ABSTRACT
The enrichment of Fusobacterium nucleatum (Fn) has been identified in CRC patients and associated with worse outcomes. However, whether Fn was involved in the metastasis of CRC was not well determined. Here, we found that the abundance of Fn was significantly increased in CRC patients with lymph nodes metastasis. To further clarify the role of Fn in CRC metastasis, we performed transwell and wound healing assays after incubating CRC cell lines with or without Fn and injected Fn-treated or untreated CRC cells into nude mice via tail vein. The results indicated that Fn infection promoted CRC cells migration in vitro, as well as lung metastasis in vivo. Interestingly, colonization of Fn was detected in metastatic lung lesions of nude mice by fluorescence in situ hybridization. Mechanistically, RNA sequencing and validation study revealed that Fn significantly upregulated the expression of long non-coding RNA Keratin7-antisense (KRT7-AS) and Keratin7 (KRT7) in CRC cells. Importantly, Fn-induced CRC lung metastasis was attenuated by the depletion of KRT7-AS. In addition, KRT7-AS facilitated CRC cells migration by upregulating KRT7. Subsequently, we found that NF-κB signaling pathway was involved in the upregulation of KRT7-AS upon Fn infection. In conclusion, Fn infection upregulated KRT7-AS/KRT7 by activating NF-κB pathway, which promoted CRC cell migration in vitro and metastasis in vivo.
Introduction
Colorectal cancer (CRC) is the fourth most common cancer and the second leading cause of cancer-related death worldwide.Citation1 Approximately 25% of patients with CRC show synchronous metastases while another 25% develop metastases throughout the course of their disease.Citation2 The prognosis of CRC patients with distant metastases is dismal with a 5-year survival rate of less than 10%.Citation3 Unfortunately, the underlying mechanisms for the metastasis of CRC have not been fully clarified.
Recent evidence suggested that host–microflora interactions might play a key role in the initiation and progression of CRC. Study indicated that gavage of stool samples from patients with CRC promoted intestinal carcinogenesis in germ-free and conventional mice, which confirmed the direct pro-tumorigenic effect of gut microbiota.Citation4 Identification of specific carcinogenic microbiota remains an important area for CRC research. Previous studies have documented that several microbiota were involved in carcinogenesis of CRC, including Peptostreptococcusanaerobius,Citation5 enterotoxigenic Bacteriodesfragiles,Citation6 Escherichia coli,Citation7 and Fusobacterium nucleatum(Fn).Citation8 However, previous studies mainly focused on the proliferative effects of microbiota, while their roles in the metastasis of CRC were not well explored.
Fn is a gram-negative anaerobic bacterium prevalent in the oral cavity which has gained significant attention for its potential role in CRC.Citation9–Citation12 The enrichment of Fn in CRC tissues compared to the adjacent normal tissues has been confirmed by several studies.Citation11,Citation13,Citation14 Epithelial barrier defects that occurred in sites of dysplasia might allow Fn to thrive in the local tumor environment.Citation15 Fn possesses key pathogenic features that may allow it to predominate. Fap2, a surface protein of Fn, could bind to tumor-expressed Gal-GalNAc, which resulted in the enrichment of Fn in CRC.Citation16 FadA, a unique virulence factor of Fn, could bind to E-cadherin and activate β-catenin, which stimulated tumor cell growth in CRC. Additionally, introduction of Fn to ApcMin/+ mice accelerated tumorigenesis.Citation17 Furthermore, the presence of Fn might be a poor prognostic biomarker in CRC. The level of Fn DNA presented in CRC tissues was correlated with higher CRC-specific mortality.Citation18 Meanwhile, Fn was associated with CRC recurrence and resistance to chemotherapy by activating the autophagy pathway.Citation19
Recently, some clinical evidence suggested that the enrichment of Fn might be related to CRC metastasis.Citation11,Citation13,Citation20–Citation22 However, the role of Fn in CRC metastasis and its underlying mechanism remains unclear. This study first clarified the relationship between Fn infection and CRC metastasis. We found Fn infection upregulated KRT7-AS/KRT7 by activating NF-κB pathway, which promoted CRC cell migration in vitro and metastasis in vivo.
Results
Fn promoted CRC cells migration and metastasis
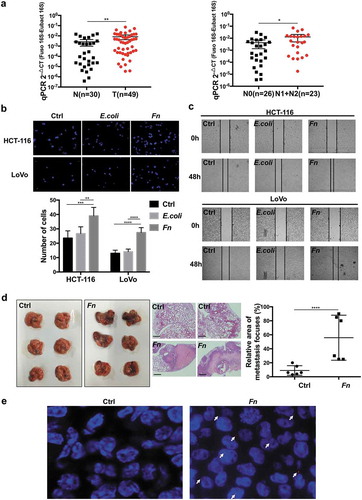
To identify the clinical significance of Fn, we performed qRT-PCR to quantify the abundance of Fn in fecal samples from CRC patients and healthy controls. The results indicated an enrichment of Fusobacterium spp. in the feces of CRC patients compared with healthy controls. Meanwhile, CRC patients with positive lymph nodes metastasis showed more abundance of Fn in feces compared to CRC patients without lymph nodes metastasis (). To evaluate the effect of Fn infection on CRC cell migration, we performed transwell and wound healing assays after incubating HCT-116 or LoVo cells with Fn for 12 h. The results indicated that Fn infection significantly enhanced HCT-116 and LoVo cells migration compared with E. coli DH5a or PBS control treatment (,c). However, heat-killed Fn was unable to promote HCT-116 and LoVo cells migration (Supplementary Figure 1a,b). To confirm the results in vitro, we injected Fn-treated or PBS-treated HCT-116 cells into nude mice via tail vein. In accordance with the results in vitro, Fn-treated cells induced more metastatic nodules in lung than PBS-treated cells (). Notably, after intravenous injection of Fn-treated cells into nude mice, Fn was detected in the metastatic lung lesions by FISH assay ().
