ABSTRACT
The canine gut microbiota is a complex microbial population that is potentially related to metabolism, immunologic activity and gastrointestinal (GI) diseases. Early studies revealed that the canine gut microbiota was dynamic, and bacterial populations in the adjacent gut segments were similar, with anaerobes predominating. Metagenomics analysis revealed that nutrient contents in the diet modulated bacterial populations and metabolites in the canine gut. Further research revealed significant correlations between dietary factors and canine gut core microbiomes. Canine GI diseases are closely correlated with gut microbiota dysbiosis and metabolic disorders. Probiotic-related therapies can effectively treat canine GI diseases. Recent studies have revealed that the canine gut microbiota is similar to the human gut microbiota, and dietary factors affect both. Studying canine intestinal microorganisms enables clarifying changes in the canine intestinal bacteria under different conditions, simulating human diseases in dog models, and conducting in-depth studies of the interactions between intestinal bacteria and disease.
Introduction
From basic research to clinical applications, the importance of the gut microbiota has rapidly evolved in recent years.Citation1 With the applications of metagenomics, metabolomics and culturomics in investigating the gut microbiota, many researchers have reported the important roles of the gut microbiota in contributing to metabolic functions, immunologic activity and neurodevelopment.Citation2,Citation3 The gut microbiota has been widely studied in humans and laboratory rodents. Canines are common laboratory animals that are widely used in different fields. To date, limited reviews have been published on the canine gut microbiota. Here, we describe findings on the canine gut microbiota from early studies as well as the effects of different foods, models of gastrointestinal diseases and probiotics on the canine gut microbiota.
Early studies on the canine gut microbiota
Canines, especially beagles, are widely used in scientific research. In as early as 1977, the gut microbiomes in various regions of the gastrointestinal (GI) tracts of beagles were assessed by culturing strict anaerobes and making observations via optical and scanning electron microscopy. All canines possess complicated microbiotas in their colons, with anaerobes predominating. One study showed that the canine microbiota became altered and increased in complexity when dogs were housed in a locked environment. These alterations were closely associated with the gastrointestinal epithelial cells and crypts of Lieberkuhn.Citation4 Authors have also predicted that less predominant microbial species in the GI tract can change over time when animals are housed in locked environments. Anaerobic bacteria are identified by Gram staining, biochemical tests, and volatile and nonvolatile fatty acids via gas chromatography. Bacterial isolates were difficult to identify in large quantities until PCR techniques emerged.
Denaturing gradient gel electrophoresis (DGGE) was first used in 1993. DGGE is used to analyze the genetic diversity of complex microbial populations such as environmental or fecal samples.Citation5 The gut microbiota comprises complex microbial populations that can be analyzed via DGGE.Citation6 Molecular fingerprinting via PCR-DGGE has revealed different diversity levels in fecal samples from different animals. Here, we present research findings on the canine gut microbiota, which were obtained using DGGE.
One study used DGGE to evaluate the differences in bacterial compositions of the intestinal compartments of canines housed in similar environments and fed identical diets. These authors found significantly more similarities in microbial diversity levels between neighboring intestinal regions than between non-neighboring regions.Citation7 Another study using canine duodenal fluid indicated that the canine small intestine had a highly diverse microbiota with marked differences in diversity between individual dogs. The results were duplicated and highly reproducible. Each dog’s duodenum, jejunum, ileum, colon and rectum had significantly different gut microbiotas, and the colon and rectum had higher bacterial diversity than did the other gut segments.Citation8 Although the gut segments significantly differed in their bacterial populations, the adjacent gut segments were more similar than were the separated gut segments.
