ABSTRACT
Diverse intestinal microbiota is frequently used in in vitro bioreactor models to study the effects of diet, chemical contaminations, or medication. However, the reproducible cultivation of fecal microbiota is challenging and the resultant communities behave highly dynamic. To approach the issue of reproducibility in in vitro models, we established an intestinal microbiota model community of reduced complexity, SIHUMIx, as a valuable model for in vitro use.
The development of the SIHUMIx community was monitored over time with methods covering the cellular and the molecular level. We used microbial flow cytometry, intact protein profiling and terminal restriction fragment length polymorphism analysis to assess community structure. In parallel, we analyzed the functional level by targeted analysis of short-chain fatty acids and untargeted metabolomics. The stability properties constancy, resistance, and resilience were approached both on the structural and functional level of the community. We show that the SIHUMIx community is highly reproducible and constant since day 5 of cultivation. Furthermore, SIHUMIx has the ability to resist and recover from a pulsed perturbation, with changes in community structure recovered earlier than functional changes.
Since community structure and function changed divergently, both levels need to be monitored at the same time to gain a full overview of the community development. All five methods are highly suitable to follow the community dynamics of SIHUMIx and indicated stability on day five. This makes SIHUMIx a suitable in vitro model to investigate the effects of e.g. medical, chemical, or dietary interventions.
Introduction
The human intestinal microbiota is essential to human health and is even considered as an extra organ, as it provides enzymes for nutrient break-down and produces essential nutrients, such as short-chain fatty acids (SCFA) and vitamins.Citation1–Citation3 It was also shown that the intestinal microbiota modulates the immune system.Citation4,Citation5 Due to the importance of the intestinal microbiota to human health and well-being, the effects of environmental stress on the intestinal microbiota can have serious consequences. For example, pesticides, but also other chemicals, like perfluoroalkyl acids or plasticizers, mainly enter the human body via the oral route thus potentially affecting the intestinal microbiota.Citation6,Citation7
Testing the impact of environmental influences (e.g., pesticides, nutrients) on the intestinal microbiota cannot be accomplished with the required throughput in animal models due to ethical and practical reasons. Thus, the development of suitable models is essential. The cultivation of intestinal bacteria under gut-like conditions is a relevant approach to gain insight into the bacterial response to environmental influences. The effect upon various treatments, e.g. of dietary compounds, the impact of pathogenic microorganisms on the intestinal microbiota or the response to chemicals has already been investigated in in vitro bioreactor systems.Citation8,Citation9 Most often fecal bacteria are cultivated in these bioreactor systems.Citation8–Citation11 However, true experimental replication is hard to achieve due to the intra-individual and longitudinal heterogeneity of fecal samples.Citation12 Additionally, a variety of methods, like denaturing gradient gel electrophoresis (DGGE),Citation13 short chain fatty acidCitation14 and next-generation 16S rRNA sequencing analysis,Citation10 have been used to define constant in vitro community states, when the community remains unchanged (often referred to as stable state). This state is essential to start the treatment, since a starting point is a pivotal element for the experimental set up.Citation15,Citation16 Depending on the method different results regarding the time of cultivation to reach such a state have been obtained.Citation16,Citation17 Proceeding from this and from an ecological point of view the term stability is ambiguous.Citation18 Grimm and Wissel (1997) point out that stability itself is not a stability property and therefore extracted main stability properties.Citation19 Some of these are constancy, resilience and resistance. E.g., (1) constancy means that a system remains essentially unchanged, (2) resistance resembles the ability of a system to remain unchanged despite a perturbation and (3) resilience describes the ability of a system to recover to a reference state after a perturbation.Citation19
To circumvent challenges regarding the community reproducibility and the identification of suitable states to introduce a treatment, we established the extended simplified human microbiota (SIHUMIx) as a model community for in vitro use. SIHUMIx comprises of eight bacterial species, Anaerostipes caccae, Bifidobacterium longum, Bacteroides thetaiotaomicron, Blautia producta, Clostridium butyricum, Clostridium ramosum, Escherichia coli and Lactobacillus plantarum, and covers the genera Firmicutes, Bacteroidetes and Proteobacteria that are dominant in human feces.Citation20 We tracked the dynamics during community adaptation with a multi-method approach, which combined standard fingerprinting and OMICs techniques. Methods like terminal restriction fragment length polymorphismCitation21 analysis, flow cytometric fingerprintingCitation22 and short chain fatty acid analysisCitation23 are widely applied in microbial community characterization. Moreover, we evaluated untargeted metabolomics and intact protein profiling,Citation24 as these methods have the potential to depict community development on the metabolic and structural level, respectively, even though these methods have not been used to follow community development yet.
Acidification of the colon lumen by a decreased luminal pH resembles a severe disturbance and has been reported from patients with active ulcerative colitis.Citation25 For those reasons, we on the one hand cultivated the SIHUMIx community unimpededly, and on the other hand perturbed the community with a decrease of pH during cultivation. The aims of our study were to clarify if the SIHUMIx community (i) behaves in a reproducible way, (ii) establishes a constant state, (iii) is sensitive and responsive to perturbations, and (iv) if such a perturbation can be traced both on the structural (FC) and metabolic level (SCFA).
Results
Development of SIHUMIx under defined chemostat conditions
In order to follow the community dynamics of SIHUMIx, pre-cultivated bacteria were inoculated into three parallel bioreactors A, B and C (day 0) and grown on complex intestinal medium (CIM, supplemental file 1: Table S1) at a dilution rate of D = 0.04 d−1. After an initial establishing phase of 24 h (day 1) the continuous cultivation was started and community structure and function were monitored daily in a 24 h interval until day 7 (). Samples were taken for flow cytometric fingerprinting analysis (FC), intact protein profiling (IPP) and t-RFLP profiling, to study possible variations on the cell, protein, and DNA level from biomass, respectively. In addition, community function was assessed with untargeted metabolomics and short chain fatty acid (SCFA) analysis from the culture supernatant.