Figure 1. Fn promoted CRC cells migration and metastasis. (a) The abundance of Fn in the feces of CRC patients (T, n = 49) versus healthy people (N, n = 30).The abundance of Fn is higher in the feces of CRC patients with lymph nodes metastasis (N1+ N2, n = 23) than those without metastasis (N0, n = 26). (b,c) HCT-116 and LoVo cells were incubated with PBS control, E. coli or Fn. The migration ability of cells was evaluated by transwell assay (b) or wound healing assay (c). In transwell assay, migrated cells were stained with DAPI and images were randomly taken under microscope. Every 6 fields were counted for each sample. (d) HCT-116 cells were incubated with Fn or PBS for 24 h and then injected into BALB/C nude mice via tail vein (n = 6 per group). The left panel shows macroscopic lungs of nude mice. The area of metastatic lesions was labeled by dashed circles. The middle panel indicates the hematoxylin-eosin (H&E) staining of lung metastasis (scale bar = 100 μm), two representative images per group. The right panel shows the statistical analysis of metastasis focuses. (e) Fn in lung metastasis tissues was detected by FISH. The colonization of Fn was detected in lung metastasis from mice injected with Fn-treated cells (1000 times magnification power). The white arrows indicate positive stain. All data are shown as mean ± SD, *p < .05, **p < .01, ***p < .001, ****p < .0001 (Mann–Whitney test was used in a, unpaired Student’s t test was used in b and d). Ctrl, control; E.coli, Escherichia coli; Fn, Fusobacterium nucleatum.
KRT7-AS/KRT7 expression is upregulated in Fn-treated CRC cells
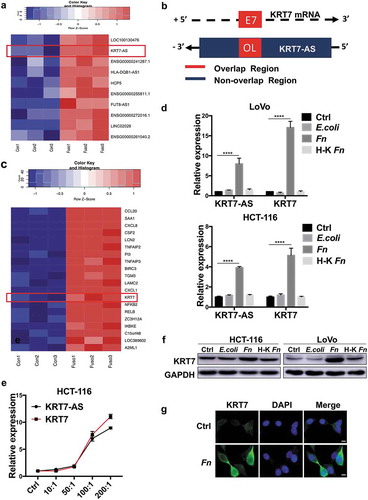
Long-non-coding RNAs (lncRNAs) were reported to play essential roles in the metastasis of CRC.Citation23,Citation24 However, the interplay between Fn infection and lncRNA has never been identified. To explore whether lncRNAs were involved in Fn infection-induced CRC metastasis, we performed RNA sequencing to evaluate the expression profiles of lncRNAs after incubating LoVo cells with Fn or PBS for 24 h. The RNA sequencing results showed that Fn treatment significantly upregulated 43 lncRNAs, while downregulated 63 lncRNAs (FDR<0.05, |log2 (fold change)|>1) (Supplementary Table 1). Among these differentially expressed lncRNA, we were particularly interested in the lncRNA Homo sapiens keratin 7 antisense RNA (KRT7-AS), which was one of the most differentially upregulated lncRNAs(FDR <0.0001, fold change = 6.06) (, Supplementary Figure 2). KRT7-AS was reported to be a single antisense RNA that was transcribed from the negative strand of the keratin 7 (KRT7) ().Citation25 Interestingly, RNA sequencing analysis also revealed that KRT7 mRNA was consistently upregulated in Fn-treated cells (FDR<0.0001, fold change = 7.5) (). To validate RNA sequencing results, we performed qRT-PCR assay after incubating HCT-116 or LoVo cells with live or heat-killed Fn, E.coli or PBS control for 24 h. Consistently, both KRT7-AS and KRT7 were significantly upregulated in live Fn-treated cells, while not in E.coli or heat-killed Fn-treated cells (). In addition, the expression of KRT7-AS and KRT7 were simultaneously increased with the increasing Fn multiplicity of infection (MOI) (). We confirmed that Fn infection increased the protein levels of KRT7 by both Western blot and immunofluorescence assays (,g). Collectively, these results showed that Fn infection upregulated both KRT7-AS and KRT7 in CRC cells.
Figure 2. KRT7-AS/KRT7 expression is upregulated in Fn-treated CRC cells. (a) Heatmap of representative differentially expressed LncRNA between Fn-treated and PBS-treated LoVo cells (n = 3 per group, fold change>2, logCPM>2, FDR<0.05). (b) The schematic representation of KRT7-AS (ensemble gene transcript ENST00000546688, on the negative DNA strand) and KRT7 mRNA (ReSeq gene NM_005556, on the positive strand). E7 indicates seventh exon of KRT7 gene. Black arrows indicates transcription direction. “+” indicates the positive strand. “-” indicates the negative strand. (c) Heatmap of representative differentially expressed mRNA between Fn-treated and PBS-treated cells (n = 3 each, fold change>2, logCPM>2, FDR <0.05). (d) HCT-116 or LoVo were incubated with live or heat-killed Fn, E.coli or PBS for 24 h. The RNA expression level of KRT7-AS and KRT7 were analyzed by qRT-PCR. (d) HCT-116 cells were incubated with Fn in different multiplicity of infection (MOI) (10:1, 50:1, 100:1, 200:1) for 24 h and the expression level of KRT7-AS and KRT7 were analyzed by qRT-PCR. (f) HCT-116 or LoVo were incubated with live or heat-killed Fn, E.coli or PBS for 24 h. KRT7 protein was analyzed by Western blot. (g) HCT-116 were incubated with Fn or PBS for 24 h, immunofluorescence assay was conducted to show KRT7 protein in HCT-116 cells (scale bar = 50 μm). All data are shown as mean ± SD, **** p < .0001 (unpaired Student’s t test). Ctrl, control; Fn, Fusobacterium nucleatum.