Percent G + C profiling is another approach to analyzing bacterial community profiles.Citation9 One study used percent G + C profiling to determine the relationship between the beagle gut microbiota and the contents of animal-derived protein and carbohydrates from feed. Beagles were divided into the high-carbohydrate (HC), high-protein (HP) and dry commercial (DC; control) groups. The HC and HP groups were fed formulated HC (starch: 438 g/kg) and HP (crude protein: 609 g/kg) diets for one week. Compared with the DC group, the HC and HP groups had decreased microbial diversity with different representative bacteria at the order level. Diversity among Fusobacteriales was significantly increased in the HP group, and the dogs in this group had diarrhea. In the HC group, the most abundant sequences belonged to the order Clostridiales.Citation10 The altered bacterial diversity may have played a role in inducing diarrhea, thus revealing the potential relationships between the gut microbiota and gastrointestinal diseases. A modified strategy combines percent G + C profiling (GC fractionation) with DGGE (GC-DGGE). GC-DGGE enhances microbiota diversity assessments and detection of minority microbiome communities in fecal samples.Citation11 Using molecular fingerprinting techniques, research on the canine GI microbiome revealed that differences in the levels of canine GI microbial diversity existed between individuals and intestinal compartments.
Effect of diets and nutrients on the canine gut microbiota
Compared with environmental factors, diet plays an important role in adjusting the GI microbiome balance in canines. Next-generation sequencing (NGS) has revolutionized the approaches in researching complex microbiota samples. Fecal samples from canine models can be analyzed via NGS to determine the relationships and interactions between hosts and the microbiota.Citation12
High-throughput sequencing has also revealed interactions between the canine gut microbiota and the diet. Different ratios of proteins and carbohydrates, prebiotics, and dietary habits are the major research directions for the canine gut microbiota. Inulin and other fibers are current research topics on the gut microbiota.Citation13 One study revealed that consuming a small amount of beet pulp as dietary fiber changed the abundance of the typical canine gut microbiome. Adding 7.5% beet pulp (60% of the total dietary fiber) to standard canine diets for 14 days significantly shifted the Firmicutes abundance and tripled the low taxonomic levels of Faecalibacterium. Eubacterium hallii, which produces butyrate, appeared after a fiber diet treatment.Citation14 These abundance changes may have been due to dietary selection for complex fermentative activity with additional dietary fiber. In another study, overweight canines were fed an approximately 1% inulin-type prebiotic diet, and the results showed that inulin-type prebiotics may modulate the gut microbiota, short-chain fatty acid (SCFAs) and bile acids (BAs) in overweight dogs. The overweight canines had significantly increased SCFAs and lower indole concentrations after prebiotic treatment. Fermentation of inulin-type prebiotics produced SCFAs, which decreased the luminal pH and prevented BA reabsorption. BAs are the major pathway for excreting cholesterol from the body.Citation15 Another study showed that continuously feeding dogs low-fat and high crude-fiber diets for 17 weeks decreased the body weights of obese dogs by an average of 18% of the initial body weight.Citation16 Thus, inulin-type prebiotics and fibers can modulate canine fecal metabolites such as SCFAs and BAs. Bacterial diversity can also be changed with prebiotic and fiber treatment. Studies on body weight loss in canines also indicate that prebiotics and fiber may play important roles in improving SCFA production, body conditions and other potential metabolic factors.
