ABSTRACT
Technological developments, including massively parallel DNA sequencing, gnotobiotics, metabolomics, RNA sequencing and culturomics, have markedly propelled the field of microbiome research in recent years. These methodologies can be harnessed to improve our in-depth mechanistic understanding of basic concepts related to consumption of probiotics, including their rules of engagement with the indigenous microbiome and impacts on the human host. We have recently demonstrated that even during probiotic supplementation, resident gut bacteria in a subset of individuals resist the mucosal presence of probiotic strains, limiting their modulatory effect on the microbiome and on the host gut transcriptional landscape. Resistance is partly alleviated by antibiotics treatment, which enables probiotics to interact with the host at the gut mucosal interface, although rather than promoting reconstitution of the indigenous microbiome and of the host transcriptional profile, they inhibit these components from returning to their naïve pre-antibiotic configurations. In this commentary, we discuss our findings in the context of previous and recent works, and suggest that incorporating the state-of-the-art methods currently utilized in microbiome research into the field of probiotics may lead to improved understanding of their mechanisms of activity, as well as their efficacy and long-term safety.
Probiotics: decades of conflicting reports
The concept of promoting human health through consumption of beneficial microorganisms has evolved during the last century, starting with Metchnikoff’s observation at the beginning of the twentieth century that fermented dairy products are associated with longevity,Citation1 through Fuller’s initial definition of probiotic therapy,Citation2 to its current common definition as coined by the FAO/WHO in 2001: “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host”.Citation3 Thousands of studies have been published under this broad definition, attempting to demonstrate a beneficial effect for probiotic microorganisms (mostly within the Lactobacillus, Bifidobacterium, Lactococcus, Streptococcus, and Saccharomyces genera) in a long list of clinical conditions, with highly mixed results. For example, several studies suggested that probiotics may prevent or limit acute gastroenteritis,Citation4-Citation7 while other studies did not find such an effect,Citation8-Citation13 including two recent multi-center prospective trials encompassing nearly 2,000 children.Citation14,Citation15 Conflicting reports also question the efficacy of probiotics in antibiotics-associated diarrhea.Citation16-Citation21 More specifically, several meta-analyses have reported a beneficial role for probiotics in prevention of Clostridium difficile infection or its associated comorbidities,Citation22-Citation26 although some also indicated considerable heterogeneity between the analyzed trials,Citation27 and lowCitation28 to intermediateCitation29 quality of evidence. Of note, when each of the analyzed trials is considered independently, the majority of works did not find a significant effect of probiotics on C. difficile-associated diarrhea (CDAD) or C. difficile infection.Citation30-Citation60 Studies that did find a significant positive effect of probiotics in these conditions were associated with a rarely encountered C. difficile infection rate of over 5%,Citation61-Citation63 and one trial even associated probiotics use with enhanced incidence of C. difficile infections.Citation64 These conflicting results greatly highlight a need for high-quality, sufficiently powered prospective trials to resolve the usefulness of probiotics in this and other indications. Conflicting findings were also reported in the context of irritable bowel syndrome (IBS), Citation65-Citation67 respiratory infections,Citation12,Citation68-Citation84 Helicobacter pylori infections,Citation85-Citation88 inflammatory bowel disease,Citation89 improving antiretroviral treatment,Citation90,Citation91 and the metabolic syndrome.Citation92,Citation93 A more convincing indication of probiotics has been recently documented in a neonatal population at high risk of sepsis, in which synbiotic supplementation (Lactobacillus plantarum plus prebiotic fructooligosaccharide) significantly prevented morbidity and mortality.Citation94 In contrast, a 2014 Cochrane meta-analysis reported a beneficial effect for various probiotic regimens in the prevention of necrotizing enterocolitis (NEC) and all-cause mortality in preterm infants, but not sepsis,Citation95 suggesting that efficacy might depend on additional factors, such as baseline risk or the addition of prebiotics. Evidence regarding probiotics efficacy in preventing NEC are equally equivocal, as a later multi-center trial with >1300 infants found no effect for B. breve BBG-001 in this indication.Citation96
This confusing reality resulted in a peculiar state, in which, on the one hand, probiotics are not approved as medical interventions by regulatory authorities such as the US Food and Drug AdministrationCitation97 and the European Food Safety Authority,Citation98 and on the other, they are globalized, popularized and integrated into foods, cosmetics, and supplements.Citation99 This discrepancy highlights the great interest of the public in probiotics, and the concurrent need to improve the state of evidence with regard to this therapeutic modality.
Circumventing old obstacles with new technology
The considerable variation in outcome between trials may be attributed to multiple factors, including the wide array of studied microorganisms in each context, heterogeneity between participants in response to probiotics, and technical differences between studies. Discrepancies associated with the treatment may be related to speciesCitation100- and strainCitation101-specific probiotic traits, resulting in differential clinical efficacy between trials comparing various probiotic microorganisms;Citation102 in the case of commercially available products, the batch-to-batch variation might result in different outcomes.Citation103 Host factors that might affect probiotics colonization and/or efficacy include diet,Citation104 age,Citation105,Citation106 antibiotics exposure,Citation107,Citation108 underlying medical conditions,Citation109,Citation110 and baseline microbiome compositionCitation111-Citation113 and function.Citation114,Citation115 Methodological advances in next-generation sequencing, metabolomics, and gnotobiotics, now enable a better understanding of concepts such as colonization resistance or permissiveness to exogenous microorganisms, bio-geographical diversity, inter-individual variability in microbiome configuration, and how these may affect the response to therapies.
In our recent studiesCitation107,Citation111 we aimed at tackling elementary questions regarding the current commercially available probiotics by employing multi-omic analyses complemented by in vivo and in vitro experimentation, while disentangling potential confounding factors that may arise from next-generation sequencing data. Our focus and design were not purposed to show clinical benefit or harm to the host, but rather to invasively and directly characterize the effects of exogenous probiotic microorganisms administration on the gut mucosa and the indigenous microbiome at various sites along the gastrointestinal tract. To that end, we subjected human volunteers to probiotics and other treatments and collected sequential stool samples before, during and after the interventions. Additionally, we performed endoscopic examinations of their gastrointestinal tract to obtain mucosal and luminal contents before and during the intervention. Stool, mucosal, and luminal samples underwent 16S rRNA and shotgun sequencing to assess the microbiome component, while mucosal biopsies underwent RNA sequencing to directly assess the host. We further utilized probiotics species-specific quantitative PCR to achieve absolute abundances of selected bacteria. The entire study design was replicated in mice.
By this combined approach we aimed to overcome several shortcomings in probiotics research: First, the vast majority of previous studies have inferred mucosal colonization of probiotics from their persistence in stool after discontinuing the treatment with the probiotic preparation, and thus were unable to determine mucosal adherence of these strains during the period of administration. We directly sampled the mucosal microbiome to bypass this limitation. Second, we set out to sample luminal and mucosal samples throughout the entirety of the human digestive tract, reachable proximally and distally by an endoscope, to account for biogeographical differences. Third, we characterized the human transcriptome in different regions of the gastrointestinal tract and evaluated its alterations in response to various treatments. Furthermore, we delineated gene expression landscapes as harbingers of response to probiotics. Fourth, we examined carefully selected 11 probiotic strains, which on the one hand, shed light on strain-dependent impacts on the host and the microbiome, and on the other hand, featured multiple representative strains of the most commonly used probiotic genera (Lactobacillus, Bifidobacterium, Lactococcus and Streptococcus), thereby enabling to assess class effects. Finally, in the most invasive study of its kind, we portrayed the host and the microbiome, both before and during probiotics exposure, thereby allowing for a person-specific evaluation.
The colonization debate
The question whether allochthonous probiotic strains are capable of colonizing the gut, and whether this colonization is transient or persists after cessation of probiotics consumption, remains highly debated. Based on the constantly expanding body of knowledge regarding interactions between the host and resident commensal and pathogenic bacteria, it is logical to assume that probiotics may require contact with, or approximation to the host epithelium to exert direct, or metabolite-mediated effects. For example, the sortase-dependent pili of Bifidobacterium bifidum mediate adherence to the host cells and tumor necrosis factor (TNF)-alpha production;Citation116 Similarly, the SpaC pili of Lactobacillus rhamnosus are required for adherence to the gut mucosa, stimulation of reactive oxygen species production, and promotion of cell proliferation and protection against intestinal injury,Citation117 and the degree of Lactobacillus reuteri strains’ adhesion to the mouse ileal mucosa is correlated with their immunomodulatory properties.Citation118 Competitive exclusion of pathogens from the gut mucosa and their displacement has been suggested to be among the antagonistic mechanisms of probiotics.Citation112,Citation119-Citation122 A beneficial effect of probiotic microorganisms on intestinal barrier integrity may also require interaction with the epithelium: Binding of L. casei GG to specific receptors on enterocytes is required to upregulate MUC2,Citation123 and adhesion of Lactobacillus strains to epithelial cells enhances production of MUC3.Citation124
However, whether such probiotics mucosal colonization universally exists remained debated. A handful of studies directly sampling the host mucosa in humansCitation125-Citation128 and porcine modelsCitation129,Citation130 suggested that probiotics universally inhabit the gut mucosa during administration or shortly thereafter, while others have shown inconsistencies across participants and sites along the gastrointestinal tract,Citation131-Citation134 however due to relatively small sample sizes, host and microbiome factors that promote or resist colonization have not been fully elucidated in these studies. Other non-invasive works, which examined probiotic strains presence in stool samples as a proxy for their persistence in the gut mucosa following cessation of their administration, have yielded similarly confusing findings, as probiotics were detected only in subsets of examined human individualsCitation112,Citation133-Citation145 and rats.Citation113 Several other recent works associated the extent to which supplemented probiotics colonized the guts of animals or humans to their ability to ameliorate colitisCitation146 (in mice), depressionCitation147 (in rats) or IBS (in humans).Citation148
Some researchers would still argue that probiotics might benefit the host without even transiently being detected at the proximity of the host-microbiome mucosal interface.Citation149,Citation150 As colonization of the human intestinal mucosa with probiotics has been quantified in situ only in a limited number of trials, and none have demonstrated an effect on the host in the absence of mucosal colonization, this hypothesis remains to be experimentally validated. Current indirect evidence for an effect exerted by probiotic microorganisms that do not actively engage the mucosa stem from trials with killed bacteria,Citation151-Citation153 or those conducted with their secretomes.Citation112,Citation154-Citation156 Most probably, even mechanisms of activity involving signaling to the host through metabolite secretion or passive sensing of surface moleculesCitation149 necessitate microbial mucosal approximation to enable effective metabolite concentrations to reach their epithelial/immune cellular targets through the mucus layer. In future trials, next-generation sequencing can be utilized to determine the physiological state of probiotic bacteria in mucosal samples, and how it affects their ability to exert an effect on the host.Citation157 Importantly, our recent studies directly assessing the presence of probiotics along the entirety of the human gastrointestinal tractCitation107,Citation111 failed to detect exogenously consumed strains among participants resistant to probiotics mucosal colonization during their consumption even in luminal samples (), rendering even putative luminal activities of probiotics, such as degradation of dietary lactoseCitation158 or deconjugation of bile saltsCitation159 unlikely in those individuals. Collectively, these results suggest that in the absence of mucosal colonization, a consistent luminal presence of probiotics is debated and limited at best, with consumed probiotics rapidly finding their way into stool.Citation111
Figure 1. Luminal presence of supplement-specific probiotics strains in human individuals.