Figure 1. Experimental-set-up and development of the SIHUMIx community. (a) The SIHUMIx community was cultivated in vitro. After a sterile run the bioreactors A, B and C were inoculated (day 0) and after 24 h establishing time continuous cultivation started. Daily samples were taken from day 1 to day 7. Bacteria pellets were analyzed by flow cytometric fingerprinting (FC), t-RFLP and intact protein profiling (IPP) to investigate community structure. Culture supernatants were used for functional analysis namely SCFA analysis and untargeted metabolomics. (b) Community development was visualized by NMDS: (1) FC, (2) IPP, (3) t-RFLP, (4) SCFA, (5) untargeted metabolomics. Grouping was based on the day of cultivation.
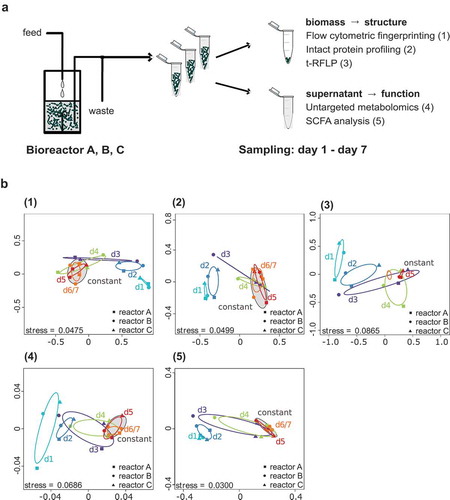
By non-metric multidimensional scaling (NMDS), we visualized upcoming community variations in the three parallel bioreactors (, Supplemental file 2: input data for NMDS analysis). All methods, FC (1), IPP (2), t-RFLP profiling (3), untargeted metabolome analysis (4), and SCFA analysis (5) revealed the same development pattern of the SIHUMIx community for all reactors. The community structure and function during the first two days of cultivation were similar and significantly different from the states of the communities at the end of the cultivation (p-value 0.03, pairwise Permanova, Supplemental file 1: Table S2, Table S3). The states of the communities on day 3 and day 4 lay between the states of the early communities and the late communities, thus mapping transitional states. Finally, the community on day 5 of all bioreactor replicates A, B and C was indistinguishable from the communities on day 6/7 (pairwise Permanova, Supplemental file 1: Table S2, Table S3).
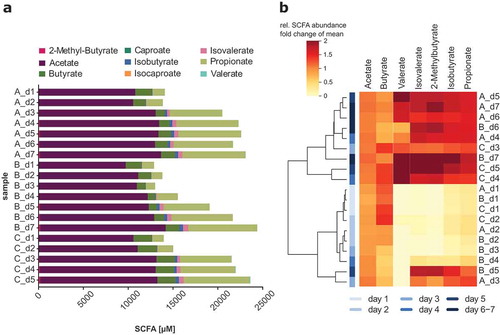
For SCFA analysis, the concentrations of non-branched SCFA acetate, propionate, butyrate, valerate and caproate and the branched SCFA isobutyrate, isovalerate, isocaproate and 2-methyl butyrate were measured. As implied from the NMDS plot (), the absolute SCFA production changed notably during the time of cultivation (, supplemental file 1: Table S4). The dominating SCFA detected in the culture supernatant of SIHUMIx were acetate, propionate and butyrate. The acetate and butyrate concentrations increased, whereas the concentration of butyrate decreased during the time of cultivation. The concentration of 2-methylbutyate, isobutyrate and isovalerate also rose during the cultivation of SIHUMIx until day 7. Moreover, we observed an increasing total SCFA concentration during the time of cultivation. Clustering analysis based on the development of SCFA production revealed that data points related to the bioreactor samples clearly arranged in two major clusters with one cluster comprising all replicates A, B and C of day 1 and day 2 and the other cluster containing all replicates of day 5 and day 6/7 (). In the first days of cultivation, the communities produced above-average levels of acetate and butyrate and below-average levels of valerate, isobutyrate, propionate, 2-methyl-butyrate, and isovalerate. During the days 5, 6 and 7 of cultivation this SCFA pattern inversed. Caproate and isocaproate were not detected or below the detection limit. The SCFA production patterns of SIHUMIx was similar in all three bioreactors.
Figure 2. Short chain fatty acid (SCFA) production of SIHUMIx in three independent, parallel bioreactors A, B and C from day 1 to day 7. (a) The absolute SCFA concentrations were determined with targeted SCFA mass spectrometry metabolomics in the culture supernatant of SIHUMIx during cultivation. (b) Heatmap and hierarchical clustering of bioreactor samples based on the absolute SCFA production of SIHUMIx. Mean normalized absolute SCFA concentrations were used for hierarchical clustering. Above mean values are colored in red, mean values in orange and below mean values in yellow. The color code in hierarchical clustering analysis shows light blue color for early days with increasing saturation for later days of cultivation.
SIHUMIx shows a high reproducibility
To assess the reproducibility of the SIHUMIx community, two batches of single strain pre-cultures were used. Bioreactors A and B were inoculated with one and bioreactor C with the other batch of pre-cultures (day 0). During NMDS analysis Bray-Curtis (BC) distances were calculated for each method (Supplemental file 3: BC distance matrices). Based on the BC distances, the average BC similarity among replicates was determined after the initial establishing phase directly after inoculation (day 1) and at a later time point (day 5) (, Supplemental file 1: Table S5). For all the individual methods but t-RFLP (p-value <0.0001), the replicate BC similarity on day 1 and day 5 was very similar (two-way ANOVA, Supplemental file 1: Table S5). Structural analyses showed a lower BC similarity (FC: 0.89 and 0.93, IPP: 0.83 and 0.75, t-RFLP: 0.67 and 0.88) than metabolic analyses (SCFA: 0.98 and 0.89, untargeted metabolomics: 0.94 and 0.96) on day 1 and day 5, respectively.