KRT7-AS promoted metastasis of CRC cells by regulating KRT7
To elucidate the interaction between KRT7-AS and KRT7, we then performed loss and gain of function assays in CRC cells. We found that knockdown of KRT7-AS in HCT-116 significantly suppressed mRNA and protein levels of KRT7 (,b). Meanwhile, overexpression of KRT7-AS in HCT-116 or LoVo cells upregulated the expression of KRT7 (,b, Supplementary Figure 3a). In contrast, knockdown or overexpression of KRT7 had no effect on the expression of KRT7-AS (,b, Supplementary Figure 3a,b).
Figure 3. KRT7-AS promoted metastasis of CRC cells by regulating KRT7. (a) RNA level of KRT7-AS and KRT7 was detected by qRT-PCR after knockdown of KRT7-AS (upper left panel) or overexpression of KRT7-AS (upper right panel), respectively, in HCT-116 cells. Expression level of KRT7-AS and KRT7 were detected by qRT-PCR after knockdown of KRT7 (lower left panel) or overexpression of KRT7 (lower right panel), respectively, in HCT-116 cells. (b) KRT7 protein level was detected by Western blot after knockdown or overexpression of KRT7-AS or KRT7. (c, d) HCT-116 cells were transfected with siRNA targeting KRT7-AS (KRT7) or KRT7-AS (KRT7) plasmids. Wound healing assays (c) and transwell assays (d) were performed. For transwell assay, migrated cells were stained with DAPI and images were randomly taken under microscope. Every 6 fields were counted for each sample. (e) HCT-116 cells stably transfected with KRT7-AS shRNA lentivirus were established and injected into nude mice via tail vein. The number of metastatic nodules in each slide was counted after H&E staining. (n = 8 mice per group). GFP signal shows metastatic cells infected by lentivirus in lungs. (f) HCT-116 cells were transfected with shKRT7-AS lentivirus alone or combined with KRT7 overexpression plasmid, and transwell assay was performed. Migrated cells were stained with DAPI and images were randomly taken under microscope. Every 6 fields were counted for each sample. All data are shown as mean ± SD, *p < .05, **p < .01, ***p < .001, ****p < .0001 (unpaired Student’s t test). NC, negative control; Fn, Fusobacterium nucleatum; vec, vector; si, siRNA.
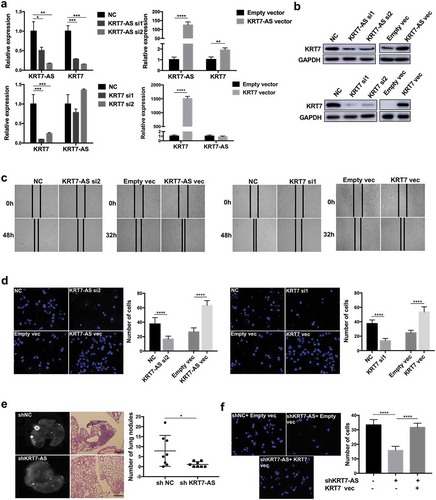
Then, we performed wound healing and transwell assays to evaluate the role of KRT7-AS and KRT7 in CRC cell migration. Knockdown of KRT7-AS or KRT7 in HCT-116 significantly inhibited cell migration, while overexpression of KRT7-AS or KRT7 in HCT-116 or LoVo promoted cell migration (,d, Supplementary Figure 3c). To confirm the function of KRT7-AS in vitro, we stably transfected HCT-116 cells with KRT7-AS shRNA or control lentivirus and then injected these infected cells into nude mice. Result indicated knockdown of KRT7-AS significantly inhibited lung metastasis of nude mice (Supplementary Figure 3d). In addition, overexpression of KRT7 restored the suppressed migration properties of HCT-116 cells induced by knockdown of KRT7-AS (). These results indicated that KRT7-AS promoted metastasis of CRC cells by upregulating KRT7.
Fn promoted metastasis of CRC by modulating KRT7-AS/KRT7
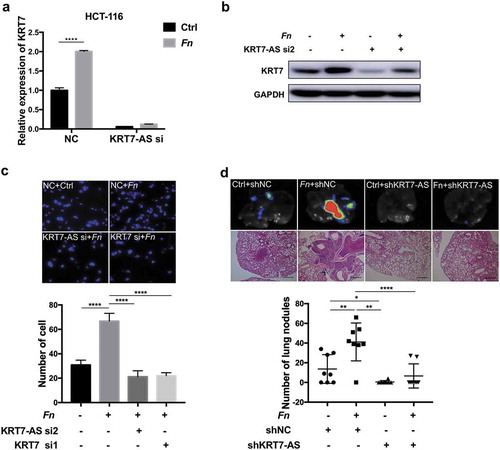
To determine whether the effect of Fn on the metastasis of CRC cells was mediated by KRT7-AS/KRT7, we treated HCT-116 cells with siRNA targeting KRT7-AS alone or together with Fn. We observed that knockdown of KRT7-AS attenuated Fn-induced upregulation of KRT7 (,b). The pro-migratory effect induced by Fn infection was abolished by knockdown of KRT7-AS or KRT7 in HCT116 cells (). To confirm these results in vitro, HCT-116 cells which were stably transfected with KRT7-AS shRNA or control lentivirus were incubated with Fn or PBS for 24 h and then injected into nude mice. Consistent with above result, Fn treatment significantly induced more lung metastatic nodules. Importantly, depletion of KRT7-AS attenuated Fn-induced CRC lung metastasis (). These results suggested that Fn promoted metastasis of CRC cells by upregulating the expression of KRT7-AS/KRT7.