Probiotics strain quantification based on mapping of metagenomic sequences to unique genes, which correspond to the strains found in the supplemented probiotics pill, in the descending colon lumen of healthy individuals treated with an 11-strain probiotic mix (N = 10) or placebo (N = 5). Permissive individuals are those, who were significantly colonized by probiotics in the lower GI mucosa, compared to resistant individuals, in whom no significant colonization of the mucosa was determined. Red, the sample contains the same probiotic strain present in the supplement; Dark gray, the sample contains a different strain of the same species. Strain identification was performed as previously described.Citation111 LAC, Lactobacillus acidophilus; LCA, Lactobacillus casei; LPA, Lactobacillus casei sbsp. paracasei; LPL, Lactobacillus plantarum; LRH, Lactobacillus rhamnosus; BLO, Bifidobacterium longum; BBI, Bifidobacterium bifidum; BBR, Bifidobacterium breve; LLA, Lactococcus lactis; STH, Streptococcus thermophilus.
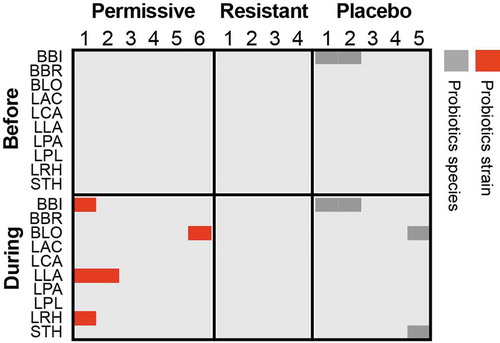
The introduction of next-generation techniques has considerably improved our ability to address the colonization question even at strain resolution to differentiate between endogenous and exogenous bacteria.Citation111,Citation114 In our recent studies we showed that regardless of global shedding in stool, even during supplementation to healthy treatment-naïve individuals, probiotic strains did not colonize the gut mucosa equally among participants, and we categorized them as probiotics ‘resisters’ or ‘permissives’ according to their level of colonization. This personalized colonization capacity may stem from host factors, as we showed that ‘resister’ and ‘permissive’ individuals differed in mucosal immune-related gene expression in some gastrointestinal organs. Additionally, it may also result from autochthonous bacteria-mediated colonization resistance, which we could demonstrate through several observations: First, a reduction or a complete lack of gut microbiome seen in antibiotics-treated or germ-free mice, respectively, exhibited improved probiotics colonization; Second, ‘humanized’ mice, transplanted with feces derived from ‘resister’ or ‘permissive’ humans, which were treated with oral probiotics, recapitulated the degree of colonization resistance to probiotics of the donor. Our data support the notion that colonization resistance patterns follow the phylogenetic exclusion principle. Of note, in many of the aforementioned studies colonization was assessed by enumeration of the probiotic bacteria in feces, and not directly by mucosal sampling, and no causative colonization-modulating effect was demonstrated for the indigenous microbiome or host gut function. We showed that probiotics affected ‘permissive’ individuals differently than ‘resisters’ in terms of their cecal and colonic transcriptional landscape. Collectively, there is sufficient evidence to conclude that probiotics adherence to the gut mucosa is adjusted by a host- and microbiome-driven colonization resistance, and that mucosal colonization may be a prerequisite for mediating some, if not most, microbiome alterations and clinical effects on the host.
Probiotics effect on the microbiome
In addition to an effect on the host exerted through direct probiotic-host cell interactions, probiotics may indirectly act through modulation of the indigenous commensal microbial community. In their 2014 consensus statement regarding the definition of probiotics, the International Scientific Association for Probiotics and Prebiotics states “modulation of perturbed microbiota” as a widespread activity, shared across probiotics genera. An even more generalized statement regarding probiotics ability to beneficially modulate the microbiota is often promoted by other lobbying and commercial entities. Nonetheless, due to the complex nature of host–microbiome interactions, in which the same bacteria or metabolite can produce context-dependent opposing effects (reviewed inCitation160), it is often difficult to attribute definitions such as “beneficial” or “detrimental” to a microbial assemblage. To achieve such definitions, a microbiome altered by probiotics should produce a beneficial effect upon transplantation to gnotobiotic animals, however, such experiments are so far surprisingly lacking. Regardless of the physiological effect, even the ability of probiotics to modulate the microbiome is contested. The majority of trials do not support the claim that probiotics can modulate the microbiome in the absence of prior perturbation, as reflected by systematic reviews that found such evidence only in 14%,Citation161 20%,Citation162 and 21%Citation108 of trials included in the analyses. Even this limited number of supportive trials should be considered with care, as in some cases, the mere presence of administered probiotics species in a stool sample was reported as microbiome alterations, and in others, a reduction in relative abundance of taxa might be a concomitant result of adding the exogenous probiotics species, rather than an actual effect.Citation163,Citation164 An additional limitation in some of the trials is the use of culture- or probe-based methods of limited scope to characterize the effect of probiotics on the microbiome, thus giving excessive weight to specific taxonomic units of choice, yet without the ability to report on global community measurements such as alpha and beta diversity. Nonetheless, even global community measurements such as 16S rDNA analysis, may be confounded by the presence of the administered probiotics, due to their limited resolution beyond the genus level. Finally, as mentioned above, stool samples may not accurately mirror an effect (or the lack of it) in the gut mucosal niche. These common limitations in studies assessing probiotics effect on the microbiome are summarized in .
Figure 2. Common biases and challenges observed in studies assessing probiotics effect on microbiome configuration. Supplemented probiotic bacteria are illustrated in green. A-div, alpha diversity; B-div, beta diversity. A, before supplementation; B, during or after supplementation.
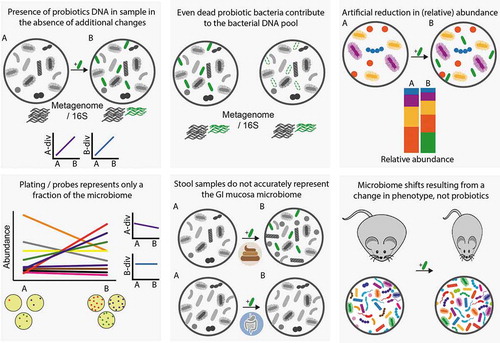
In our work, we sought to address these limitations by employing a multi-omic, invasive bio-geographical analysis of two homeostatic hosts: mice and humans. Indeed, the microbiome of mice supplemented with an 11-strain probiotic mixture displayed a bio-geographical discrepancy, as probiotics significantly elevated the alpha and beta diversity of the lower gastrointestinal (LGI) tract mucosal, but not the fecal, microbiome. In humans, probiotics affected both the composition and the function of the fecal microbiome, as well as the total bacterial load, compared to baseline or placebo, with several species, genes and pathways altered even one month following cessation of probiotics supplementation. In contrast, no significant effect was observed in the LGI lumen and mucosa. Functional alterations to the fecal microbiome were also reported in several trials, though these may be attributed to the probiotics species themselves.Citation114,Citation165-Citation167 Importantly, the extent to which probiotics affected the microbiome was associated with their colonization capacity. In our study, changes to the fecal microbiome composition, function, diversity, and bacterial load were significantly more pronounced in ‘permissive’ individuals, which displayed significant probiotics colonization in their gut mucosa. Similarly, both the mucosal microbiome composition and bacterial load were more affected in ‘permissive’ individuals.Citation111 This observation is in line with a report by Zhang et al., in which individuals colonized with Lactococcus lactis given as a component of a probiotic food supplement had a greater effect on fecal microbiome beta diversity, compared to colonization-resistant individuals.Citation113 To conclude, probiotics may have a limited effect on the microbiome composition and potentially function, which may depend on the ability of the administered microorganisms to colonize the host, at least transiently.
Further support to the importance of probiotics colonization in inducing microbiome alterations stems from trials in antibiotics-perturbed subjects. In our study,Citation107 we administered probiotics after the cessation of antibiotics and an invasive assessment of antibiotics-associated dysbiosis, thereby enabling to accurately quantify the impact of probiotics on the host and the microbiome without having the probiotics strains present and confounding during baseline measurements. Surprisingly, while antibiotics-induced depletion of the indigenous microbiome enhanced the colonization of probiotics in the LGI mucosa of humans (but only marginally in mice), they persistently inhibited the restoration of the pre-antibiotics microbial community composition, function, diversity and bacterial load in both mice and humans, as well as the human naïve gut transcriptional landscape, compared to both autologous fecal microbiota transplantation (FMT) and spontaneous recovery. Our study’s design suggests that this probiotics-induced inhibition of microbiome and host reversion to their naïve state would probably be even more pronounced, if probiotics were allowed to colonize the host also during the administration of antibiotics.