Figure 3. Reproducibility of SIHUMIx. (a) To analyze the reproducibility of SIHUMIx the Bray-Curtis (BC) similarity of bioreactor replicates A, B and C on day 1 and on day 5 (constant state) were determined. Calculations were based on different methods: flow cytometric fingerprinting analysis (FC), intact protein profiling (IPP), t-RFLP, SCFA analysis, and untargeted metabolomics. Data are shown as the mean ± standard deviation of triplicates. (b) Community structure was in detail analyzed by metaproteomics revealing the relative species abundances of the communities on day 1 and day 5 in bioreactors A and B. (c) SCFA production of the SIHUMIx communities was analyzed for the replicate bioreactors A, B and C on day 1 and day 5, respectively.
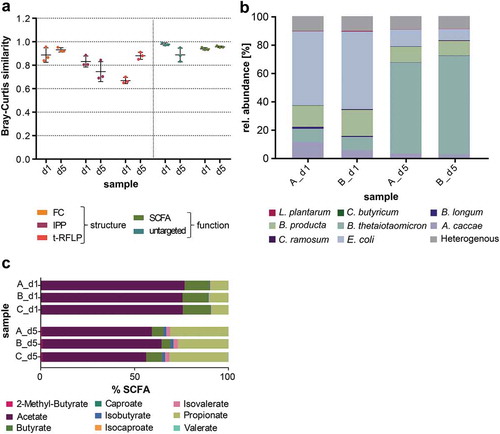
The reproducibility of SIHUMIx was proved by metaproteomics and SCFA analysis. Microbial composition of SIHUMIx was analyzed based on the abundance of species-specific proteins.Citation26,Citation27 The relative species abundances of SIHUMIx from bioreactor A and B on day 1 and day 5, respectively, were very similar (, Supplemental file 1: Table S6). The communities on day 1 mainly comprised of E. coli, B. producta, A. caccae and B. thetaiotaomicron. The remaining strains, C. butyricum, C. ramosum, B. longum and L. plantarum, contributed to less than 1% relative species abundance. On day 5 to day 7, the communities had changed considerably and comprised of more than ~60% B. thetaiotaomicron, ~10% B. producta and E. coli and ~2% A. caccae. Proteins of all other strains (C. ramosum, C. butyricum, L. plantarum, and B. longum) were detected, but they contributed to less than 2% species abundance (Supplemental file 1: Table S6). Furthermore, SCFA analysis revealed highly similar relative SCFA concentrations within the replicate bioreactors but the concentrations changed over time (). On day 1, the most abundant SCFAs in the supernatant were acetate, butyrate, and propionate in decreasing order. On day 5, the realtive SCFA composition was clearly different from that on day 1. The concentrations of acetate and butyrate decreased, but the concentration of propionate, isobutyrate, and isovalerate increased clearly on day 5 (Supplemental file 1: Table S7). Our results prove that SIHUMIx is a highly reproducible community in our bioreactor system, since community adaptation on taxonomy and SCFA levels were similar for all inocula.
SIHUMIx shows constant growth state starting on day 5
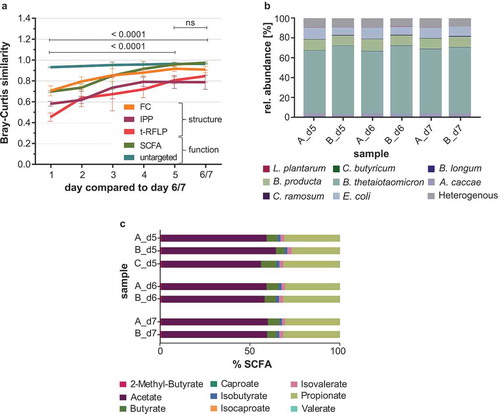
To evaluate the ability of SIHUMIx to reach a constant state, the Bray-Curtis (BC) distance values were used as described previously.Citation10 Therefore, the BC distances were calculated based on the analysis of cell abundances per cell population (technically termed as gate in FC method, Supplemental file 1: Figure S1), relative abundances of proteins, metabolites or SCFAs (IPP, untargeted and SCFA analysis, respectively) and relative abundance of terminal restriction fragments (t-RFLP, Supplemental file 3: BC distance matrices). The average BC similarity of all replicate samples (A, B, and C) compared to the replicates on days 6 and 7 (A and B) from day 1 until day 6 and 7 were visualized (). For all methods, the BC similarities followed the same trend: On day 1 and day 2, BC similarities were significantly lower compared to day 6/7 (pairwise PERMANOVA, p-values < 0.0001, Supplemental file 1: Table S8 and S9) but BC similarity increased steadily over time until day 5 (Supplemental file 1: Table S8 and S9). The average BC similarity on day 5 and day 6/7 compared to day 6/7 was nearly identical and revealed community similarity on day 5. The BC similarity on day 5 compared to day 6/7 was as high as the replicate BC similarity on day 5, respectively (Supplemental file 1: Table S5). Constancy, respectively the structural and metabolic stability, of SIHUMIx was further analyzed by metaproteomics and SCFA profiles. On day 5, day 6, and day 7 the relative species abundances were very similar () and SIHUMIx comprised mainly of B. thetaiotaomicron, B. producta, E. coli and A. caccae. The other four strains, C. butyricum, C. ramosum, B. longum and L. plantarum, contributed to less than 1% (Supplemental file 1: Table S6). SCFA analysis likewise revealed similar SCFA concentrations on day 5, day 6, and day 7 (). The major SCFAs in the supernatant were acetate, propionate, butyrate, isovalerate, and isobutyrate in decreasing order, whereas 2-methyl butyrate and isovalerate were produced only in very low concentrations (Supplemental file 1: Table S4 and Table S7).