Figure 4. Fn promoted metastasis of CRC by modulating KRT7-AS/KRT7. (a,b) HCT-116 cells were transfected with siRNA targeting KRT7-AS and then incubated with Fn. qRT-PCR (a) and western blot (b) indicated that knockdown of KRT7-AS suppressed the Fn-induced upregulation of KRT7. (c) HCT-116 cells were transfected with siRNA targeting KRT7-AS or KRT7 and then incubated with Fn or PBS. Transwell assay was performed. Migrated cells were stained with DAPI and images were randomly taken under microscope. Every 6 fields were counted for each sample. (d) HCT-116 cells were stably transfected with KRT7-AS shRNA or control lentivirus and then incubated with Fn or PBS for 24 h. Then, above-prepared cells were injected into nude mice via tail vein (n = 8 each group). The number of metastatic nodules in lung was counted in each slide after H&E staining (scale bar = 100 μm). GFP signal shows metastatic HCT-116 cells infected by lentivirus in lungs. All data are shown as mean ± SD, *p < .05, **p < .01, ****p < .0001 (unpaired Student’s t-test). NC, negative control; Fn, Fusobacterium nucleatum; si, siRNA.
Fn regulated KRT7-AS through NF-κB signaling pathway
The mechanism for the upregulation of KRT7-AS upon Fn infection remained unclear. Therefore, we aimed to find certain pathway regulated by Fn and identify the upstream regulators of KRT7-AS. KEGG analysis based on RNA sequencing results demonstrated that NF-κB pathway was activated by Fn infection (). We validated this result by performing Western blot after incubating HCT-116 or LoVo cells with live or heat-killed Fn, E.coli or PBS control for 24 h. Consistently, live Fn treatment dramatically upregulated phospho-p65 (activated NF-κB subunit), and downregulated IκB-α (NF-κB inhibitor) compared with heat-killed Fn, E.coli or PBS control (). Similarly, we observed increased nuclear localization of NF-κB p65 in Fn-treated CRC cells ().
Figure 5. Fn regulated KRT7-AS through NF-κB signaling pathway. (a) The KEGG analysis based on RNA sequencing result shows activation of NF-κB induced by Fn infection. (b) HCT-116 or LoVo cells were incubated with live or heat-killed Fn, E.coli or PBS for 24 h. The protein level of phospho-p65, p65, and IκB-α was measured by Western blot. (c) HCT-116 or LoVo were incubated with Fn or PBS control for 2 h. Immunofluorescence assay was performed to detect the p65 in the nucleus. DAPI was used to stain cell nucleus (scale bar = 50 μm). (d) Western blot shows the efficiency of two siRNA targeting p65. (e) HCT116 or LoVo was transfected with siRNA targeting p65. After 48 h, the qRT-PCR was performed to analyze the expression of KRT7-AS. (f) HCT-116 cells were treated with BAY-117082 (Selleck, USA) in different concentrations (5 μM, 10 μM, 20 μM) for 2 h (upper panel). HCT-116 cells were treated with 20 μM BAY-117082 for 2 h followed by Fn incubation for 4 h (lower panel). The qRT-PCR was performed to analyze the expression of KRT7-AS. (g) HEK293T and HCT-116 cells were transfected with luciferase reporter plasmid along with p65 siRNA or negative control. After 48 h, the luciferase activity was measured. The transfection efficiency data were normalized by dividing the Firefly luciferase activities with that of Renilla luciferase. All data are shown as mean ± SD, *p < .05, **p < .01, ***p < .001, ****p < .0001 (unpaired Student’s t-test). Ctrl, control; NC, negative control. Fn, Fusobacterium nucleatum; si, siRNA.
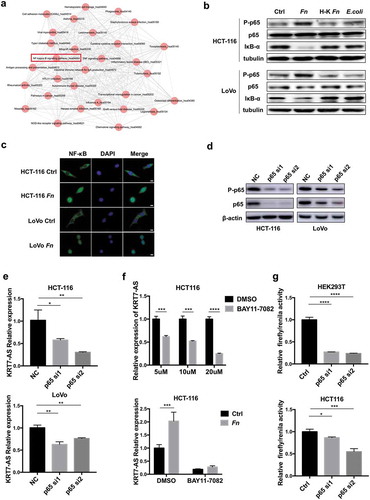
We next used two software packages (TESS and FIMO)Citation26,Citation27 to predict transcriptional factor binding to the promotor region of KRT7-AS. Interestingly, the consensus binding sequence (−2470-2461 bp 5ʹ-GGGAACCCCA-3ʹ) for NF-κB p65 in the promotor region of KRT7-AS was predicted. Knockdown of NF-κB p65 in HCT-116 or LoVo significantly decreased the expression of KRT7-AS as well as KRT7 (,e, Supplementary Figure 3e). Administration with an NF-κB inhibitor, BAY-117082, at different concentrations to HCT-116 cells also significantly decreased expression of KRT7-AS and KRT7 (, Supplementary Figure 3f). In addition, the upregulation of KRT7-AS by Fn infection was attenuated when HCT-116 cells were pretreated with NF-κB inhibitor, BAY-117082 (). Next, we constructed luciferase reporter plasmid containing promotor region of KRT7-AS to determine the interaction between p65 and KRT7-AS promoter. The luciferase reporter assay showed that knockdown of p65 significantly decreased the transcriptional activity of KRT7-AS (). These results demonstrated that Fn infection increased KRT7-AS expression through activating NF-κB signaling pathway.
The expression of KRT7-AS/KRT7 in CRC patients
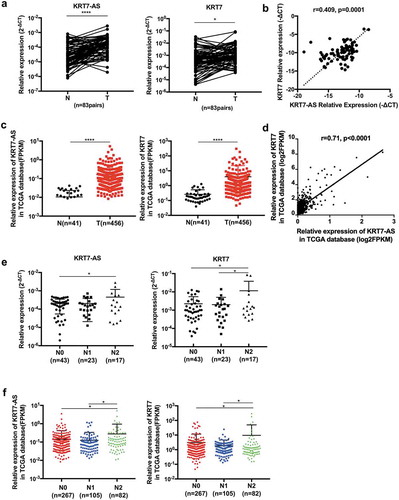
To identify the clinical significance of KRT-AS and KRT7, we performed qRT-PCR to quantify the expression of KRT7-AS and KRT7 using fresh frozen tissues from CRC patients. The expression of KRT7-AS and KRT7 was significantly increased in CRC tissues compared with adjacent normal tissues (n = 83) (). Consistently published data from The Cancer Genome Atlas (TCGA) also demonstrated that KRT7-AS and KRT7 expression were significantly elevated in colon cancer tissues compared with normal tissues (). Furthermore, the KRT7-AS expression was positively associated with KRT7 expression both in our tested CRC tissues and TCGA database (,d). Though the expression of KRT7-AS and KRT7 were not increased in N1 patients compared with N0 patients, both our tested CRC tissues and TCGA database consistently showed that the expression of KRT7-AS and KRT7 were significantly increased in CRC tissue of N2 patients. These results indicated that both KRT7-AS and KRT7 were positively associated with more advanced N stage in CRC patients (,f).