In vitro, we demonstrated that this inhibition might be mediated by soluble factors secreted by Lactobacillus spp.Citation107 Surprisingly, while many probiotic species were reported to possess anti-bacterial properties through the production of acidCitation168,Citation169 or bacteriocins,Citation130,Citation170,Citation171 these have been so far described solely in the context of pathogens, despite no mechanistic explanation to support a pathogen-specific, commensal-tolerant activity. In line with this notion, several studies provided data that do not support a beneficial effect for probiotics in the recovery of the microbiome from antibiotics or even its inhibition. In mice, a four-strain probiotic mixture did not result in an improved alpha-diversity when given either during or following antibiotics, and was in fact associated with lower diversity compared to no treatment.Citation172 In humans, a combination of L. acidophilus and B. bifidum given during antibiotics resulted in a lower restoration to baseline of anaerobes, facultative anaerobes and specifically Bacteroides compared to placebo or probiotics given following antibiotics,Citation173 and administration of S. boulardii CNCM I-745 during antibiotics did not restore the microbiome to an antibiotics-naïve configuration, and reduced bacterial diversity compared to no intervention.Citation174 Importantly, prolonged antibiotics-associated dysbiosis and reduced diversity may expose the host to long-term complications due to infectious and other non-communicable diseases.Citation175-Citation192
To conclude, while antibiotics may alleviate colonization resistance to probiotics and enable their supposedlybeneficial ‘place-holder’ effect against pathogen colonization (an effect that remains to be thoroughly validated in humans), this may result in inhibition, rather than restoration of the indigenous gut microbiome and host gut function. As prolonged dysbiosis may have considerable health implications, this previously underappreciated trade-off merits further clinical exploration. We suggest a framework that combines longitudinal whole community composition and function characterization, utilizing both relative and absolute quantifications and prospective long-term and sufficiently powered clinical assessment.
Perspective
The reports discussed in this commentary point to several aspects in which methodological advances might improve our understanding of probiotics-host-microbiome interactions. First, a strain-level resolution achieved with shotgun sequencing enables to distinguish the supplemented organism from closely related strains already present in the sample. Synthesizing studies that utilized this approach lead to the conclusion that intestinal colonization with probiotics was observed only in some individuals, both in the long term and even during the supplementation itself. Strain-level resolution and functional microbiome analysis also point to the prior presence of phylogenetically related species, or functional redundancy of the resident microbiome with that contributed by the supplemented probiotic, as factors inversely correlated with colonization. While global shedding in stool masks heterogeneity in gut mucosal colonization, fecal samples may still be used to predict permissiveness to colonization. Transplantations of these fecal samples into germ-free mice enable to validate a causative role for the resident microbiome in dictating colonization resistance, and may be further utilized to identify specific crucial factors required for allochthonous colonization. Importantly, due to the complexity of sampling the GI mucosa, it would be useful to develop computational algorithms that predict mucosal colonization based on sequencing of fecal samples.
Advancing from limited culture- and probe-based techniques, the combined knowledge from 16S rDNA, metagenomics, and metabolomics now enables better elucidation of probiotics colonization, and direct or microbiome-mediated impacts (or their lack thereof) on the human host, paving the way for improved efficacy and safety of probiotics.
Author contributions
All authors have researched data for the article, made substantial contribution to the discussion of content, and wrote, reviewed and edited the manuscript before submission.
Disclosure of potential conflicts of interest
EE is a paid consultant at DayTwo and BiomX. None of his work on probiotics is related to, funded or endorsed by, shared or discussed with or licensed to any commercial entity.
Acknowledgments
We thank the members of the Elinav laboratory for discussions and apologize to authors whose work was not included due to space constraints. J.S. is the recipient of the Strauss Institute research fellowship. N.Z. is supported by the Gilead Sciences International Research Scholars Program in Liver Disease. E.E. is supported by Y. and R. Ungar, the Abisch Frenkel Foundation for the Promotion of Life Sciences, the Gurwin Family Fund for Scientific Research, the Leona M. and Harry B. Helmsley Charitable Trust, the Crown Endowment Fund for Immunological Research, the estate of J. Gitlitz, the estate of L. Hershkovich, the Benoziyo Endowment Fund for the Advancement of Science, the Adelis Foundation, J.L. and V. Schwartz, A. and G. Markovitz, A. and C. Adelson, the French National Center for Scientific Research (CNRS), D.L. Schwarz, the V.R. Schwartz Research Fellow Chair, L. Steinberg, J.N. Halpern, A. Edelheit, grants funded by the European Research Council, a Marie Curie Integration grant, the German-Israeli Foundation for Scientific Research and Development, the Israel Science Foundation, the Minerva Foundation, the Rising Tide Foundation, the Helmholtz Foundation, and the European Foundation for the Study of Diabetes. E.E. is a senior fellow of the Canadian Institute of Advanced Research (CIFAR) and an international scholar of the Bill and Melinda Gates Foundation and Howard Hughes Medical Institute (HHMI).
References
- Metchnikoff II. The prolongation of life: optimistic studies. New York (NY): Springer Publishing Company; 2004.
- Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989;66:365–378.
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. Expert consensus document. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–514. doi:10.1038/nrgastro.2014.66.
- Allen SJ, Martinez EG, Gregorio GV, Dans LF. Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst Rev. 2010. doi:10.1002/14651858.CD003048.pub3. Cd003048.
- Feizizadeh S, Salehi-Abargouei A, Akbari V. Efficacy and safety of saccharomyces boulardii for acute diarrhea. Pediatrics. 2014;134:e176–191. doi:10.1542/peds.2013-3950.
- Szajewska H, Skorka A, Ruszczynski M. Meta-analysis: G-BD. Lactobacillus GG for treating acute gastroenteritis in children–updated analysis of randomised controlled trials. Aliment Pharmacol Ther. 2013;38:467–476. doi:10.1111/apt.12403.
- Van Niel CW, Feudtner C, Garrison MM, Christakis DA. Lactobacillus therapy for acute infectious diarrhea in children: a meta-analysis. Pediatrics. 2002;109:678–684.
- Pereg D, Kimhi O, Tirosh A, Orr N, Kayouf R, Lishner M. The effect of fermented yogurt on the prevention of diarrhea in a healthy adult population. Am J Infect Control. 2005;33:122–125. doi:10.1016/j.ajic.2004.11.001.
- Freedman SB, Pasichnyk D, Black KJL, Fitzpatrick E, Gouin S, Milne A, Hartling L, Choonara I. Gastroenteritis therapies in developed countries: systematic review and meta-analysis. PLoS One. 2015;10:e0128754. doi:10.1371/journal.pone.0128754.
- Laursen RP, Ellis BH, Lantos JD. Probiotics and child care absence due to infections: a randomized controlled trial. Pediatrics. 2017;140. doi:10.1542/peds.2017-0735.
- Szajewska H, Ruszczynski M, Kolacek S. Meta-analysis shows limited evidence for using lactobacillus acidophilus LB to treat acute gastroenteritis in children. Acta Paediatrica (Oslo, Norway: 1992). 2014;103:249–255. doi:10.1111/apa.12487.
- Hojsak I, Tokic Pivac V, Mocic Pavic A, Pasini AM, Kolacek S. Bifidobacterium animalis subsp. lactis fails to prevent common infections in hospitalized children: a randomized, double-blind, placebo-controlled study. Am J Clin Nutr. 2015;101:680–684. doi:10.3945/ajcn.114.102004.
- Salazar-Lindo E, Miranda-Langschwager P, Campos-Sanchez M, Chea-Woo E, Sack RB. Lactobacillus casei strain GG in the treatment of infants with acute watery diarrhea: a randomized, double-blind, placebo controlled clinical trial [ISRCTN67363048]. BMC Pediatr. 2004;4:18. doi:10.1186/1471-2431-4-18.
- Schnadower D, Tarr PI, Casper TC, Gorelick MH, Dean JM, O’Connell KJ, Mahajan P, Levine AC, Bhatt SR, Roskind CG, et al. Lactobacillus rhamnosus GG versus placebo for acute gastroenteritis in children. N Engl J Med. 2018;379:2002–2014. doi:10.1056/NEJMoa1802598.
- Freedman SB, Williamson-Urquhart S, Farion KJ, Gouin S, Willan AR, Poonai N, Hurley K, Sherman PM, Finkelstein Y, Lee BE, et al. Multicenter trial of a combination probiotic for children with gastroenteritis. N Engl J Med. 2018;379:2015–2026. doi:10.1056/NEJMoa1802597.
- Goldenberg JZ, Lytvyn L, Steurich J, Parkin P, Mahant S, Johnston BC. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev. 2015:CD004827. doi:10.1002/14651858.CD004827.pub4. .
- Szajewska H, Kolodziej M. Systematic review with meta-analysis: lactobacillus rhamnosus GG in the prevention of antibiotic-associated diarrhoea in children and adults. Aliment Pharmacol Ther. 2015;42:1149–1157. doi:10.1111/apt.13404.
- Hempel, S, Newberry SJ, Maher AR, Wang Z, Miles JN Shanman R, Johnsen B Shekelle PG. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: a systematic review and meta-analysis. JAMA. 2012;307:1959–1969. doi:10.1001/jama.2012.3507.
- Jafarnejad S, Shab-Bidar S, Speakman JR, Parastui K, Daneshi-Maskooni M, Djafarian K. Probiotics reduce the risk of antibiotic-associated diarrhea in adults (18–64 years) but not the elderly (>65 years): a meta-analysis. Nutr Clin Pract. 2016;31:502–513. doi:10.1177/0884533616639399.