Figure 4. Development of SIHUMIx toward the constant state. (a) The average Bray-Curtis similarity (bioreactors A, B, C) on a given day of cultivation compared to the other bioreactors on day 6 and day 7 is shown for flow cytometric fingerprinting (FC), intact protein profiling (IPP), t-RFLP, SCFA analysis, and untargeted metabolomics. Data are shown as mean ± standard deviation. Comparison of community Bray-Curtis similarity by one-way ANOVA. (b) Species abundances in the SIHUMIx communities on day 5, day 6, and day 7 of bioreactors A and B based on metaproteomics. (c) Relative SCFA composition produced by the SIHUMIx communities in replicate bioreactors A, B and C on day 5, day 6, and day 7.
SIHUMIx shows resistance and resilience after pulsed perturbation
Two bioreactors, D and E, were used to examine the dynamics of SIHUMIx when exposed to a pH-caused pulsed perturbation. Therefore, on day 4 the pH was set to 5.5 and reset to 6.5 after 24 h on day 5 (). Community dynamics were visualized in NMDS plots () on the basis of (1) flow cytometric fingerprinting (FC) and (2) SCFA analysis that represent changes on the structural and functional level, respectively (). For comparison the constant state of SIHUMIx samples from bioreactors A, B, and C were included (gray ellipse, ). The reference space defines the normal fluctuation of the constant SIHUMIx community and the threshold (tr, gray dashed line, ) indicates the maximum fluctuation of SIHUMIx. The reference state resembles the center of the reference space and represents the mean of all samples used to build the reference space. Structural and functional deviations from the reference state in the disturbed bioreactors were followed. In addition, pH recording is shown to indicate the pH drop (red line). Maximal deviations were used to compute community resistance (RS) values. RS is the ability of a community to remain essentially unchanged in spite of a perturbation. Furthermore, resilience (RE) and elasticity (EL) were calculated. RE is the capability of a community to return to the reference state after a perturbation, and EL is the time it takes for a community to recover to the constant state.
Figure 5. Experimental set-up and development of SIHUMIx before and after a pH drop. (a) The SIHUMIx community was cultivated in vitro and two replicate bioreactors D and E were inoculated (day 0). After 24 h the medium feed was turned on. On day 4, the pH was reduced from 6.5 to 5.5. After sampling on day 5, pH was reset to 6.5. Samples were taken on day 1, 2, 3, 5, 7, 9, 13 and 14. (b) Community structure was analyzed by (1) flow cytometric fingerprinting and community function by (2) SCFA analysis. The reference space of the SIHUMIx community resembles the unimpeded community of bioreactors A, B, C on days 5–7. (c) The deviation from the reference state (the center of the reference space) is shown for the communities in bioreactor D and E. The threshold of the reference space indicates the maximal deviation from the reference state in the constant community state. Communities with a deviation below the threshold are similar to the constant community, communities with above-threshold deviation are different from the constant community. The calculations are based on (1) flow cytometric fingerprinting analysis and (2) SCFA analysis.
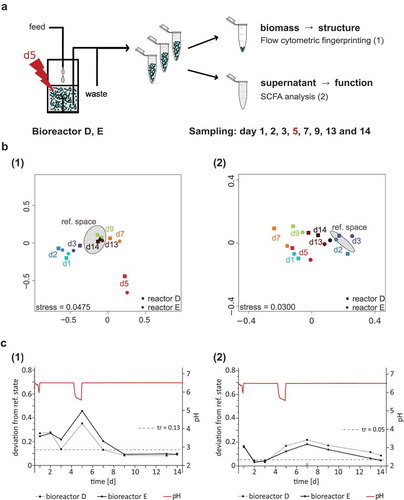
After inoculation, the communities in bioreactors D and E were different compared to those from bioreactors A, B and C with regard to (1) community structure and (2) community function (Supplemental file: Figure S2). Nevertheless, these communities developed toward the constant state with respect to both community structure and function until day 3 (). This development was disturbed and immediately mirrored in structural and functional changes (). Generally, deviation values for the functional analysis were smaller than those for structural analysis. This became apparent in a smaller threshold value for the reference space (). Nevertheless, since the pH drop on day 5 community structure and function have been influenced
The community structure showed the strongest deviation from the reference state on day 5. The RS of bioreactor D was slightly higher than the RS of bioreactor E, with RSD = 0.64 and RSE = 0.54, respectively. After the pH drop, the community structure developed toward the constant state of the unimpeded community (ref. state) and the deviation values decreased. Samples from both bioreactors were fully recovered to the constant state on day 9, when the deviation values declined below the threshold. During the recovery process, the community in bioreactor E showed a similar resilience (RLE = 0.66) compared to bioreactor D (RLD = 0.62). The constant community state was kept until the end of the experiment on day 14. For bioreactor D the EL value was 0.018 d−1 and for bioreactor E 0.019 d−1, respectively.
For community function, changes were also visible directly on day 5, but deviation values peaked on day 7. Here, the RS values were RSD = 0.78 and RSE = 0.82, respectively. After day 7 community functions developed toward the constant state. The community in bioreactor E recovered on day 14 and thus showed functional resilience (RLE = 0.60) to a higher degree compared to the community in bioreactor D (RLD = 0.43). Bioreactor D did not recover until the end of the experiment on day 14. Compared to the recovery of the community structure, community function recovered at a slower velocity.
