori infection by the probiotic strains Lactobacillus johnsonii MH‐68 and L. salivarius ssp. salicinius AP‐32. Helicobacter. 2012;17(6):466–477. doi: 10.1111/j.1523-5378.2012.00992.x.
- Sgouras D, Maragkoudakis P, Petraki K, Martinez-Gonzalez B, Eriotou E, Michopoulos S, Kalantzopoulos G, Tsakalidou E, Mentis A. In vitro and in vivo inhibition of helicobacter pylori by Lactobacillus casei strain Shirota. Appl Environ Microbiol. 2004;70(1):518–526.
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736.
- Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11(1):119. doi: 10.1186/1471-2105-11-119.
- Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11–16. doi: 10.1093/nar/gkh152.
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153.
- Tomida S, Nguyen L, Chiu B-H, Liu J, Sodergren E, Weinstock GM, Li H. Pan-genome and comparative genome analyses of propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. MBio. 2013;4(3):e00003–13. doi: 10.1128/mBio.00003-13.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158.
- van Heel AJ, de Jong A, Montalbán-López M, Kok J, Kuipers OP. BAGEL3: automated identification of genes encoding bacteriocins and (non-) bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013;41(W1):W448–W453. doi: 10.1093/nar/gkt391.

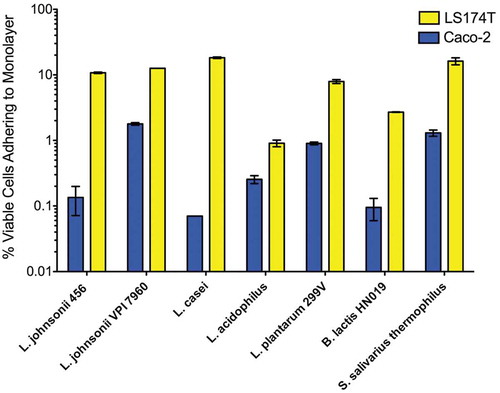
![Figure 4. Co-culture with L. johnsonii 456 significantly inhibits growth of pathogenic strains. Data expressed as means and SEM. Relevant statistically significant differences are indicated [* = p < 0.05 (t test); n = 6]. Experiment was repeated twice.](/cms/asset/40d00d9f-24e8-4fae-9427-4bd56ef6258f/kgmi_a_1547612_f0004_oc.jpg)