Different formulated commercial foods and natural foods with different nutrient sources and ratios (such as proteins and carbohydrates) are widely used in canine diets. Daily diets with different ingredients also alter the abundance and diversity of the canine gut microbiota. High-protein, low-carbohydrate (HP-LC) diets can modulate body weight in obese canines.Citation17 Canine models fed HP-LC diets had lower Bacteroidetes/Firmicutes ratios and higher Bacteroides/Prevotella ratios than did those fed low-protein HC diets, and the abundances of Clostridium hiranonis, Ruminococcus gnavus, and Clostridium perfringens were increased. Enriched microbial gene networks are also associated with weight maintenance.Citation18
Another study showed that HP diets increased the abundance of butyrate-producing bacteria and enhanced the activity in canine gut microbiomes independent of body condition. Canines fed HP diets had higher concentrations of fecal ammonia, isovalerate, isobutyrate, phenol, indole, serum indoxyl sulfate and plasma 3-OH isovalerylcarnitine, and these dogs had diarrhea. High concentrations of these metabolites are correlated with high protein intake.Citation19–Citation21 Compared with lean canines, increased dietary protein content more strongly affected the gut microbiotas in obese canines.Citation22 No differences were found in gut bacterial richness between canines fed high-fat, low-starch and low-fat, and high-starch diets. However, lower relative abundances of Prevotella, Solobacterium, and Coprobacillus were found in the high-fat diet group as well as higher concentrations of fecal BAs after 7 weeks of feeding. Moderate variations in these diets did not alter the dogs’ gut microbiotas.Citation23
One study replaced protein and starch with navy beans in canine diets to investigate the effect of different nutrient sources on canines. In the navy bean diet, which included 25% navy bean powder, macronutrients, and micronutrients, the total calories were adjusted to match the standard diet. After feeding canines the navy bean or standard diet for 4 weeks, the gut microbial populations did not significantly differ between dogs fed either diet. Compared with the baseline, the navy bean-diet group had increased Firmicutes and decreased Actinobacteria and Fusobacteria.Citation24
Dietary models with different nutritional concentrations have revealed that macronutrient ratios are key factors in modulating the canine gut microbiota (). Distinct bacterial diversity and populations can be observed in the gut microbiotas and fecal metabolites of dogs fed different diets. If different diets contain the same levels of macronutrients and micronutrients, the effect on the bacterial population in the canine gut microbiota may be minimal.
Table 1. Diet-related studies of the canine gut microbiota and metabolites.
Feeding dogs natural vs commercial foods also affects the microbiota population and abundance. Meat is the major protein supplement in natural dog foods. One study fed dogs natural and commercial diets for 1 year and revealed biases in the microbial abundances and populations in the gut microbiota. Canine natural diets containing approximately 90% raw meat were divided into 4 groups with different meats: kangaroo, beef, chicken and duck. The other two groups were supplemented with commercial foods with balanced nutrition. Despite the various meat sources, all canines fed natural diets had higher gut microbiome diversity than did the commercial-feed groups. Beta diversity analysis showed that the canine gut microbiota diversity differed significantly between the dietary types. Clostridium perfringens and Fusobacterium varium in the canine gut microbiota were more abundant in dogs fed the natural diets. In this study, the natural diet had 30–52% crude protein and 11–50% total fat content regardless of meat type, while the commercial canine foods had 18–21% crude protein and 8–10% total fat content.Citation25 Another study of the canine gut microbiota compared the interaction between dogs fed a canine commercial extruded diet and dogs fed a mixed natural diet with 70% beef. After feeding for 14 days, the diets of the two groups were reversed, and the dogs were fed for another 14 days, but dogs fed the commercial extruded diet were shifted to the natural diet and vice versa. The dietary interactions showed that the natural diet promoted more balanced bacterial growth in the canine gut microbiota.Citation26 Similar research showed that changing from commercial dry food to a natural diet with minced beef influenced the canine gut microbiome. The concentration of the minced beef in the natural diet was increased from 25% to 75% of the dogs’ total energy requirement in 3 weeks. The results showed that the changes in the canine gut microbiome were reversible. The minced beef diet influenced the fecal microbiota concentration and SCFA profiles in the canine fecal samples, and the isovaleric acid concentration increased as the protein consumption increased. Recent research on the human microbiota revealed that higher concentrations of fecal isovaleric acid were correlated with depression.Citation29 Reintroducing the dogs to the initial commercial dry food significantly reversed these changes.Citation27 The differences in protein and energy digestion in canines revealed that the key bacterial families, Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae, played important roles in the relationships between the canine gut microbiota, nutrient digestibility and metabolism.