Discussion
Bioreactor systems are useful tools to investigate effects on the intestinal microbiota in vitro.Citation12,Citation28 Most often complex fecal samples are used to inoculate bioreactors simulating the human intestine, but these complex inocula lack true biological replication.Citation12,Citation16 It has recently been shown that despite the use of the same homogenous inoculum complex bacterial communities can develop toward completely different communities.Citation29 Besides, community structure can keep fluctuating over time in complex bacterial communities.Citation29,Citation30 A variety of methods has been developed to analyze the community dynamics. In principle, the community structure and/or the community function are used to define an unchanged, constant state in complex communities. As stability is no stability property itself, we described the community development on the basis of the stability properties constancy, resistance and resilience.Citation19
The major aim of this study was to establish an in vitro model community for bioreactor use that reproducibly develops toward a constant community state with regard to both community structure and function. Furthermore, we wanted to determine, if different methods similarly depict the community development. Therefore, we cultivated the SIHUMIx community under defined chemostat conditions and analyzed the community development on the structural and functional level with five different analytical methods.Citation20 Even though the SIHUMIx community comprised of only eight bacterial species SIHUMIx shares the major SCFA with complex fecal communities.Citation31 SIHUMIx produces high levels of acetate, propionate and butyrate and low levels of branched SCFA. Additionally, SIHUMIx covers the major phyla Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. Nevertheless contrasting fecal communities, the model community SIHUMIx is far less diverse. Under balanced state conditions, SIHUMIx was dominated by B. thetaiotaomicron (phylum Bacteroides). Besides, E. coli (phylum Proteobacteria), B. producta and A. caccae (phylum Firmicutes) were found in decreasing order with a relative species abundance >3%. The remaining bacterial strains were present with relative species abundances below 1%. Since we aimed to simulate the colonic environment as close as possible in vitro, we used a complex culture medium similar to other culture media used to cultivate fecal microbiota.Citation13,Citation29,Citation32 The culture medium contains mucin and since B. thetaiotaomicron is a known mucin degrader this species gains a growth advantage.Citation33 Microbial growth of the model community SIHUMI (SIHUMIx without C. butyricum) has been modeled in the absence and presence of mucin.Citation34 Bauer et al. show that in the presence of mucin B. thetaiotaomicron will be the dominant species in the community. Nevertheless, it has recently been shown that all SIHUMIx bacterial strains remain in the community during continuous cultivation of SIHUMIx for up to 14 days, even though four strains from the SIHUMIx community are low abundant with less than 1% relative species abundance [Schäpe et al. 2019 in microorganisms, accepted for publication].
With respect to community structure analyses, the range of methods used in bioreactor research is considerably large. Analyses like microbial flow cytometry (FC),Citation35 metaproteomics,Citation36 and DNA-based methods, such as 16S rRNA gene analysis, terminal restriction fragment length polymorphism (t-RFLP) analysis,Citation21 next-generation sequencingCitation10 or denaturing gradient gel electrophoresis (DGGE),Citation13 have been published. We wanted to analyze the community structure on different cellular levels and therefore applied FC, intact protein profiling (IPP) and t-RFLP to analyze the cell-, protein- and DNA-related structural development. T-RFLP profiling is frequently used to track community dynamics in bacterial communities and the choice of the target gene allows the analysis of distinct microbial domains or phyla.Citation37 FC has the ability to resolve changes down to the single cell level and is therefore highly sensitive. Furthermore, FC provides the option of next-generation 16S rRNA analysis after sorting of interesting cell populations.Citation38 In contrast to metaproteomics, the workflow of IPP is much faster and, even though no biological information is generated, it provides a protein-based fingerprint.Citation39
Community function of gut microbiota is routinely assessed using SCFA analysis and provides insights into the metabolic activity and thereby delivers specific information on the fermentation process.Citation16,Citation17,Citation40 In some studies, besides SCFA, other metabolites have been measured.Citation14,Citation41 For that reason, we decided to include a more general approach in our analyses, specifically untargeted metabolomics.
To demonstrate the constant growth state of SIHUMIx, we compared the Bray-Curtis (BC) similarity of the communities on each day of cultivation to the communities on day 6/7. With proceeding adaptation, the BC similarity increased and reached a maximum on day 5. Then, the BC similarity on day 5 compare to day 6/7 was identical to the BC similarity of all samples on day 6/7. Thus, our results demonstrate that the SIHUMIx community starts to adapt directly after inoculation and then reaches a constant state with regard to both community structure (bacteria (FC), protein (IPP), and DNA (t-RFLP) composition) and community metabolism (SCFA analysis, untargeted metabolomics). Furthermore, compared to the in vitro cultivation of fecal communities the adaptation of SIHUMIx is very fast. This might result from the low diversity and the high abundance of B. thetaiotaomicron. McDonald et al. (2013) observed a structure-related constant state after 30 to 36 days of cultivation of fecal communities in their structural analysis with DGGE.Citation13 Liu et al. (2018) followed the development of their fecal community in the TWIN-SHIME by 16S rRNA and SCFA analyses.Citation17 In their study, the community structure became constant on day 10, whereas the communities’ SCFA metabolism became constant not before day 17. Furthermore, in contrast to the complex fecal community, SIHUMIx develops simultaneously on the structural and metabolic level during adaptation.Citation17
During the adaptation of SIHUMIx, all five methods similarly depicted the community development and indicated that the SIHUMIx community reached a constant state on day 5 with regard to community structure, as well as with regard to function. Since FC, t-RFLP and SCFA analyses have already been applied to follow complex community dynamics, we expected them to record the development of SIHUMIx reliably. However, to our knowledge, IPP and untargeted metabolomics have not been used to follow community dynamics, yet. Our results prove that both methods are suitable. For complex fecal communities, their suitability has to be validated.