- Olek A, Woynarowski M, Ahrén IL, Kierkuś J, Socha P, Larsson N, Önning G. Efficacy and safety of lactobacillus plantarum DSM 9843 (LP299V) in the prevention of antibiotic-associated gastrointestinal symptoms in children-randomized, double-blind, placebo-controlled study. J Pediatr. 2017;186:82–86. doi:10.1016/j.jpeds.2017.03.047.
- Allen SJ, Wareham K, Wang D, Bradley C, Hutchings H, Harris W, Dhar A, Brown H, Foden A, Gravenor MB, et al. Lactobacilli and bifidobacteria in the prevention of antibiotic-associated diarrhoea and clostridium difficile diarrhoea in older inpatients (PLACIDE): a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2013;382:1249–1257. doi:10.1016/S0140-6736(13)61218-0.
- Pattani R, Palda VA, Hwang SW, Shah PS. Probiotics for the prevention of antibiotic-associated diarrhea and Clostridium difficile infection among hospitalized patients: systematic review and meta-analysis. Open Med. 2013;7:e56–67.
- Avadhani A, Miley H. Probiotics for prevention of antibiotic-associated diarrhea and Clostridium difficile-associated disease in hospitalized adults–a meta-analysis. J Am Acad Nurse Pract. 2011;23:269–274. doi:10.1111/j.1745-7599.2011.00617.x.
- Shen NT, Maw A, Tmanova LL, Pino A, Ancy K, Crawford CV, Simon MS, Evans AT. Timely use of probiotics in hospitalized adults prevents clostridium difficile infection: a systematic review with meta-regression analysis. Gastroenterol. 2017;152:1889–1900 e1889. doi:10.1053/j.gastro.2017.02.003.
- Goldenberg JZ, Ma SS, Saxton JD, Martzen MR, Vandvik PO, Thorlund K, Guyatt GH, Johnston BC. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev. 2013:Cd006095. doi:10.1002/14651858.CD006095.pub3. .
- McFarland LV. Probiotics for the primary and secondary prevention of C. difficile infections: a meta-analysis and systematic review. Antibiotics (Basel). 2015;4:160–178. doi:10.3390/antibiotics4020160.
- Goldenberg JZ, Yap C, Lytvyn L, Lo CKF, Beardsley J, Mertz D, Johnston, BC. Probiotics for the prevention of clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev. 2017;12:Cd006095. doi:10.1002/14651858.CD006095.pub4.
- Szajewska H, Canani RB, Guarino A, Hojsak I, Indrio F, Kolacek S, Orel R, Shamir R, Vandenplas Y, van Goudoever JB, et al. Probiotics for the prevention of antibiotic-associated diarrhea in children. J Pediatr Gastroenterol Nutr. 2016;62:495–506. doi:10.1097/mpg.0000000000001081.
- Goldenberg JZ, Mertz D, Johnston BC. Probiotics to prevent clostridium difficile infection in patients receiving antibiotics. Jama. 2018;320:499–500. doi:10.1001/jama.2018.9064.
- Selinger CP, Bell A, Cairns A, Lockett M, Sebastian S, Haslam N. Probiotic VSL#3 prevents antibiotic-associated diarrhoea in a double-blind, randomized, placebo-controlled clinical trial. J Hosp Infect. 2013;84:159–165. doi:10.1016/j.jhin.2013.02.019.
- Bravo MV, Bunout D, Leiva L, Barrera G, Hirsch S. [Effect of probiotic Saccharomyces boulardii on prevention of antibiotic-associated diarrhea in adult outpatients with amoxicillin treatment]. Rev Med Chil. 2008;136:981–988. doi:/S0034-98872008000800004.
- Cindoruk M, Erkan G, Karakan T, Dursun A, Unal S. Efficacy and safety of Saccharomyces boulardii in the 14-day triple anti-Helicobacter pylori therapy: a prospective randomized placebo-controlled double-blind study. Helicobacter. 2007;12:309–316. doi:10.1111/j.1523-5378.2007.00516.x.
- Koning CJ, Jonkers DM, Stobberingh EE, Mulder L, Rombouts FM, Stockbrügger RW. The effect of a multispecies probiotic on the intestinal microbiota and bowel movements in healthy volunteers taking the antibiotic amoxycillin. Am J Gastroenterol. 2008;103:178–189. doi:10.1111/j.1572-0241.2007.01547.x.
- Shimbo I, Yamaguchi T, Odaka T, Nakajima K, Koide A, Koyama H, Saisho H. Effect of clostridium butyricum on fecal flora in helicobacter pylori eradication therapy. World J Gastroenterol. 2005;11:7520–7524.
- Siitonen S, Vapaatalo H, Salminen S, Gordin A, Saxelin M, Wikberg R, Kirkkola AL. Effect of lactobacillus GG yoghurt in prevention of antibiotic associated diarrhoea. Ann Med. 1990;22:57–59.
- Georgieva M, Pancheva R, Rasheva N, Usheva N, Ivanova L, Koleva K. Use of the probiotic lactobacillus reuteri dsm 17938 in the prevention of antibiotic-associated infections in hospitalIzed Bulgarian children: a randomized, controlled trial. J IMAB–Annu Proc Sci Pap. 2015;21:895–900. doi:10.5272/jimab.
- Thomas MR, Litin SC, Osmon DR, Corr AP, Weaver AL, Lohse CM. Lack of effect of lactobacillus GG on antibiotic-associated diarrhea: a randomized, placebo-controlled trial. Mayo Clinic Proc. 2001;76:883–889. doi:10.4065/76.9.883.
- McFarland LV, Surawicz CM, Greenberg RN, Elmer GW, Moyer KA, Melcher SA, Bowen KE, Cox JL. Prevention of beta-lactam-associated diarrhea by saccharomyces boulardii compared with placebo. Am J Gastroenterol. 1995;90:439–448.
- Safdar N, Barigala R, Said A, McKinley L. Feasibility and tolerability of probiotics for prevention of antibiotic-associated diarrhoea in hospitalized US military veterans. J Clin Pharm Ther. 2008;33:663–668. doi:10.1111/j.1365-2710.2008.00980.x.
- Pozzoni P, Riva A, Bellatorre AG, Amigoni M, Redaelli E, Ronchetti A, Stefani M, Tironi R, Molteni EE, Conte D, et al. Saccharomyces boulardii for the prevention of antibiotic-associated diarrhea in adult hospitalized patients: a single-center, randomized, double-blind, placebo-controlled trial. Am J Gastroenterol. 2012;107:922–931. doi:10.1038/ajg.2012.56.
- Surawicz CM, Elmer GW, Speelman P, McFarland LV, Chinn J, van Belle G. Prevention of antibiotic-associated diarrhea by saccharomyces boulardii: a prospective study. Gastroenterol. 1989;96:981–988.
- Plummer SF, Garaiova I, Sarvotham T, Cottrell SL, Le Scouiller S, Weaver MA, Tang J, Dee P, Hunter J. Effects of probiotics on the composition of the intestinal microbiota following antibiotic therapy. Int J Antimicrob Agents. 2005;26:69–74. doi:10.1016/j.ijantimicag.2005.04.004.
- Can M, Besirbellioglu BA, Avci IY, Beker CM, Pahsa A. Prophylactic saccharomyces boulardii in the prevention of antibiotic-associated diarrhea: a prospective study. Med Sci Monit. 2006;12:Pi19–22.
- Beausoleil M, Fortier N, Guénette S, L’ecuyer A, Savoie M, Franco M, Lachaine J, Weiss K. Effect of a fermented milk combining lactobacillus acidophilus Cl1285 and Lactobacillus casei in the prevention of antibiotic-associated diarrhea: a randomized, double-blind, placebo-controlled trial. Can J Gastroenterol. 2007;21:732–736.
- Wong S, Rutherfurd SM, Olson TD, Purba AS, Drummond LN, Boland MJ, Moughan PJ. A Lactobacillus casei Shirota probiotic drink reduces antibiotic-associated diarrhoea in patients with spinal cord injuries: a randomised controlled trial. Br J Nutr. 2014;111:672–678. doi:10.1017/s0007114513002973.
- Ehrhardt S, Guo N, Hinz R, Schoppen S, May J, Reiser M, Schroeder MP, Schmiedel S, Keuchel M, Reisinger EC. et al. Saccharomyces boulardii to prevent antibiotic-associated diarrhea: a randomized, double-masked, placebo-controlled trial. Open Forum Infect Dis. 2016;3:ofw011. doi:10.1093/ofid/ofw011.
- Arvola T, Megevand A, Page HD. Prophylactic Lactobacillus GG reduces antibiotic-associated diarrhea in children with respiratory infections: a randomized study. Pediatrics. 1999;104:e64.
- Duman DG, Bor S, Ozütemiz O, Sahin T, Oğuz D, Iştan F, Vural T, Sandkci M, Işksal F, Simşek I, et al. Efficacy and safety of saccharomyces boulardii in prevention of antibiotic-associated diarrhoea due to helicobacterpylori eradication. Eur J Gastroenterol Hepatol. 2005;17:1357–1361.
- Kotowska M, Albrecht P, Szajewska H. Saccharomyces boulardii in the prevention of antibiotic-associated diarrhoea in children: a randomized double-blind placebo-controlled trial. Aliment Pharmacol Ther. 2005;21:583–590. doi:10.1111/j.1365-2036.2005.02356.x.
- Lewis SJ, Potts LF, Barry RE. The lack of therapeutic effect of saccharomyces boulardii in the prevention of antibiotic-related diarrhoea in elderly patients. J Infect. 1998;36:171–174.
- Lonnermark E, Friman V, Lappas G, Sandberg T, Berggren A, Adlerberth I. Intake of Lactobacillus plantarum reduces certain gastrointestinal symptoms during treatment with antibiotics. J Clin Gastroenterol. 2010;44:106–112. doi:10.1097/MCG.0b013e3181b2683f.