In another study, dogs were divided and fed kibble or meat diets. The correlation heatmap and relevance network plot showed that Clostridiaceae was the central node in the canine gut microbiota, nutrient digestibility and macronutrient composition. Clostridiaceae, dietary protein content, and protein digestibility were positively correlated, while fecal protein content was negatively correlated with Clostridiaceae. The Clostridiaceae abundance also increased when dogs were fed a meat diet.Citation28 Erysipelotrichaceae are important bacteria in the canine digestive tract and are associated with high-fat diets in humans and rodent models.Citation30–Citation32 Erysipelotrichaceae were positively correlated with markers associated with carbohydrate digestion, such as dietary carbohydrates, fiber content and volatile fatty acid (VFA) production. In addition, Erysipelotrichaceae were negatively correlated with protein metabolism markers. Bacteroidaceae, as with other members of Bacteriodetes, played a role in nutrient digestion in canines, producing several VFAs, including succinate, acetate and propionate. Propionate is produced from succinate,Citation33 and acetate and propionate can activate both GPR41 and GPR43. GPR41 and GPR43 are a pair of mammalian G protein-coupled receptors thought to be potential targets for metabolic disorders such as obesity.Citation34,Citation35 Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae were at the center of the relationship between the canine gut microbiota and nutrient digestibility, and changes in the canine gut microbiota were related to diet. Canines fed natural diets had more complex bacterial populations in their gut microbiotas than did those fed formulated commercial diets. Thus, differences in macronutrients between natural diets and commercial food strongly suggest that nutrients affect the canine gut microbiota. The canine gut microbiota also affects nutrient digestibility, and some bacteria appear more important in modulating the microbiome. Significant changes in the canine gut microbiota have been observed with both short-term and long-term feeding, and reversibility exists with short-term feeding. Canine fecal metabolites also differ between dogs fed natural food versus commercial food. Although diets with different macronutrient ratios exert significantly different effects on the canine gut microbiota, the effect of micronutrients in natural foods as well as whether micronutrients play specific roles in modulating the canine gut microbiota remains uncertain.
Canine gut microbiota and GI diseases
Inflammatory bowel disease (IBD) refers to a diverse group of chronic gastrointestinal diseases.Citation36 IBD is an autoimmune condition that is difficult to diagnose, and gut microbiome dysbiosis has been proposed as an induced factor in its pathogenesis.Citation37 Dysbiosis of the human gut microbiome exerts similar changes to those seen with gut dysbiosis in canines and other mammals. Research on canine IBD is valuable because it enables making predictions from the canine gut microbiota to the human gut microbiota.Citation38 Here, we discuss the relationships between the canine gut microbiota, metabolism, other factors in canine IBD, and other GI diseases. summarizes some recent studies of GI diseases and the canine gut microbiota.
Table 2. GI disease-related studies of the canine gut microbiota.
One study proposed that microbiotas enriched with Gram-negative bacteria in obese canines may be correlated with chronic inflammation. These authors fed the same diet to dogs for 6 months. Canines in the obese group were fed food ad libitum, while dogs in the lean group were fed food with restricted nutrients. The obese group had significantly increased body weight, higher leptin concentrations, and decreased adiponectin and 5-hydroytryptamine (5-HT) in the cerebrospinal fluid. Proteobacteria were predominant in the gut microbiotas of the obese canines (76%), while Firmicutes (85%) were predominant in the lean canines. Enriched Gram-negative bacteria can influence intestinal lipopolysaccharide (LPS) levels. Decreased 5-HT concentrations in obese canines can increase their appetites and cause a cycle leading to bacterial dysbiosis with increased Gram-negative bacteria and enriched LPS. LPS is associated with chronic inflammation.Citation46 This research revealed the relationships between IBD, diet and bacterial dysbiosis.