It was observed that during cultivation microbial communities from the same homogenous inoculum can develop into different communities with regard to community structure.Citation29 This was not the case for SIHUMIx, which adapted to the system to a similar state in each bioreactor. To prove that for SIHUMIx, we used different batches of bacteria to inoculate replicate bioreactors. SIHUMIx unravels a highly reproducible development toward the same constant state. Surprisingly, directly after inoculation the communities from the disturbed bioreactors (D and E) were different compared to the unimpeded bioreactors. The only difference was that the pre-cultures for bioreactor A, B and C were grown for 72 h, whereas those for bioreactor D and E were cultivated for 48 h. However, the communities reached the same constant community state. This shows that the constant state of SIHUMIx is highly reproducible, but to achieve maximal reproducibility during the adaptation there is a need for standardized inocula. Generally, the BC similarity among replicates was slightly lower for structural analyses compared to functional analyses. Even though it is well known that community function is more stable than community structure,Citation42 the difference in the range of the BC similarity for structural and functional analyses is likely to result from the sample preparation. The medium background might have elevated the level of replicate BC similarity for metabolome analyses to ~0.95 in comparison to structural analyses that have ~0.80 BC similarity (BC similarity of 1 represents equality), since for structural analyses, bacteria pellets were used to prepare the cell suspensions, protein and DNA extracts. For targeted metabolomics, culture supernatants were used since SCFA are secreted into the medium.Citation43 Nevertheless, compared to the literature a BC similarity of 0.8 is already high.Citation10,Citation17 The highly similar and reproducible community development is also visible in the relative species abundances and SCFA concentrations.
It has been found that acidification of the colon occurs in patients with active ulcerative colitis and resembles a severe disturbance.Citation25 Thus, we validated the responsiveness of the SIHUMIx community to a pH drop for 24 h. Since community structure and function can develop differently,Citation16,Citation17 we tracked the community dynamics both on a structural and functional level. Since all methods are appropriate, we applied the most informative methods for structural and functional analysis, which are FC and SCFA analysis, respectively. Both methods clearly revealed the consequences of the pH drop and showed the similar behavior of the replicate bioreactors. Community structure responded and recovered faster than community function. This is in accordance with published studies that also found that community structure stabilizes earlier than community function.Citation16,Citation17 Our data disclose that the SIHUMIx community, in comparison to the complex community of Liu et al., shows a higher resistance against the perturbation.Citation29 They introduced a long-term temperature perturbation, whereas we exposed the SIHUMIx community only for 24 h to a decreased pH. Nevertheless, the SIHUMIx community possesses the ability to recover to the constant state and therefore is highly resilient. However, to compare the resistance and resilience of both communities indisputably, the communities have to be exposed to the same perturbation.
SCFA analysis, untargeted metabolomics, FC, IPP, and t-RFLP were evaluated and proven appropriate to trace the development of the SIHUMIx community. Our results suggest that various methods are suitable to follow community dynamics, but it is important to analyze community development both on the structural and functional level at the same time.
Reproducibility with regard to structural and functional development and the ability to reach a reproducible constant state within five days are the major advantages of SIHUMIx. The constant community state of SIHUMIx shares features of a complex community with regard to both community structure and function. The community shows high resistance, even though it responds to perturbations, and has the ability to recover (resilience). These properties allow the investigation of effects on the community that result from environmental factors, like pesticides or plasticizers, but also dietary or medical interventions. Moreover, the usage of microbial model communities, like SIHUMIx, might help to replace animal models in order to evaluate treatment effects. At the same time, the cultivation of small model communities opens the opportunity to study microbial interactions on the species level. Furthermore, the contribution of individual species to resultant changes in the communities’ performance is possible as well as the expansion of SIHUMIx to a more complex, but still defined model community. Thus, our results show that SIHUMIx is a suitable model for in vitro testing.
Materials and methods
A detailed description of materials and methods is given in the online repository of this journal (Supplemental file 4 – Methods).
Single stage bioreactor – experimental set-up
The cultivation was carried through in a Multifors 2 bioreactor (Infors). The reactors were inoculated with 1 × 109 bacterial cells per strain.
For the establishment of the SIHUMIx community as in vitro model, three parallel and independent 250 mL culture vessels (A, B, and C) were modified as chemostat. To investigate the effect of a pH drop on the SIHUMIx community, two parallel and independent bioreactors (D and E) were run for 14 days. The pH was reduced to 5.5 on day 4 and the community was exposed to the low pH for 24 h. After sampling on day 5, the pH was reset to the original pH of 6.5 until the end of the cultivation.
Sampling and sample analyses
Samples were taken in a 24 h interval starting the day after inoculation (d1). For t-RFLP analysis, IPP, SCFA analysis and untargeted metabolomics, samples were centrifuged (5,000 x g, 5 min, and 4°C). Cell pellets without supernatant were stored directly at −20°C for t-RFLP and IPP, whereas the supernatants for metabolome analysis were stored at −80°C. The cells for flow cytometric fingerprinting were harvested by centrifugation (3,200 x g, 10 min, and 4°C) and prepared directly.
Details for flow cytometric fingerprinting, intact protein profiling, metaproteome analysis, targeted short chain fatty acid analysis, untargeted metabolome analysis and terminal restriction fragment length polymorphism are given in the supplementary online repository of this journal.