- Miller M, Florencio S, Eastmond J, Reynolds S. Results of 2 prospective randomized studies of Lactobacillus GG to prevent C. difficile infection in hospitalized adults receiving antibiotics. Intersci Conf Antimicrob Agents Chemother. 2008;48:578–579.
- Nord CE, Lidbeck A, Orrhage K, Sjostedt S. Oral supplementation with lactic acid-producing bacteria during intake of clindamycin. Clin Microbiol Infect. 1997;3:124–132.
- Sampalis J, Psaradellis E, Rampakakis E. Efficacy of BIO K+ CL1285 in the reduction of antibiotic-associated diarrhea - a placebo controlled double-blind randomized, multi-center study. Arch Med Sci: AMS. 2010;6:56–64. doi:10.5114/aoms.2010.13508.
- Ruszczynski M, Radzikowski A, Szajewska H. Clinical trial: effectiveness of lactobacillus rhamnosus (strains E/N, Oxy and Pen) in the prevention of antibiotic-associated diarrhoea in children. Aliment Pharmacol Ther. 2008;28:154–161. doi:10.1111/j.1365-2036.2008.03714.x.
- Sullivan A, Johansson A, Svenungsson B, Nord CE. Effect of lactobacillus F19 on the emergence of antibiotic-resistant microorganisms in the intestinal microflora. J Antimicrob Chemother. 2004;54:791–797. doi:10.1093/jac/dkh406.
- Wenus C, Goll R, Loken EB, Biong AS, Halvorsen DS, Florholmen J. Prevention of antibiotic-associated diarrhoea by a fermented probiotic milk drink. Eur J Clin Nutr. 2008;62:299–301. doi:10.1038/sj.ejcn.1602718.
- Wullt M, Johansson Hagslatt ML, Odenholt I, Berggren A. Lactobacillus plantarum 299v enhances the concentrations of fecal short-chain fatty acids in patients with recurrent clostridium difficile-associated diarrhea. Dig Dis Sci. 2007;52:2082–2086. doi:10.1007/s10620-006-9123-3.
- Lawrence SJ, Korzenik JR, Mundy LM. Probiotics for recurrent Clostridium difficile disease. J Med Microbiol. 2005;54:905–906. doi:10.1099/jmm.0.46096-0.
- Barker AK, Ding B, Ye M, Wang P, Bi Y, Wu S, Xu X, Guo Q, Wang M. A randomized controlled trial of probiotics for clostridium difficile infection in adults (PICO). J Antimicrob Chemother. 2017;72:3177–3180. doi:10.1093/jac/dkx254.
- Gao XW, Mubasher M, Fang CY, Reifer C, Miller LE. Dose-response efficacy of a proprietary probiotic formula of lactobacillus acidophilus CL1285 and lactobacillus casei LBC80R for antibiotic-associated diarrhea and clostridium difficile-associated diarrhea prophylaxis in adult patients. Am J Gastroenterol. 2010;105:1636–1641. doi:10.1038/ajg.2010.11.
- Hickson M, D’Souza AL, Muthu N, Rogers TR, Want S, Rajkumar C, Bulpitt CJ. Use of probiotic lactobacillus preparation to prevent diarrhoea associated with antibiotics: randomised double blind placebo controlled trial. BMJ. 2007;335:80. doi:10.1136/bmj.39231.599815.55.
- Rafiq R, Pandey D, Mahgoubosman S, et al. Prevention of Clostridium difficile diarrhoea with probiotics in hospitalized patients treated with antibiotics. Gastroenterology. 2007;132 (suppl 2):A187.
- Carvour ML, Wilder SL, Ryan KL, Walraven C, Qeadan F, Brett M, Page K. Predictors of Clostridium difficile infection and predictive impact of probiotic use in a diverse hospital-wide cohort. Am J Infect Control. 2018. doi:10.1016/j.ajic.2018.07.014.
- Moayyedi P, Ford AC, Talley NJ, Cremonini F, Foxx-Orenstein AE, Brandt LJ, Quigley EMM. The efficacy of probiotics in the treatment of irritable bowel syndrome: a systematic review. Gut. 2010;59:325–332. doi:10.1136/gut.2008.167270.
- McKenzie Y, Thompson J, Gulia P, Lomer M. British dietetic association systematic review of systematic reviews and evidence‐based practice guidelines for the use of probiotics in the management of irritable bowel syndrome in adults (2016 update). J Hum Nutr Diet. 2016;29:576–592. doi:10.1111/jhn.12386.
- Ford AC, Harris LA, Lacy BE, Quigley EMM, Moayyedi P. Systematic review with meta-analysis: the efficacy of prebiotics, probiotics, synbiotics and antibiotics in irritable bowel syndrome. Aliment Pharmacol Ther. 2018;48:1044–1060. doi:10.1111/apt.15001.
- Leyer GJ, Li S, Mubasher ME, Reifer C, Ouwehand AC. Probiotic effects on cold and influenza-like symptom incidence and duration in children. Pediatrics. 2009;124:e172–179. doi:10.1542/peds.2008-2666.
- Rerksuppaphol S, Rerksuppaphol L. Randomized controlled trial of probiotics to reduce common cold in schoolchildren. Pediatr Int. 2012;54:682–687. doi:10.1111/j.1442-200X.2012.03647.x.
- Hatakka K, Savilahti E, Pönkä A, Meurman JH, Poussa T, Näse L, Saxelin M, Korpela R. Effect of long term consumption of probiotic milk on infections in children attending day care centres: double blind, randomised trial. BMJ. 2001;322:1327. doi:10.1136/bmj.322.7298.1327.
- Wang Y, Deng D, Xiang L, Cheng P, Liao L, Deng X, Yan J, Lin F. Probiotics for prevention and treatment of respiratory tract infections in children: a systematic review and meta-analysis of randomized controlled trials. Medicine. 2016;95:e4509. doi:10.1097/md.0000000000004509.
- Hojsak I, Snovak N, Abdović S, Szajewska H, Mišak Z, Kolaček S. Lactobacillus GG in the prevention of gastrointestinal and respiratory tract infections in children who attend day care centers: a randomized, double-blind, placebo-controlled trial. Clin Nutr. 2010;29:312–316. doi:10.1016/j.clnu.2009.09.008.
- Merenstein D, Murphy M, Fokar A, Hernandez RK, Park H, Nsouli H, Sanders ME, Davis BA, Niborski V, Tondu F, et al. Use of a fermented dairy probiotic drink containing lactobacillus casei (DN-114 001) to decrease the rate of illness in kids: the DRINK study. A patient-oriented, double-blind, cluster-randomized, placebo-controlled, clinical trial. Eur J Clin Nutr. 2010;64:669–677. doi:10.1038/ejcn.2010.65.
- Maldonado J, Cañabate F, Sempere L, Vela F, Sánchez AR, Narbona E, López-Huertas E, Geerlings A, Valero AD, Olivares M, et al. Human milk probiotic Lactobacillus fermentum CECT5716 reduces the incidence of gastrointestinal and upper respiratory tract infections in infants. J Pediatr Gastroenterol Nutr. 2012;54:55–61. doi:10.1097/MPG.0b013e3182333f18.
- Ozen M, Kocabas Sandal G, Dinleyici EC. Probiotics for the prevention of pediatric upper respiratory tract infections: a systematic review. Expert Opin Biol Ther. 2015;15:9–20. doi:10.1517/14712598.2015.980233.
- Nocerino R, Paparo L, Terrin G, Pezzella V, Amoroso A, Cosenza L, Cecere G, De Marco G, Micillo M, Albano F, et al. Cow’s milk and rice fermented with Lactobacillus paracasei CBA L74 prevent infectious diseases in children: a randomized controlled trial. Clin Nutr. 2017;36:118–125. doi:10.1016/j.clnu.2015.12.004.
- King S, Tancredi D, Lenoir-Wijnkoop I, Gould K, Vann H, Connors G, Sanders ME, Linder JA, Shane AL, Merenstein D. Does probiotic consumption reduce antibiotic utilization for common acute infections? A systematic review and meta-analysis. Eur J Public Health. 2018. doi:10.1093/eurpub/cky185.
- de Vrese M, Winkler P, Rautenberg P, Harder T, Noah C, Laue C, Ott S, Hampe J, Schreiber S, Heller K, et al. Effect of Lactobacillus gasseri PA 16/8, bifidobacterium longum SP 07/3, B. bifidum MF 20/5 on common cold episodes: a double blind, randomized, controlled trial. Clin Nutr. 2005;24:481–491. doi:10.1016/j.clnu.2005.02.006.
- King S, Glanville J, Sanders ME, Fitzgerald A, Varley D. Effectiveness of probiotics on the duration of illness in healthy children and adults who develop common acute respiratory infectious conditions: a systematic review and meta-analysis. Br J Nutr. 2014;112:41–54. doi:10.1017/s0007114514000075.
- Smith TJ, Rigassio-Radler D, Denmark R, Haley T, Touger-Decker R. Effect of lactobacillus rhamnosus LGG(R) and bifidobacterium animalis ssp. lactis BB-12(R) on health-related quality of life in college students affected by upper respiratory infections. Br J Nutr. 2013;109:1999–2007. doi:10.1017/s0007114512004138.
- West NP, Pyne DB, Cripps AW, Hopkins WG, Eskesen DC, Jairath A, Christophersen CT, Conlon MA, Fricker PA. Lactobacillus fermentum (PCC(R)) supplementation and gastrointestinal and respiratory-tract illness symptoms: a randomised control trial in athletes. Nutr J. 2011;10:30. doi:10.1186/1475-2891-10-30.
- Gleeson M, Bishop NC, Struszczak L. Effects of Lactobacillus casei Shirota ingestion on common cold infection and herpes virus antibodies in endurance athletes: a placebo-controlled, randomized trial. Eur J Appl Physiol. 2016;116:1555–1563. doi:10.1007/s00421-016-3415-x.