Another study characterized the bacterial dysbiosis among canine idiopathic IBD, acute non-hemorrhagic diarrhea and acute hemorrhagic diarrhea (AHD). Compared with healthy dogs, those with AHD had the most significant changes in gut microbiota abundance and diversity. The abundances of Faecalibacterium, Ruminococcaceae, Turicibacter spp. and Blautia spp. were significantly decreased, while Sutterella and Clostridium perfringens were significantly increased. qPCR assay results confirmed that only canines with AHD had significant increases in Clostridium perfringens. Some populations of decreased bacteria were believed to be important producers of SCFAs. Faecalibacterium spp. and Fusobacteria were decreased in dogs with active IBD. Faecalibacterium spp. were decreased in the canine gut microbiotas in dogs with both acute diarrhea and IBD. Decreases in Faecalibacterium prausnitzii often occur in human IBD.Citation39 Studies have shown that Faecalibacterium prausnitzii secretes anti-inflammatory peptides in vitro,Citation47 which may demonstrate the relationships between GI disorders and microbiota dysbiosis. IBD influenced the canine gut microbiota profiles and metabolism.
The canine chronic enteropathy clinical activity index (CCECAI) is linked with the clinical scores of canines with IBD. In this study, the total CCECAI score was divided into 5 levels by IBD symptoms as follows: clinically insignificant (0–3), mild (4–5), moderate (6–8), severe (9–11), or very severe (≥12). CCECAI was significantly negatively correlated with the total fecal SCFA concentration and the Lactobacillus abundance in the canine gut microbiome. Dogs characterized as having severe IBD had low proportions of Lactobacillus, higher plasma concentrations of valine, free carnitine and total acylcarnitines, decreased plasma concentrations of citrulline and short-chain acylcarnitine-to-free carnitine ratios and decreased folate in the serum. Canines with IBD also had higher 3-hydroxyisovalerylcarnitine concentrations and isovalerylcarnitine/leucine ratios compared with those of the healthy group, and these increased levels may be associated with the catabolism of branched-chain amino acids.Citation40 Valine has been implicated in lipid and fatty acid metabolism.Citation48,Citation49 The metabolism of carnitine in the gut microbiota can produce trimethylamine (TMA), and further metabolism of TMA to trimethylamine-N-oxide (TMAO) can accelerate atherosclerosis.Citation50 Human studies have shown that acylcarnitines are associated with insulin resistance,Citation51,Citation52 and short-chain acylcarnitine can be hydrolyzed to carnitine.Citation53 Citrulline is an amino acid and validated marker of enterocyte function and the gastrointestinal barrier, which is produced by enterocytes of the small bowel mucosa. Decreased concentrations of citrulline can reduce the enterocyte mass such as that in short bowel syndrome.Citation54 Accumulations of 3-hydroxyisovalerylcarnitine and isovalerylcarnitine in higher concentrations are associated with metabolic deficiency in leucine metabolism and with protein absorption from foods.Citation55–Citation57 Lower folate concentrations in the serum are linked to abnormal functioning of the proximal small intestinal mucosa, which can reduce folate absorption.Citation58 Tissue damage and alterations in the host metabolism can significantly change the short-chain acylcarnitines, amino acids in the plasma, and other metabolic profiles in canines with IBD.Citation40 Further evidence showed dysbiosis of other metabolites and canine gut microbes in canines with IBD. Compared with healthy canines, canines with IBD had significantly lower gut microbiota diversity. Canine serum metabolites also differed significantly. Canines with IBD had higher concentrations of 3-hydroxybutyrate, hexuronic acid, ribose, and gluconic acid lactone than did healthy canines.Citation41 The higher concentrations in the ketone body of 3-hydroxybutyrate may be correlated with a high energy demand in the host.Citation59 Hexuronic acid is also known as vitamin C and is considered an antioxidant because it protects against oxidative stress and plays an important role in collagen and carnitine synthesis.Citation60 Ribose is required for biological systems and provides further metabolism. Gluconic acid lactone and ribose both protect the cells from oxidative stress.Citation41 Although clinical symptoms of canine IBD were improved after medical therapy, no significant changes occurred in the serum metabolite profiles or gut microbiotas. Enriched bacterial functions of the secretory system, transcription factors and an unknown pathway were overrepresented in canines with IBD, while amino acid metabolism was enriched in healthy canines. Canines with IBD and other GI diseases showed gut microbiota dysbiosis and altered indexes, such as 5-HT, in the serum. Dysbiosis was also found in the fecal SCFA concentrations in canines with IBD; thus, a model of SCFA dysbiosis can be used to evaluate the clinical symptoms of canine IBD.