Authors’ contributions
JLK (Jannike Lea Krause) and SSS (Stephanie Serena Schaepe) performed the bioreactor experiments and evaluated the data. KFW (Katarina Fritz-Wallace), BE (Beatrice Engelmann) performed the metabolome analyses and were supported by URK (Ulrike Rolle-Kampczyk). JLK, FS (Florian Schattenberg) and ZL (Zishu Liu) performed the flow cytometric analyses with support from SM (Susann Mueller). JLK, did the t-RFLP analyses supported by SK (Sabine Kleinsteuber). SSS did metproteomics with support from NJ (Nico Jehmlich). MvB (Martin von Bergen) and GH (Gunda Herberth) conceptualized the study. All authors contributed to and approved the manuscript.
Availability of data and materials
Raw cytometric data can be found at flow repository ID: FR-FCM-ZYVG under: https://flowrepository.org.
Disclosure of potential conflicts of interest
The authors report no conflict of interest.
Supplemental Material
Download Zip (1,018 KB)Acknowledgments
We thank the German Federal Environmental Foundation (DBU) for financial support of Jannike Lea Krause. We thank Prof. Dr. Michael Blaut (German Institute of Human Nutrition, Potsdam-Rehbruecke) for providing the SIHUMIx bacteria. We are thankful for technical assistance from Jeremy Knespel, Eva-Annamaria Stier, Kathleen Eismann and Nicole Gröger and for culture medium supply from Martina Kolbe.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi:10.1126/science.1104816.
- Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4:137rv7–137rv7. doi:10.1126/scitranslmed.3004184.
- O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi:10.1038/sj.embor.7400731.
- Levy M, Blacher E, Elinav E. Microbiome, metabolites and host immunity. Curr Opin Microbiol. 2017;35:8–15. doi:10.1016/j.mib.2016.10.003.
- Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi:10.1038/nature18847.
- Oates L, Cohen M. Assessing diet as a modifiable risk factor for pesticide exposure. Int J Environ Res Public Health. 2011;8:1792–1804. doi:10.3390/ijerph8061792.
- EFSA. Opinion of the scientific panel on contaminants in the food chain on Perfluorooctane Sulfonate (PFOS), Perfluorooctanoic Acid (PFOA) and their salts. EFSA J. 2008; 1–131.
- Joly C, Gay-Quéheillard J, Léké A, Chardon K, Delanaud S, Bach V, Khorsi-Cauet H. Impact of chronic exposure to low doses of chlorpyrifos on the intestinal microbiota in the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®) and in the rat. Environ Sci Pollut Res. 2013;20:2726–2734. doi:10.1007/s11356-012-1283-4.
- Reygner J, Joly Condette C, Bruneau A, Delanaud S, Rhazi L, Depeint F, Abdennebi-Najar L, Bach V, Mayeur C, Khorsi-Cauet H. Changes in composition and function of human intestinal microbiota exposed to chlorpyrifos in oil as assessed by the SHIME® model. Int J Environ Res Public Health. 2016;13:1088. doi:10.3390/ijerph13111088.
- Auchtung JM, Robinson CD, Britton RA Cultivation of stable, reproducible microbial communities from different fecal donors using minibioreactor arrays (MBRAs). Microbiome [Internet]. 2015;3:42. [cited 2018 Mar 3]. http://www.microbiomejournal.com/content/3/1/42.
- Tanner SA, Zihler Berner A, Rigozzi E, Grattepanche F, Chassard C, Lacroix C. In vitro continuous fermentation model (PolyFermS) of the swine proximal colon for simultaneous testing on the same gut microbiota. PLoS One. 2014;9:e94123. doi:10.1371/journal.pone.0094123.
- Payne AN, Zihler A, Chassard C, Lacroix C. Advances and perspectives in in vitro human gut fermentation modeling. Trends Biotechnol. 2012;30:17–25. doi:10.1016/j.tibtech.2011.06.011.
- McDonald JAK, Schroeter K, Fuentes S, Heikamp-deJong I, Khursigara CM, de Vos WM, Allen-Vercoe E. Evaluation of microbial community reproducibility, stability and composition in a human distal gut chemostat model. J Microbiol Methods. 2013;95:167–174. doi:10.1016/j.mimet.2013.08.008.
- Carman RJ, Woodburn MA. Effects of low levels of ciprofloxacin on a chemostat model of the human colonic microflora. Regul Toxicol Pharmacol. 2001;33:276–284. doi:10.1006/rtph.2001.1473.
- Possemiers S, Verthé K, Uyttendaele S, Verstraete W. PCR-DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol. 2004;49:495–507. doi:10.1016/j.femsec.2004.05.002.
- Van den Abbeele P, Grootaert C, Marzorati M, Possemiers S, Verstraete W, Gerard P, Rabot S, Bruneau A, El Aidy S, Derrien M, et al. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for bacteroidetes and clostridium cluster IX. Appl Environ Microbiol. 2010;76:5237–5246. doi:10.1128/AEM.00759-10.
- Liu L, Firrman J, Tanes C, Bittinger K, Thomas-Gahring A, Wu GD, Van den Abbeele P, Tomasula PM. Establishing a mucosal gut microbial community in vitro using an artificial simulator. PLoS One. 2018;13:e0197692. doi:10.1371/journal.pone.0197692.
- Grimm V, Schmidt E, Wissel C. On the application of stability concepts in ecology. Ecol Model. 1992;63:143–161. doi:10.1016/0304-3800(92)90067-O.
- Grimm V, Wissel C. Babel, or the ecological stability discussions: an inventory and analysis of terminology and a guide for avoiding confusion. Oecologica. 1997;109:323–334. doi:10.1007/s004420050090.
- Becker N, Kunath J, Loh G, Blaut M. Human intestinal microbiota: Characterization of a simplified and stable gnotobiotic rat model. Gut Microbes. 2011;2:25–33. doi:10.4161/gmic.2.1.14651.