- Kekkonen RA, Vasankari TJ, Vuorimaa T, Haahtela T, Julkunen I, Korpela R. The effect of probiotics on respiratory infections and gastrointestinal symptoms during training in marathon runners. Int J Sport Nutr Exerc Metab. 2007;17:352–363.
- Gleeson M, Bishop NC, Oliveira M, McCauley T, Tauler P, Lawrence C. Effects of a lactobacillus salivarius probiotic intervention on infection, cold symptom duration and severity, and mucosal immunity in endurance athletes. Int J Sport Nutr Exerc Metab. 2012;22:235–242.
- Lu C, Sang J, He H, Wan X, Lin Y, Li L, Li Y, Yu C. Probiotic supplementation does not improve eradication rate of Helicobacter pylori infection compared to placebo based on standard therapy: a meta-analysis. Sci Rep. 2016;6:23522. doi:10.1038/srep23522.
- Lu M, Lin Y-L, Lu S-W, Huang C-W, Wang Y-T, Chung Y-C. Efficacy of probiotic supplementation therapy for helicobacter pylori eradication: a meta-analysis of randomized controlled trials. PLoS One. 2016;11:e0163743. doi:10.1371/journal.pone.0163743.
- McFarland LV, Huang Y, Wang L, Malfertheiner P. Systematic review and meta-analysis: multi-strain probiotics as adjunct therapy for helicobacter pylori eradication and prevention of adverse events. U Eur Gastroenterol J. 2016;4:546–561. doi:10.1177/2050640615617358.
- Wang F, Feng J, Chen P, Liu X, Ma M, Zhou R, Chang Y, Liu J, Li J, Zhao Q. Probiotics in Helicobacter pylori eradication therapy: systematic review and network meta-analysis. Clin Res Hepatol Gastroenterol. 2017;41:466–475. doi:10.1016/j.clinre.2017.04.004.
- Dong J, Teng G, Wei T, Gao W, Wang H. Methodological quality assessment of meta-analyses and systematic reviews of probiotics in inflammatory bowel disease and pouchitis. PLoS One. 2016;11:e0168785. doi:10.1371/journal.pone.0168785.
- Miller H, Ferris R, Phelps BR. The effect of probiotics on CD4 counts among people living with HIV: a systematic review. Benef Microbes. 2016;7:345–351. doi:10.3920/bm2015.0163.
- Serrano-Villar S, de Lagarde M, Vázquez-Castellanos J, Vallejo A, Bernadino JI, Madrid N, Matarranz M, Díaz-Santiago A, Gutiérrez C, Cabello A, et al. Effects of immunonutrition in advanced human immunodeficiency virus disease: a randomized placebo-controlled clinical trial (promaltia study). Clin Infect Dis. 2019;68:120–130. doi:10.1093/cid/ciy414.
- Million M, Angelakis E, Paul M, Armougom F, Leibovici L, Raoult D. Comparative meta-analysis of the effect of Lactobacillus species on weight gain in humans and animals. Microb Pathog. 2012;53:100–108. doi:10.1016/j.micpath.2012.05.007.
- Zhang Q, Wu Y, Fei X. Effect of probiotics on body weight and body-mass index: a systematic review and meta-analysis of randomized, controlled trials. Int J Food Sci Nutr. 2015;67:571–580. doi:10.1080/09637486.2016.1181156.
- Panigrahi P, Parida S, Nanda NC, Satpathy R, Pradhan L, Chandel DS, Baccaglini L, Mohapatra A, Mohapatra SS, Misra PR, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 2017;548:407–412. doi:10.1038/nature23480.
- AlFaleh K, Anabrees J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev. 2014:Cd005496. doi:10.1002/14651858.CD005496.pub4. .
- Costeloe K, Hardy P, Juszczak E, Wilks M, Millar MR. Bifidobacterium breve BBG-001 in very preterm infants: a randomised controlled phase 3 trial. Lancet. 2016;387:649–660. doi:10.1016/s0140-6736(15)01027-2.
- Saldanha LG. Food US. and Drug Administration regulations governing label claims for food products, including probiotics. Clin Infect Dis. 2008;46(Suppl 2):S119–121. discussion S144–151. doi:10.1086/523328.
- Rijkers GT, de Vos WM, Brummer R-J, Morelli L, Corthier G, Marteau P. Health benefits and health claims of probiotics: bridging science and marketing. Br J Nutr. 2011;106:1291–1296. doi:10.1017/S000711451100287X.
- Hoffmann DE, Fraser CM, Palumbo F, Ravel J, Rowthorn V, Schwartz J. Probiotics: achieving a better regulatory fit. Food Drug Law J. 2014;69:237–272, ii.
- Macho Fernandez E, Caberto CP, Lum-Jones A, Seifried A, Wilkens LR, Schumacher FR, Monroe KR, Lim U, Tiirikainen M, Kolonel LN, et al. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut. 2011;60:1050–1059. doi:10.1136/gut.2010.232918.
- Lin YP, Thibodeaux CH, Pena JA, Ferry GD, Versalovic J. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm Bowel Dis. 2008;14:1068–1083. doi:10.1002/ibd.20448.
- Sniffen JC, McFarland LV, Evans CT, Goldstein EJC. Choosing an appropriate probiotic product for your patient: an evidence-based practical guide. PLoS One. 2018;13:e0209205. doi:10.1371/journal.pone.0209205.
- Biagioli M, Laghi L, Carino A, Cipriani S, Distrutti E, Marchianò S, Parolin C, Scarpelli P, Vitali B, Fiorucci S. Metabolic variability of a multispecies probiotic preparation impacts on the anti-inflammatory activity. Front Pharmacol. 2017;8:505. doi:10.3389/fphar.2017.00505.
- Aceti A, Maggio L, Beghetti I, Gori D, Barone G, Callegari M, Fantini M, Indrio F, Meneghin F, Morelli L, et al. Probiotics prevent late-onset sepsis in human milk-fed, very low birth weight preterm infants: systematic review and meta-analysis. Nutrients. 2017;9:904. doi:10.3390/nu9080904.
- He F, Ouwehand AC, Isolauri E, Hosoda M, Benno Y, Salminen S. Differences in composition and mucosal adhesion of bifidobacteria isolated from healthy adults and healthy seniors. Curr Microbiol. 2001;43:351–354.
- Andriantsoanirina V, Teolis AC, Xin LX, Butel MJ, Aires J. Bifidobacterium longum and Bifidobacterium breve isolates from preterm and full term neonates: comparison of cell surface properties. Anaerobe. 2014;28:212–215. doi:10.1016/j.anaerobe.2014.07.002.
- Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, Zur M, Regev-Lehavi D, Ben-Zeev Brik R, Federici S, et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174:1406–1423.e1416. doi:10.1016/j.cell.2018.08.047.
- McFarland LV. Use of probiotics to correct dysbiosis of normal microbiota following disease or disruptive events: a systematic review. BMJ Open. 2014;4:e005047. doi:10.1136/bmjopen-2014-005047.
- Roessler A, Friedrich U, Vogelsang H, Bauer A, Kaatz M, Hipler UC, Schmidt I Jahreis G. The immune system in healthy adults and patients with atopic dermatitis seems to be affected differently by a probiotic intervention. Clin Exp Allergy. 2008;38:93–102. doi:10.1111/j.1365-2222.2007.02876.x.
- Pelto L, Isolauri E, Lilius EM, Nuutila J, Salminen S. Probiotic bacteria down-regulate the milk-induced inflammatory response in milk-hypersensitive subjects but have an immunostimulatory effect in healthy subjects. Clin Exp Allergy. 1998;28:1474–1479.
- Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, Kotler E, Zur M, Regev-Lehavi D, Brik RB-Z, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388–1405.e1321. doi:10.1016/j.cell.2018.08.041.
- Davoren MJ, Liu J, Castellanos J, Rodriguez-Malave NI, Schiestl RH. A novel probiotic, Lactobacillus johnsonii 456, resists acid and can persist in the human gut beyond the initial ingestion period. Gut Microbes. 2018:1–23. doi:10.1080/19490976.2018.1547612.
- Zhang C, Derrien M, Levenez F, Brazeilles R, Ballal SA, Kim J, Degivry M-C, Quéré G, Garault P, van Hylckama Vlieg JET, et al. Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J. 2016;10:2235–2245. doi:10.1038/ismej.2016.13.
- Maldonado-Gomez MX, Martínez I, Bottacini F, O’Callaghan A, Ventura M, van Sinderen D, Hillmann B, Vangay P, Knights D, Hutkins R, et al. Stable engraftment of bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe. 2016;20:515–526. doi:10.1016/j.chom.2016.09.001.
- Ferrario C, Taverniti V, Milani C, Fiore W, Laureati M, De Noni I, Stuknyte M, Chouaia B, Riso P, Guglielmetti S. Modulation of fecal Clostridiales bacteria and butyrate by probiotic intervention with Lactobacillus paracasei DG varies among healthy adults. J Nutr. 2014;144:1787–1796. doi:10.3945/jn.114.197723.
- Turroni F, Serafini F, Foroni E, Duranti S, O’Connell Motherway M, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, et al. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci U S A. 2013;110:11151–11156. doi:10.1073/pnas.1303897110.
- Ardita CS, Mercante JW, Kwon YM, Luo L, Crawford ME, Powell DN, Jones RM, Neish AS, Elkins CA. Epithelial adhesion mediated by pilin SpaC is required for lactobacillus rhamnosus GG-induced cellular responses. Appl Environ Microbiol. 2014;80:5068–5077. doi:10.1128/aem.01039-14.
- Gao K, Liu L, Dou X, Wang C, Liu J, Zhang W, Wang H. Doses lactobacillus reuteri depend on adhesive ability to modulate the intestinal immune response and metabolism in mice challenged with lipopolysaccharide. Sci Rep. 2016;6:28332. doi:10.1038/srep28332.