Canines with chronic enteropathy had symptoms of SCFA dysbiosis. Research on 49 healthy canines and 73 canines with chronic enteropathy showed higher total SCFA concentrations in healthy canines, while canines with chronic enteropathy had lower acetate and propionate concentrations. Higher dysbiosis indices and decreased bacterial diversity and richness were observed in the gut microbiotas of canines with chronic enteropathy,Citation42 thus revealing the clinical importance of canines with chronic enteropathy having altered concentrations of fecal SCFAs and gut microbes. Microbiota dysbiosis and other symptoms also existed in canines with acute diarrhea (AD). Evaluations of the gut microbiota, fecal SCFAs, serum, and urine metabolite profiles revealed differences between healthy canines and canines with AD. Canines with AD also presented significantly lower bacterial diversity and altered microbial communities with different representative bacteria. Clostridium was significantly increased in canines with AD. Fecal propionic acid decreased and was associated with decreased Faecalibacterium in canines with AD. Total fecal SCFAs and branched-chain fatty acids showed no significant changes. The predicted functional gene content of the microbiome also revealed that genes for the methyl-accepting chemotaxis protein and the transposase enzyme were overrepresented. Serum kynurenic acid concentrations were significantly decreased in canines with AD. Urine concentrations of 2-methyl-1H-indole and 5-methoxy-1H-indole-3-carbaldehyde were also significantly decreased in dogs with AD.Citation45
Further research associated canines with idiopathic acute hemorrhagic diarrhea syndrome (AHDS) with fecal markers and microbiota. Idiopathic AHDS is characterized by acute onset of bloody diarrhea and severe dehydration and can result in high mortality rates if left untreated. In this study, 3 assayed markers were selected to evaluate idiopathic AHDS. Calprotectin and S100A12 were the markers of inflammation, and α1-proteinase inhibitor was the marker of gastrointestinal protein loss. All 3 markers had significantly higher concentrations in canines with idiopathic AHDS. After 3 days of treatment, these markers decreased significantly.Citation44 Canine gut microbiota dysbiosis appeared to last longer than did the markers of inflammation and protein loss. Canines with AHDS had gut microbiota dysbiosis and abundances of specific bacteria, including Ruminococcaceae, Faecalibacterium spp., Bifidobacterium spp., and Proteobacteria. This microbiota dysbiosis did not resolve immediately after treatment. Bacterial diversity did not differ in the canine gut microbiota except a mild decrease in Ruminococcaceae 3 days after treatment.
Another study using qPCR established a mathematical model termed the dysbiosis index (DI) in canine chronic inflammatory enteropathy (CE) to evaluate the gut microbiota dysbiosis level. Eight indexes of the universal bacteria, Faecalibacterium, Turicibacter, Streptococcus, Escherichia coli, Blautia, Fusobacterium, and Clostridium hiranonis, were measured via qPCR to assess the DI. The DI in canines with different CE levels was defined as a mathematical formula using these 8 indexes to compare the bacteria with those of healthy canines.Citation43 Canines with CE differed significantly from healthy canines in all 8 bacterial target indexes. Escherichia coli and Streptococcus were significantly higher in the diseased group, while the other indexes were significantly lower. The CCECAI was significantly correlated with DI. qPCR-based indexes of this model showed that the model had a 74% sensitivity and 95% specificity to separate CE canines and healthy canines. This model also provided a new approach to assess whether the canine gut microbiota was normalized in response to CE treatment. In canines with other GI diseases, SCFA dysbiosis and gut microbiota dysbiosis are common. Therapy for GI diseases shows that the canine gut microbiota does not return to normal shortly after treatment, and a canine GI disease model can be established and evaluated using specific bacteria in the canine gut microbiota.