- Sträuber H, Schröder M, Kleinsteuber S. Metabolic and microbial community dynamics during the hydrolytic and acidogenic fermentation in a leach-bed process. Energy Sustain Soc. 2012;2:13. doi:10.1186/2192-0567-2-13.
- Koch C, Müller S. Personalized microbiome dynamics – Cytometric fingerprints for routine diagnostics. Mol Aspects Med. 2018;59:123–134. doi:10.1016/j.mam.2017.06.005.
- Wissenbach DK, Oliphant K, Rolle-Kampczyk U, Yen S, Höke H, Baumann S, Haange SB, Verdu EF, Allen-Vercoe E, von Bergen M. Optimization of metabolomics of defined in vitro gut microbial ecosystems. Int J Med Microbiol. 2016;306:280–289. doi:10.1016/j.ijmm.2016.03.007.
- Jehmlich N, Schmidt F, Taubert M, Seifert J, von Bergen M, Richnow H-H VC. Comparison of methods for simultaneous identification of bacterial species and determination of metabolic activity by protein-based stable isotope probing (Protein-SIP) experiments. Rapid Commun Mass Spectrom. 2009;23:1871–1878. doi:10.1002/rcm.4084.
- Nugent SG, Kumar D, Rampton DS, Evans DF. Intestinal luminal pH in inflammatory bowel disease: possible determinants and implications for therapy with aminosalicylates and other drugs. Gut. 2001;48:571–577. doi:10.1136/gut.48.4.571.
- Ke X, Walker A, Haange S-B, Lagkouvardos I, Liu Y, Schmitt-Kopplin P, von Bergen M, Jehmlich N, He X, Clavel T, et al. Synbiotic-driven improvement of metabolic disturbances is associated with changes in the gut microbiome in diet-induced obese mice. Mol Metab. 2019;22:96–109. doi:10.1016/j.molmet.2019.01.012.
- Kleiner M, Thorson E, Sharp CE, Dong X, Liu D, Li C, Strous M. Assessing species biomass contributions in microbial communities via metaproteomics. Nat Commun. 2017;8:1558. doi:10.1038/s41467-017-01544-x.
- Macfarlane GT, Macfarlane S. Models for intestinal fermentation: association between food components, delivery systems, bioavailability and functional interactions in the gut. Curr Opin Biotechnol. 2007;18:156–162. doi:10.1016/j.copbio.2007.01.011.
- Liu Z, Cichocki N, Hübschmann T, Süring C, Ofiţeru ID, Sloan WT, Grimm V, Müller S. Neutral mechanisms and niche differentiation in steady-state insular microbial communities revealed by single cell analysis: Non-equilibria systems. Environ Microbiol. 2019;21:164–181. doi:10.1111/emi.2019.21.issue-1.
- Fernández A, Huang S, Seston S, Xing J, Hickey R, Criddle C, Tiedje J. How stable is stable? Function versus community composition. Appl Environ Microbiol. 1999;65:3697–3704.
- Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-gavilán CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol [Internet]. 2016;7:185. [cited 2019 Feb 13]. http://journal.frontiersin.org/Article/10.3389/fmicb.2016.00185/abstract.
- Child MW, Kennedy A, Walker AW, Bahrami B, Macfarlane S, Macfarlane GT. Studies on the effect of system retention time on bacterial populations colonizing a three-stage continuous culture model of the human large gut using FISH techniques: the effect of system retention time on bacterial populations. FEMS Microbiol Ecol. 2006;55:299–310. doi:10.1111/fem.2006.55.issue-2.
- Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi:10.1038/nrmicro2746.
- Bauer E, Zimmermann J, Baldini F, Thiele I, BacArena KC. Individual-based metabolic modeling of heterogeneous microbes in complex communities. PLoS Comput Biol. 2017;13:e1005544. doi:10.1371/journal.pcbi.1005544.
- Brognaux A, Han S, Sørensen SJ, Lebeau F, Thonart P, Delvigne F. A low-cost, multiplexable, automated flow cytometry procedure for the characterization of microbial stress dynamics in bioreactors. Microb Cell Factories. 2013;12:100. doi:10.1186/1475-2859-12-100.
- Kuhn R, Benndorf D, Rapp E, Reichl U, Palese LL, Pollice A. Metaproteome analysis of sewage sludge from membrane bioreactors. Proteomics. 2011;11:2738–2744. doi:10.1002/pmic.v11.13.
- Schütte UME, Abdo Z, Bent SJ, Shyu C, Williams CJ, Pierson JD, Forney LJ. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl Microbiol Biotechnol. 2008;80:365–380. doi:10.1007/s00253-008-1565-4.
- Koch C, Günther S, Desta AF, Hübschmann T, Müller S. Cytometric fingerprinting for analyzing microbial intracommunity structure variation and identifying subcommunity function. Nat Protoc. 2013;8:190–202. doi:10.1038/nprot.2012.149.
- Usbeck JC, Wilde C, Bertrand D, Behr J, Vogel RF. Wine yeast typing by MALDI-TOF MS. Appl Microbiol Biotechnol. 2014;98:3737–3752. doi:10.1007/s00253-014-5586-x.
- Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio. 2019;10:e02566–18. doi:10.1128/mBio.02566-18.
- Macfarlane GT, Macfarlane S, Gibson GR. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol. 1998;35:180–187. doi:10.1007/s002489900072.
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214.
- LeBlanc JG, Chain F, Martín R, Bermúdez-Humarán LG, Courau S, Langella P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Factories [Internet]. 2017;16:79. [cited 2018 Dec 18]. http://microbialcellfactories.biomedcentral.com/articles/10.1186/s12934-017-0691-z.