Figure 6. The expression of KRT7-AS/KRT7 in CRC patients. (a) The qRT-PCR analysis of the expression of KRT7-AS and KRT7 in CRC tissues and their adjacent normal tissues (n = 83 pairs). (b) Correlation analysis of the expression of KRT7-AS with KRT7 in our tested CRC tissues (n = 83, spearman r = 0.409, p < .0001). (c) Data in TCGA database showed the expression levels of KRT7-AS and KRT7 in colon cancer compared with those of normal controls (n = 456 vs.41). (d) Correlation analysis of the expression of KRT7-AS with KRT7 in TCGA database (n = 456, spearman r = 0.71, p < .0001). (e, f) The expression of KRT7-AS and KRT7 among CRC patients with different N stage (N0, N1, N2) in our tested CRC tissue (e) and TCGA database (f). All data are shown as mean ± SD, *p < .05, **p < .01, ***p < .001, ****p < .0001 (Wilcoxon matched-pairs signed-rank test was used in a, Mann–Whitney test was used in c, spearman correlation analysis was used in b and d, one-way ANOVA was used in e and f). Fn, Fusobacterium nucleatum.
Discussion
In the present study, we observed that the abundance of Fn was significantly increased in CRC patients with lymph nodes metastasis than those without lymph nodes metastasis. Furthermore, we reported that Fn infection enhanced CRC cells migration in vitro and increased lung metastasis in vivo. Several previous studies also indicated that the abundance of Fn was associated with TNM stage and survival outcomes in CRC patients.Citation11,Citation13,Citation18,Citation20,Citation21 Interestingly, Bullman et al. recently found identical Fusobacterium strains in human primary CRC tissues and paired metastatic tumors in liver, suggesting these bacteria may migrate with colorectal cancer cells to distant sites.Citation22 Inconsistent with this assumption, we also found the existence of Fn in metastatic lung lesions from mice which were intravenously injected with Fn-treated cells. These findings indicated that Fn not only plays a pro-tumorigenic role in CRC but also promotes distant metastasis.
The mechanisms for pro-metastatic effect upon Fn infection are not fully understood. Long-non-coding RNAs (lncRNAs) were reported to play essential roles in the metastasis of CRC.Citation23,Citation24 The crosstalk between Fn and lncRNA has never been clarified. We hypothesized that lncRNA might be involved in the progression of CRC upon Fn infection. Interestingly, we discovered a great number of differentially expressed lncRNAs induced by Fn infection, among which KRT7-AS is one of the most differentially upregulated lncRNA. Prior study has demonstrated that overexpression of KRT7-AS in gastric cancer cells promoted cell growth, S-phase entry, and migration.Citation25 However, its role in CRC was unknown. We found that KRT7-AS was associated with advanced N stage and Fn infection promoted CRC metastasis in a KRT7-AS dependent manner.
Previous study indicated that KRT7-AS upregulated protein and mRNA levels of KRT7 by stabilizing KRT7 mRNA in gastric cancer.Citation25 In our study, we also found the similar interaction between KRT7-AS and KRT7 in CRC cells. KRT7 belongs to type II cytokeratin, which is the component of cytoskeleton and epithelial intermediate filaments.Citation28,Citation29 In addition to its role in maintaining the structural integrity of the cells, intermediate filaments, such as KRT7, have interesting properties that may promote motile activities.Citation30 KRT7 was expressed in approximately 10% of all colorectal tumors and associated with high extent of tumor budding and worse outcomes.Citation31 Reyhan et al. demonstrated that KRT7 expression was more common in CRC with lymph nodes metastasis compared to those without lymph nodes metastasis.Citation32 Though KRT7 overexpression has been observed in CRC, the biological significance is unknown.Citation25,Citation31 We found that KRT7 was a downstream target of KRT7-AS and could regulate the migration of CRC cells. In this study, we attempted to clarify the mechanism for the upregulation of KRT7-AS by Fn infection. We found that Fn infection activated NF-κB pathway. The activated NF-κB P-p65 might upregulated KRT7-AS by enhancing the transcriptional activity of KRT7-AS.
In conclusion, our study confirmed an association between Fn and CRC metastasis and uncovered a novel mechanism by which Fn promoted CRC metastasis. This raises important questions on whether the depletion of Fn can prevent the early-stage CRC patients from an exacerbation and whether treatment regimens should be tailored based on the presence or absence of Fn in the metastatic spread of CRC patients.
Materials and methods
Bacterial strain and cell culture
Fn was purchased from American type culture collection (ATCC, Fusobacterium nucleatumsubsp.nucleatum ATCC 25586). Fn was grown in Columbia blood agar (bioMerieux Biotech Co., Shanghai, China) in an anaerobic jar (MITSUBISHI Gas Chemical Co., Japan) at 37℃. Heat-killed Fn was prepared by heating the bacteria at 95°C for 1 h. The E. coli strain DH5a (Takara, Japan) was cultured in Luria-Bertani medium. CRC cell lines (HCT-116, LoVo) were purchased from ATCC. HCT-116 were cultured in Maccoy 5A (Genom, China) supplemented with 10% fetal bovine serum (FBS), while LoVo was cultured in F-12K (Genom, China) supplemented with 10% FBS. All CRC cell lines were cultured at 37℃ in humidified 5% CO2 atmosphere.