- Bernet MF, Brassart D, Neeser JR, Servin AL. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut. 1994;35:483–489.
- Lee YK, Lim CY, Teng WL, Ouwehand AC, Tuomola EM, Salminen S. Quantitative approach in the study of adhesion of lactic acid bacteria to intestinal cells and their competition with enterobacteria. Appl Environ Microbiol. 2000;66:3692–3697.
- Kim Y, Kim SH, Whang KY, Kim YJ, Oh S. Inhibition of Escherichia coli O157: h7attachment by interactions between lactic acid bacteria and intestinal epithelial cells. J Microbiol Biotechnol. 2008;18:1278–1285.
- Gueimonde M, Margolles A, de Los Reyes-Gavilan CG, Salminen S. Competitive exclusion of enteropathogens from human intestinal mucus by bifidobacterium strains with acquired resistance to bile–a preliminary study. I J Food Microbiol. 2007;113:228–232. doi:10.1016/j.ijfoodmicro.2006.05.017.
- Mattar AF, Teitelbaum DH, Drongowski R, Yongyi F, Harmon C, Coran A. Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model. Pediatr Surg Int. 2002;18:586–590. doi:10.1007/s00383-002-0855-7.
- Mack DR, Ahrne S, Hyde L, Wei S, Hollingsworth MA. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut. 2003;52:827–833.
- Fujimura S, Tsuchiya C, Sugita H. Detection of Lactobacillus gasseri OLL2716 strain administered with yogurt drink in gastric mucus layer in humans. Lett Appl Microbiol. 2006;43:578–581. doi:10.1111/j.1472-765X.2006.02017.x.
- Valeur N, Engel P, Carbajal N, Connolly E, Ladefoged K. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl Environ Microbiol. 2004;70:1176–1181.
- Johansson ML, Molin G, Jeppsson B, Nobaek S, Ahrné S, Bengmark S. Administration of different Lactobacillus strains in fermented oatmeal soup: in vivo colonization of human intestinal mucosa and effect on the indigenous flora. Appl Environ Microbiol. 1993;59:15–20.
- Shibahara-Sone H, Gomi A, Iino T, Kano M, Nonaka C, Watanabe O, Miyazaki K, Ohkusa T. Living cells of probiotic Bifidobacterium bifidum YIT 10347 detected on gastric mucosa in humans. Benef Microbes. 2016;7:319–326. doi:10.3920/bm2015.0138.
- Yang Y, Galle S, Le MH, Zijlstra RT, Ganzle MG. Feed fermentation with reuteran- and levan-producing lactobacillus reuteri reduces colonization of weanling pigs by enterotoxigenic escherichia coli. Appl Environ Microbiol. 2015;81:5743–5752. doi:10.1128/aem.01525-15.
- Riboulet-Bisson E, Sturme MHJ, Jeffery IB, O’Donnell MM, Neville BA, Forde BM, Claesson MJ, Harris H, Gardiner GE, Casey PG, et al. Effect of Lactobacillus salivarius bacteriocin Abp118 on the mouse and pig intestinal microbiota. PLoS One. 2012;7:e31113. doi:10.1371/journal.pone.0031113.
- Crittenden R, Saarela M, Mättö J, Ouwehand AC, Salminen S, Pelto L, Vaughan EE, Vos WMD, Wright AV, Fondén R, et al. Lactobacillus paracasei subsp. paracasei F19: survival, ecology and safety in the human intestinal tract-A survey of feeding studies within the PROBDEMO project. Microb Ecol Health Dis. 2002;14:22–26. doi:10.1080/089106002760003314.
- Goossens DA, Jonkers DM, Russel MG, Stobberingh EE, Stockbrugger RW. The effect of a probiotic drink with Lactobacillus plantarum 299v on the bacterial composition in faeces and mucosal biopsies of rectum and ascending colon. Aliment Pharmacol Ther. 2006;23:255–263. doi:10.1111/j.1365-2036.2006.02749.x.
- Alander M, Satokari R, Korpela R, Saxelin M, Vilpponen-Salmela T, Mattila-Sandholm T, von Wright A. Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl Environ Microbiol. 1999;65:351–354.
- Gianotti L, Morelli L, Galbiati F, Rocchetti S, Coppola S, Beneduce A, Gilardini C, Zonenschain D, Nespoli A, Braga M. A randomized double-blind trial on perioperative administration of probiotics in colorectal cancer patients. World J Gastroenterol. 2010;16:167–175.
- Charbonneau D, Gibb RD, Quigley EM. Fecal excretion of Bifidobacterium infantis 35624 and changes in fecal microbiota after eight weeks of oral supplementation with encapsulated probiotic. Gut Microbes. 2013;4:201–211. doi:10.4161/gmic.24196.
- Alander M, Mättö J, Kneifel W, Johansson M, Kögler B, Crittenden R, Mattila-Sandholm T, Saarela M. Effect of galacto-oligosaccharide supplementation on human faecal microflora and on survival and persistence of bifidobacterium lactis Bb-12 in the gastrointestinal tract. Int Dairy J. 2001;11:817–825. doi:10.1016/S0958-6946(01)00100-5.
- Firmesse O, Mogenet A, Bresson JL, Corthier G, Furet JP. Lactobacillus rhamnosus R11 consumed in a food supplement survived human digestive transit without modifying microbiota equilibrium as assessed by real-time polymerase chain reaction. J Mol Microbiol Biotechnol. 2008;14:90–99. doi:10.1159/000106087.
- Rochet V, Rigottier-Gois L, Sutren M, Krementscki M-N, Andrieux C, Furet J-P, Tailliez P, Levenez F, Mogenet A, Bresson J-L, et al. Effects of orally administered Lactobacillus casei DN-114 001 on the composition or activities of the dominant faecal microbiota in healthy humans. Br J Nutr. 2006;95:421–429.
- Garrido D, Suau A, Pochart P, Cruchet S, Gotteland M. Modulation of the fecal microbiota by the intake of a Lactobacillus johnsonii La1-containing product in human volunteers. FEMS Microbiol Lett. 2005;248:249–256. doi:10.1016/j.femsle.2005.05.045.
- Goossens D, Jonkers DMAE, Russel MGVM, Stobberingh E, Van Den Bogaard AEJM, Stockbrügger R. The effect of Lactobacillus plantarum 299v on the bacterial composition and metabolic activity in faeces of healthy volunteers: a placebo-controlled study on the onset and duration of effects. Aliment Pharmacol Ther. 2003;18:495–505.
- Smith TJ, Anderson D, Margolis LM, Sikes A, Young AJ. Persistence of Lactobacillus reuteri DSM17938 in the human intestinal tract: response to consecutive and alternate-day supplementation. J Am Coll Nutr. 2011;30:259–264.
- Jacobsen CN, Rosenfeldt Nielsen V, Hayford AE, Møller PL, Michaelsen KF, Paerregaard A, Sandström B, Tvede M, Jakobsen M. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol. 1999;65:4949–4956.
- Sierra S, Muto M, Yaeshima T, Iwatsuki K, Aihara H, Ohashi Y, Fujisawa T. Intestinal and immunological effects of daily oral administration of Lactobacillus salivarius CECT5713 to healthy adults. Anaerobe. 2010;16:195–200. doi:10.1016/j.anaerobe.2010.02.001.
- Frese SA, Hutkins RW, Walter J. Comparison of the colonization ability of autochthonous and allochthonous strains of lactobacilli in the human gastrointestinal tract. AdvMicrobiol. 2012;2:399. doi:10.4236/aim.2012.23051.
- Tannock GW, Munro K, Harmsen HJ, Welling GW, Smart J, Gopal PK. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl Environ Microbiol. 2000;66:2578–2588.
- Suwal S, Wu Q, Liu W, Liu Q, Sun H, Liang M, Gao J, Zhang B, Kou Y, Liu Z, et al. The probiotic effectiveness in preventing experimental colitis is correlated with host gut microbiota. Front Microbiol. 2018;9:2675. doi:10.3389/fmicb.2018.02675.
- Abildgaard A, Kern T, Pedersen O, Hansen T, Wegener G Lund S. The antidepressant-like effect of probiotics and their faecal abundance may be modulated by the cohabiting gut microbiota in rats. Eur Neuropsychopharmacol. 2018. doi:10.1016/j.euroneuro.2018.10.011.
- Hod K, Dekel R, Aviv Cohen N, Sperber A, Ron Y, Boaz M, Berliner S, Maharshak N. The effect of a multispecies probiotic on microbiota composition in a clinical trial of patients with diarrhea‐predominant irritable bowel syndrome. Neurogastroenterol & Motil. 2018:e13456. doi:10.1111/nmo.2018.30.issue-12.
- Lebeer S, Bron PA, Marco ML, Van Pijkeren J-P, O’Connell Motherway M, Hill C, Pot B, Roos S, Klaenhammer T. Identification of probiotic effector molecules: present state and future perspectives. Curr Opin Biotechnol. 2018;49:217–223. doi:10.1016/j.copbio.2017.10.007.
- Sanders M, Merenstein D, Merrifield C, Hutkins R. Probiotics for human use. Nutr Bulletin. 2018;43:212–225. doi:10.1111/nbu.2018.43.issue-3.
- van Baarlen P, Troost FJ, van Hemert S, van der Meer C, de Vos WM, de Groot PJ, Hooiveld GJEJ, Brummer R-JM, Kleerebezem M. Differential NF-kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc Natl Acad Sci U S A. 2009;106:2371–2376. doi:10.1073/pnas.0809919106.
- Warda AK, Rea K, Fitzgerald P, Hueston C, Gonzalez-Tortuero E, Dinan TG, Hill C. Heat-killed lactobacilli alter both microbiota composition and behaviour. Behav Brain Res. 2018;362:213–223. doi:10.1016/j.bbr.2018.12.047.