Patients and public involvement
Fecal samples were collected from 49 previously untreated CRC patients and 30 healthy people from Sir Run Run Shaw Hospital, Zhejiang University School of Medicine. Fresh cancer and adjacent normal tissues were obtained from 83 CRC patients. In addition, published data of 456 colon carcinomas (with 41 normal tissues) from The Cancer Genome Atlas (TCGA) were analyzed. Written informed consents were obtained from all participants. The study was approved by the Institutional Review Board of Sir Run Run Shaw hospital.
RNA extraction and quantitative real-time PCR
Total RNAs were extracted from CRC cell lines or fresh frozen tissues using Trizol reagent (Ambion, USA). 1 μg of total RNAs were reverse transcribed to cDNA using PrimeScript RT Reagent Kit (Takara, Japan). Quantitive real-time PCR was performed in ROCHE LightCycler®480 System (Rotor gene 6000 Software, Sydney, Australia). Each reaction was assayed in triplicate in 10 μl reactions containing SYBR Premix Ex Taq (Takara, Japan), primers and template DNA. The Ct values obtained from samples were compared using 2−∆∆Ct method. GAPDH or U6 served as internal reference genes. The following primer sets were used:
KRT7-AS-F,5ʹ-TCCAACGCCTATGTTCCAGTTC-3ʹ,
KRT7-AS-R,5ʹ-ACATTGTGCCACGGACATCTTG-3ʹ;Citation25
KRT7-F,5ʹ-CGGCATCATCGCTGAGGTCAA-3ʹ,
KRT7-R,5ʹ-GCCTGGAGGGTCTCAAACTTGG-3ʹ;Citation25
U6-F,5ʹ-CTCGCTTC·GGCAGCACA-3ʹ,
U6-R,5ʹ-AACGCTTCACGAATTTGCGT-3ʹ;
GAPDH-F, 5ʹ-GCACCGTCAAGGCTGAGAAC-3ʹ,
GAPDH-R, 5ʹ-TGGTGAAGACGCCAGTGGA-3ʹ.
Fn quantification
Fn quantification was performed as described previously.Citation17,Citation19 DNA from fecal samples were extracted using QIAGEN stool kit (QIAGEN, Germany). Fn quantification was performed by real-time PCR. Relative abundance was calculated by 2−∆Ct method. Universal Eubacteria 16s was used as reference gene. The following primer sets were used:
Fusobacterium spp-F, 5ʹ-CGGGTGAGTAACGCGTAAAG-3ʹ,
Fusobacterium spp-R, 5ʹ-ACATTGTGCCACGGACATCTTG −3ʹ;Citation33
universal Eubacteria 16s-F, 5ʹ-CGGCAACGAGCGCAACCC-3ʹ,
universal Eubacteria 16s-R, 5ʹ-CCATTGTAGCACGTGTGTAGCC-3ʹ.Citation34
Western blotting
Total proteins were extracted from CRC cell lines or fresh frozen tissues using RIPA lysis buffer and quantified using BCA Protein Assay Kit (Beyotime, China). Protein was electrophoresed through 10% SDS polyacrylamide gels and then transferred to PVDF membranes. The membranes were blocked with 5% fat-free milk for 2 h and incubated with primary antibodies at 4℃ overnight. The following primary antibodies were used: KRT7 (1:1000 CST#4466), p65 (1:1000 CST#8242), P-p65 (1:1000 CST#3033) and IκB-α (1:1000 CST#9294). Membranes were then incubated with second antibodies labeled with HRP at room temperature on the following day and the signal was detected using an ECL kit (Fdbio science, China). GAPDH was used as a reference gene.
Immunofluorescence (IF)
CRC cells were seeded on chamber slides and then fixed with 4% paraformaldehyde for 10 min. The slides were incubated with 0.2% TritonX-100 (Beyotime, China) for 15 min and blocked in 3% BSA for 0.5 h. After washing by PBS for three times, the cells were incubated with primary antibodies against KRT7 (1:100 CST#4465) or NF-κB (1:400, CST #8242) at 4℃ overnight in wet chamber. On the following day, the slides were washed by PBST for three times and incubated with secondary antibodies at room temperature for 1 h. The cells were then stained with DAPI.
Transwell migration assays
CRC cells were incubated with Fn or E. coli DH5a at multiplicity of infection (MOI) of 100:1 for 12 h. Cells treated with PBS were set as negative control. The infected cells were resuspended in 200 μl medium without serum and were seeded into upper chambers of transwell chambers (Corning, USA) while lower chambers were filled with medium containing 10% FBS. After 20 h, the cells remaining in the upper chamber were removed while cells that had migrated across the transwell membrane were stained with DAPI. Images were randomly taken under fluorescence microscope. Every 6 fields were counted for each sample.
Wound healing assays
CRC cells were seeded in 6-well plates and left to grow until confluent. After wounding with yellow pipette tips, the cells of each well were incubated with Fn or E. coli DH5a at MOI of 100:1. Cell images were taken at 6, 24, 36 and 48 h, respectively.
Small interfering RNAs (siRNAs), lentivirus shRNA and plasmid
Two different siRNAs which exclusively targeted KRT7-AS RNA
(siRNA1,5ʹ-CUGAAUGUGUGCTGAGGAUCAATT-3ʹ;
siRNA2, 5ʹ-GAGGAUGAAAUGAGAUAAUTT-3ʹ)Citation25
and siRNAs targeted against KRT7 mRNA non-overlapping region
(siRNA1, 5ʹ-UGGAGGACUUCAAGAAUAATT-3ʹ;
siRNA2, 5ʹ-ACAAGCUGCUGGAGACCAATT-3ʹ)Citation25
were used. In addition, two different siRNA were used to target NF-κB p65
(siRNA1, 5ʹ-GCTGATGTGCACCGACAAGTT-3ʹ;
siRNA2, CAGAUACAGACGAUCGUCATT).
A non-targeting sequence (5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ) was used as negative control (NC). All siRNAs were synthesized by GenePharma Co. China. Lentivirus shRNAs were constructed by Genechem Co. China. Briefly, the siRNA targeting KRT7-AS (5ʹ-GAGGATGAAATGAGATAAT-3ʹ) was synthesized and a non-targeting sequence 5ʹ-TTCTCCGAACGTGTCACGT-3ʹ was set as negative control (NC). The DNA fragments containing loop-structure for specific short-hairpin RNA were cloned into GV248 (hU6-MCS-Ubiquitin-EGFP-IRES-puromycin). KRT7-AS and KRT7 overexpression plasmids were constructed by Genechem Co. China. Genes were cloned into pCDNA 3.1(+).
Cell transfection
The siRNAs were transfected into cells using Lipofectamine™ RNAiMAX (Thermo Fisher Scientific, Massachusetts, USA) in opti-MEM (Genom, China) according to the manufacturer’s protocol. Plasmids were transfected into cells using FuGENE HD transfection reagent (Promega, USA) in opti-MEM according to the manufacturer’s protocol.
RNA sequencing
LoVo cells were incubated with Fn (MOI = 100:1) or PBS for 24 h. Total RNAs were extracted from LoVo treated by Fn or PBS using Trizol reagent, and ribosomal RNA was removed using the Ribo-ZeroTMkit (Epicenter, Madison, WI, USA). Subsequently, the purified RNAs were subjected to first strand and second strand cDNA synthesis followed by adaptor ligation and enrichment with a low-cycle according to instructions of NEBNext® Ultra™ RNA Library Prep Kit for Illumina (San Diego, CA, USA). The purified library products were evaluated using the Agilent 2200 TapeStation and Qubit®2.0 (Life Technologies, USA). The libraries were paired-end sequenced (PE150, Sequencing reads were 150 bp) at Guangzhou RiboBio Co., Ltd. (China) using Illumina HiSeq 3000 platform. Raw fastq sequences were treated with Trimmomatic tools (v 0.36) to remove trailing sequences below a phred quality score of 20 and to achieve uniform sequence lengths for downstream clustering processes. Paired-end reads were aligned to the mouse reference genome mm10 and reads numbers mapped to each gene were counted by using Gfold (V1.1.2). The whole samples expression levels were presented as RPKM (expected number of Reads PerKilobase of transcript sequence per Million base pairs sequenced). The statistically significant differentially expressed genes were obtained by an adjusted p-value threshold of <0.05 and |log2(fold change)| > 1 using the edger (v 3.10.0) software. All the p-values were adjusted by FDR (False discovery rate) in multiple comparisons. To look for meaningful Fn-targeted lncRNA and mRNA, we filtered out lncRNA and mRNA with very low expression level (logCPM<2) among all the differentially expressed lncRNA and mRNA and obtained top 10 upregulated lncRNA and top 20 upregulated mRNA to do heatmap. All differentially expressed genes were used for KEGG ontology enrichment analyses. For KEGG enrichment analysis, a p value <.05 was used as the threshold to determine the significant enrichment of the gene sets. All differentially expressed lncRNA and mRNA list as well as KEGG pathways were shown in Supplementary Tables 1–3.
Lung metastasis in vivo
BALB/c female nude mice aged 5 weeks were purchased from Shanghai SLAC Laboratory Animal Co. China. HCT-116 cells were incubated with Fn at MOI of 100:1 for 24 h. Cell resuspension solutions were injected via tail vein, 106 cells in 100 μl per mouse. Two months later, the mice were sacrificed and their lungs were surgically excised, photographed, and measured. Subsequently, lung tissues were fixed in 10% formalin and embedded in paraffin for hematoxylin and eosin (H&E) staining to confirm histology. All nude mice were raised in a specific-pathogen-free condition. All animal procedures were performed in compliance with ethical standards and approved by the Animal Care Committee of Zhejiang University.
Fluorescence In Situ Hybridization (FISH)
Detection of Fn was performed by FISH on formalin-fixed paraffin-embedded (FFPE) section of lungs tissue from nude mice as described previously.Citation35,Citation36 The sequence of Fn targeted probe (5ʹ-CGCAATACAGAGTTGAGCCCTGC-3ʹ)Citation11 was synthesized by Guangzhou EXON Biological Technology Co. China.
Luciferase reporter assay
The DNA fragment of KRT7-AS promotor was amplified and cloned into the pGL3-Enhancer Vector (Taihe Biotechnology Co. China). The reporter plasmid was co-transfected with p65 siRNA or negative control siRNA into HEK293T or HCT-116 cells. Luciferase assays were performed 48 h after transfection by using Dual-Luciferase Reporter Assay System (Promega). Renilla luciferase activity was used as a reference. The transfection efficiency data were normalized by dividing the Firefly luciferase activities with that of Renilla luciferase.
Statistical analysis
Experimental results were evaluated using paired or unpaired Student’s t-test for normally distributed data and Mann–Whitney U test or Wilcoxon Sign-Rank test for non-parametric data. One-way ANOVA and Tukey’s multiple comparison were used for test among three groups. The Correlation analysis was performed using Spearman rank correlation test. Data were shown as mean ± standard deviation (SD). All p values were two-tailed and p values less than 0.05 were considered statistically significant. All statistical analyses were conducted using SPSS 22.0 or GraphPad Prism 7.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Authors’ contributions
Li.W., J.S. and W.Z. designed the experiment and supervised the study. T.S. and S.C. performed the experiments, analyzed the data and wrote the manuscript. Y.Z., J.H., Q.G., and L.W. performed the experiments. Li.W. and A.L. reviewed and revised the manuscript.
Supplemental Material
Download MS Word (6.8 MB)Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492.
- Bruckner HW, Motwani BT. Chemotherapy of advanced cancer of the colon and rectum. Semin Oncol. 1991;18:443–461.
- Dahan L, Sadok A, Formento JL, Seitz JF, Kovacic H. Modulation of cellular redox state underlies antagonism between oxaliplatin and cetuximab in human colorectal cancer cell lines. Br J Pharmacol. 2009;158(2):610–620. doi:10.1111/j.1476-5381.2009.00341.x.
- Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, Xiao X, Kwong TNY, Tsoi H, Wu WKK, et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. 2017;153(6):1621–33 e6. doi:10.1053/j.gastro.2017.08.022.
- Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, Chan AWH, Chan FKL, Sung JJY, Yu J. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology. 2017;152(6):1419–33 e5. doi:10.1053/j.gastro.2017.01.009.
- Thiele Orberg E, Fan H, Tam AJ, Dejea CM, Destefano Shields CE, Wu S, Chung L, Finard BB, Wu X, Fathi P, et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2017;10(2):421–433. doi:10.1038/mi.2016.53.
- Zhang S, Fu J, Dogan B, Scherl EJ, Simpson KW. 5-Aminosalicylic acid downregulates the growth and virulence of Escherichia coli associated with IBD and colorectal cancer, and upregulates host anti-inflammatory activity. J Antibiot (Tokyo). 2018;71(11):950–961. doi:10.1038/s41429-018-0081-8.
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi:10.1016/j.chom.2013.07.012.
- Suehiro Y, Sakai K, Nishioka M, Hashimoto S, Takami T, Higaki S, Shindo Y, Hazama S, Oka M, Nagano H, et al. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann Clin Biochem. 2017;54(1):86–91. doi:10.1177/0004563216643970.
- Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, Chen T, Wu Y. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer. 2016;139(6):1318–1326. doi:10.1002/ijc.30168.
- Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappab, and up-regulating expression of MicroRNA-21. Gastroenterology. 2017;152(4):851–66e24. doi:10.1053/j.gastro.2016.11.018.
- Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, Lam TYT, Zhang L, Wu JCY, Chan FKL, et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66(8):1441–1448. doi:10.1136/gutjnl-2016-312766.
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi:10.1101/gr.126516.111.
- Ray K. Colorectal cancer: fusobacterium nucleatum found in colon cancer tissue–could an infection cause colorectal cancer? Nat Rev Gastroenterol Hepatol. 2011;8(12):662. doi:10.1038/nrgastro.2011.208.
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491(7423):254–258. doi:10.1038/nature11465.
- Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20(2):215–225. doi:10.1016/j.chom.2016.07.006.
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi:10.1016/j.chom.2013.07.007.
- Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980. doi:10.1136/gutjnl-2015-310101.
- Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–63 e16. doi:10.1016/j.cell.2017.07.008.
- Li YY, Ge QX, Cao J, Zhou YJ, Du YL, Shen B, Wan YJ, Nie YQ. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016;22(11):3227–3233. doi:10.3748/wjg.v22.i11.3227.
- Yan X, Liu L, Li H, Qin H, Sun Z. Clinical significance of Fusobacterium nucleatum, epithelial-mesenchymal transition, and cancer stem cell markers in stage III/IV colorectal cancer patients. Onco Targets Ther. 2017;10:5031–5046. doi:10.2147/OTT.S145949.
- Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science (New York, NY). 2017;358(6369):1443–1448. doi:10.1126/science.aal5240.
- Huang H, Cai L, Li R, Ye L, Chen Z. A novel lncRNA LOC101927746 accelerates progression of colorectal cancer via inhibiting miR-584-3p and activating SSRP1. Biochem Biophys Res Commun. 2019. doi:10.1016/j.bbrc.2018.12.174.
- Yan Y, Wang Z, Qin B. A novel long noncoding RNA, LINC00483 promotes proliferation and metastasis via modulating of FMNL2 in CRC. Biochem Biophys Res Commun. 2018. doi:10.1016/j.bbrc.2018.12.090.
- Huang B, Song JH, Cheng Y, Abraham JM, Ibrahim S, Sun Z, Ke X, Meltzer SJ. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene. 2016;35(37):4927–4936. doi:10.1038/onc.2016.25.
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi:10.1093/bioinformatics/btr064.
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS, SUITE: MEME. tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. WebServer issue. doi:10.1093/nar/gkp335.
- Owens DW, Lane EB. The quest for the function of simple epithelial keratins. Bioessays. 2003;25(8):748–758. doi:10.1002/bies.10316.
- Magin TM, Vijayaraj P, Leube RE. Structural and regulatory functions of keratins. Exp Cell Res. 2007;313(10):2021–2032. doi:10.1016/j.yexcr.2007.03.005.
- Helfand BT, Chang L, Goldman RD. Intermediate filaments are dynamic and motile elements of cellular architecture. J Cell Sci. 2004;117(Pt 2):133–141. doi:10.1242/jcs.00936.
- Harbaum L, Pollheimer MJ, Kornprat P, Lindtner RA, Schlemmer A, Rehak P, Langner C. Keratin 7 expression in colorectal cancer – freak of nature or significant finding? Histopathology. 2011;59(2):225–234. doi:10.1111/j.1365-2559.2011.03694.x.
- Bayrak R, Yenidunya S, Haltas H. Cytokeratin 7 and cytokeratin 20 expression in colorectal adenocarcinomas. Pathol Res Pract. 2011;207(3):156–160. doi:10.1016/j.prp.2010.12.005.
- Li J, Lin S, Vanhoutte PM, Woo CW, Xu A. Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in apoe-/- mice. Circulation. 2016;133(24):2434–2446. doi:10.1161/CIRCULATIONAHA.115.019645.
- Denman SE, McSweeney CS. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol Ecol. 2006;58(3):572–582. doi:10.1111/j.1574-6941.2006.00190.x.
- Yu YN, Yu TC, Zhao HJ, Sun TT, Chen HM, Chen HY, An HF, Weng YR, Yu J, Li M, et al. Berberine may rescue Fusobacterium nucleatum-induced colorectal tumorigenesis by modulating the tumor microenvironment. Oncotarget. 2015;6(31):32013–32026. doi:10.18632/oncotarget.5166.
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi:10.1101/gr.126573.111.