- Sugahara H, Yao R, Odamaki T, Xiao JZ. Differences between live and heat-killed bifidobacteria in the regulation of immune function and the intestinal environment. Benef Microbes. 2017;8:463–472. doi:10.3920/bm2016.0158.
- He X, Alston CL, Hopton S, He L, Hargreaves IP, Falkous G, Oláhová M, McFarland R, Turnbull DM, Rocha MC, et al. Lactobacillus rhamnosus GG supernatant enhance neonatal resistance to systemic Escherichia coli K1 infection by accelerating development of intestinal defense. Sci Rep. 2017;7:43305. doi:10.1038/srep43305.
- Wang L, Cao H, Liu L, Wang B, Walker WA, Acra SA, Yan F. Activation of epidermal growth factor receptor mediates mucin production stimulated by p40, a Lactobacillus rhamnosus GG-derived protein. J Biol Chem. 2014;289:20234–20244. doi:10.1074/jbc.M114.553800.
- Wang Y, Liu Y, Sidhu A, Ma Z, McClain C, Feng W. Lactobacillus rhamnosus GG culture supernatant ameliorates acute alcohol-induced intestinal permeability and liver injury. Am J Physiol Gastrointest Liver Physiol. 2012;303(1):G32–G41. doi:10.1152/ajpgi.00024.2012.
- Korem T, Zeevi D, Suez J, Weinberger A, Avnit-Sagi T, Pompan-Lotan M, Matot E, Jona G, Harmelin A, Cohen N, et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science. 2015;349:1101–1106. doi:10.1126/science.aac4812.
- Sanders ME, Benson A, Lebeer S, Merenstein DJ, Klaenhammer TR. Shared mechanisms among probiotic taxa: implications for general probiotic claims. Curr Opin Biotechnol. 2018;49:207–216. doi:10.1016/j.copbio.2017.09.007.
- Begley M, Hill C, Gahan CG. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol. 2006;72:1729–1738. doi:10.1128/aem.72.3.1729-1738.2006.
- Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2018. doi:10.1038/s41575-018-0061-2.
- Kristensen NB, Bryrup T, Allin KH, Nielsen T, Hansen TH, Pedersen O. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med. 2016;8:52. doi:10.1186/s13073-016-0300-5.
- Khalesi S, Bellissimo N, Vandelanotte C, Williams S, Stanley D, Irwin C. A review of probiotic supplementation in healthy adults: helpful or hype? Eur J Clin Nutr. 2018. doi:10.1038/s41430-018-0135-9.
- Vandeputte D, Kathagen G, D’hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551:507–511. doi:10.1038/nature24460.
- Korpela K, Pitre FE, Pagé AP, Marleau J, Guidi Nissim W, St-Arnaud M, Labrecque M, Joly S, Yergeau E, Brereton NJB. Probiotic supplementation restores normal microbiota composition and function in antibiotic-treated and in caesarean-born infants. Microbiome. 2018;6:182. doi:10.1186/s40168-018-0567-4.
- McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. doi:10.1126/scitranslmed.3002701.
- Veiga P, Pons N, Agrawal A, Oozeer R, Guyonnet D, Brazeilles R, Faurie JM, van Hylckama Vlieg JE, Houghton LA, Whorwell PJ, et al. Changes of the human gut microbiome induced by a fermented milk product. Sci Rep. 2014;4:6328. doi:10.1038/srep06328.
- Eloe-Fadrosh EA, Brady A, Crabtree J, Drabek EF, Ma B, Mahurkar A, Ravel J, Haverkamp M, Fiorino A-M, Botelho C, et al. Functional dynamics of the gut microbiome in elderly people during probiotic consumption. MBio. 2015;6. doi:10.1128/mBio.00231-15.
- Asahara T, Shimizu K, Nomoto K, Hamabata T, Ozawa A, Takeda Y. Probiotic bifidobacteria protect mice from lethal infection with Shiga toxin-producing Escherichia coli O157:H7. Infect Immun. 2004;72:2240–2247.
- Tejero-Sarinena S, Barlow J, Costabile A, Gibson GR, Rowland I. In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: evidence for the effects of organic acids. Anaerobe. 2012;18:530–538. doi:10.1016/j.anaerobe.2012.08.004.
- Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3:777–788. doi:10.1038/nrmicro1273.
- Fayol-Messaoudi D, Berger CN, Coconnier-Polter MH, Lievin-Le Moal V, Servin AL. pH-, Lactic acid-, and non-lactic acid-dependent activities of probiotic lactobacilli against salmonella enterica serovar typhimurium. Appl Environ Microbiol. 2005;71:6008–6013. doi:10.1128/aem.71.10.6008-6013.2005.
- Grazul H, Kanda LL, Gondek D. Impact of probiotic supplements on microbiome diversity following antibiotic treatment of mice. Gut Microbes. 2016;7:101–114. doi:10.1080/19490976.2016.1138197.
- Madden JA, Dhuna V, Singh J, Kamboj SS, Nijjar KK, Agrewala JN. Effect of probiotics on preventing disruption of the intestinal microflora following antibiotic therapy: a double-blind, placebo-controlled pilot study. Int Immunopharmacol. 2005;5:1091–1097. doi:10.1016/j.intimp.2005.02.006.
- Kabbani TA, Pallav K, Dowd SE, Villafuerte-Galvez J, Vanga RR, Castillo NE, Hansen J, Dennis M, Leffler DA, Kelly CP. Prospective randomized controlled study on the effects of saccharomyces boulardii CNCM I-745 and amoxicillin-clavulanate or the combination on the gut microbiota of healthy volunteers. Gut Microbes. 2017;8:17–32. doi:10.1080/19490976.2016.1267890.
- Brown K, Godovannyi A, Ma C, Zhang Y, Ahmadi-Vand Z, Dai C, Gorzelak MA, Chan Y, Chan JM, Lochner A, et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J. 2016;10:321–332. doi:10.1038/ismej.2015.114.
- Candon S, Perez-Arroyo A, Marquet C, Valette F, Foray A-P, Pelletier B, Milani C, Ventura M, Bach J-F, Chatenoud L, et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS One. 2015;10:e0125448. doi:10.1371/journal.pone.0125448.
- Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi:10.1038/nature07336.
- Boursi B, Mamtani R, Haynes K, Yang YX. The effect of past antibiotic exposure on diabetes risk. Eur J Endocrinol. 2015;172:639–648. doi:10.1530/eje-14-1163.
- Mikkelsen KH, Knop FK, Frost M, Hallas J, Pottegard A. Use of antibiotics and risk of type 2 diabetes: a population-based case-control study. J Clin Endocrinol Metab. 2015;100:3633–3640. doi:10.1210/jc.2015-2696.
- Ajslev TA, Andersen CS, Gamborg M, Sorensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes (Lond). 2011;35:522–529. doi:10.1038/ijo.2011.27.
- Azad MB, Bridgman SL, Becker AB, Kozyrskyj AL. Infant antibiotic exposure and the development of childhood overweight and central adiposity. Int J Obes (Lond). 2014;38:1290–1298. doi:10.1038/ijo.2014.119.
- Bailey LC, Forrest CB, Zhang P, Richards TM, Livshits A, DeRusso PA. Association of antibiotics in infancy with early childhood obesity. JAMA Pediatr. 2014;168:1063–1069. doi:10.1001/jamapediatrics.2014.1539.
- Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi:10.1038/nature11400.
- Murphy R, O’Donnell TA, Frisby CL, Li H, Wittert GA, Page AJ. Antibiotic treatment during infancy and increased body mass index in boys: an international cross-sectional study. Int J Obes (Lond). 2014;38:1115–1119. doi:10.1038/ijo.2013.218.
- Saari A, Virta LJ, Sankilampi U, Dunkel L, Saxen H. Antibiotic exposure in infancy and risk of being overweight in the first 24 months of life. Pediatrics. 2015;135:617–626. doi:10.1542/peds.2014-3407.
- Arvonen M, Virta LJ, Pokka T, Kroger L, Vahasalo P. Repeated exposure to antibiotics in infancy: a predisposing factor for juvenile idiopathic arthritis or a sign of this group’s greater susceptibility to infections? J Rheumatol. 2015;42:521–526. doi:10.3899/jrheum.140348.
- Droste JH, Wieringa MH, Weyler JJ, Nelen VJ, Vermeire PA, Van Bever HP. Does the use of antibiotics in early childhood increase the risk of asthma and allergic disease? Clin Exp Allergy. 2000;30:1547–1553.
- Hoskin-Parr L, Teyhan A, Blocker A, Henderson AJ. Antibiotic exposure in the first two years of life and development of asthma and other allergic diseases by 7.5 yr: a dose-dependent relationship. Pediatr Allergy Immunol. 2013;24:762–771. doi:10.1111/pai.12153.
- Johnson CC, Poulsen LK, Platzer MH, Pedersen MH, Boltz-Nitulescu G, Skov PS, Jensen-Jarolim E. Antibiotic exposure in early infancy and risk for childhood atopy. J Allergy Clin Immunol. 2005;115:1218–1224. doi:10.1016/j.jaci.2005.04.020.
- Ong MS, Umetsu DT, Mandl KD. Consequences of antibiotics and infections in infancy: bugs, drugs, and wheezing. Ann Allergy Asthma Immunol. 2014;112:441–445 e441. doi:10.1016/j.anai.2014.01.022.
- Risnes KR, Belanger K, Murk W, Bracken MB. Antibiotic exposure by 6 months and asthma and allergy at 6 years: findings in a cohort of 1,401 US children. Am J Epidemiol. 2011;173:310–318. doi:10.1093/aje/kwq400.
- Virta L, Auvinen A, Helenius H, Huovinen P, Kolho KL. Association of repeated exposure to antibiotics with the development of pediatric Crohn’s disease–a nationwide, register-based finnish case-control study. Am J Epidemiol. 2012;175:775–784. doi:10.1093/aje/kwr400.