Canine gut microbiota dysbiosis often occurs with canine GI diseases. Treatments for GI diseases that modulate the gut microbiota, such as probiotic therapy and fecal microbiota transplantation, have been used to treat some human GI diseases, and researchers have asked whether similar therapies can be used to treat canine GI diseases.
Probiotic-related studies in canines
Probiotic therapy has been used in human GI diseases and may have potential use in treating canine GI diseases.Citation61 Canine-derived probiotics for treating canine GI disease comply with ethical standards and have been shown to have health benefits in treating canine GI diseases. One study revealed that canine-derived Lactobacillus fermentum CCM 7421 significantly modulated canine diarrhea. Administering 107–109 colony-forming units of the probiotic, Lactobacillus fermentum, to canines for 4–14 days normalized canine metabolites, including SCFAs. Using probiotic strains also modulated serum biochemical and immune parameters regardless of the duration of administration. Clostridia populations in the gut microbiotas of dogs with diarrhea were decreased, and lactic acid bacteria were increased after treatment.Citation62
Another study evaluated standard therapy and standard therapy + multi-strain probiotics on the mucosal microbiota for 8 weeks in canines with IBD. The probiotic strains, Lactobacillus plantarum DSM 24730, Streptococcus thermophiles DSM 24731, Bifidobacterium breve DSM 24732, Lactobacillus paracasei DSM 24733, Lactobacillus delbrueckii subsp. bulgaricus DSM 24734, Lactobacillus acidophilus DSM 24735, Bifidobacterium longum DSM 24736, and Bifidobacterium infantis DSM 24737, were combined in specific concentrations. Fluorescence in situ hybridization methods showed that the mucosal microbiotas from canines with IBD in the standard therapy and standard therapy + probiotics groups did not significantly differ in bacterial numbers. Both treatments increased the mucosal bacterial species and total bacterial numbers. Standard therapy showed increased Bifidobacterium spp., while standard therapy + mixed probiotics showed increased Lactobacillus spp. in the canine mucosal microbiota. Canines with IBD in the standard therapy + probiotic group had increased tight-junction protein expression. The probiotic therapy was also associated with upregulated expression of the tight-junction proteins, E-cadherin, occludin, and zonulin,Citation63 showing that probiotic therapy may benefit mucosal homeostasis.
Other probiotic strains can also modulate the composition and function of the gut microbiota in canines with recurrent diarrhea. The probiotic strains, Lactobacillus casei Zhang, Lactobacillus plantarum P-8, and Bifidobacterium animalis subsp. lactis V9, were mixed. Canines with recurrent diarrhea were given probiotic therapy for 60 days, and fecal samples were analyzed using metagenomics. After probiotic therapy, diversity and bacterial populations in the canine gut microbiota were significantly altered. Beneficial bacteria were increased in the canine gut microbiota. The abundances of Lactobacillus acidophilus, Lactobacillus johnsonii, Lactobacillus reuteri and Butyricicoccus pullicaecorum were significantly increased, and opportunistic pathogenic bacteria, such as Clostridium perfringens and Stenotrophomonas maltophilia, were significantly decreased. Metagenomic analysis revealed that probiotic therapy was significantly associated with upregulated pathways related to amino acid metabolism, metabolism of cofactors and vitamins, biosynthesis of secondary metabolites, and glycan biosynthesis and metabolism. Pathways associated with virulence of pathogenic bacteria and cell signaling were downregulated after probiotic therapy.Citation64 Therefore, probiotic therapy may play an important role in treating canine GI diseases.
Isolating new probiotic strains from canine feces is also important. Culturing potential probiotic strains may promote canine health. One study isolated a series of potential probiotic strains of Lactobacillus from canine fecal samples, and the effects were evaluated. Properties of the strains, including resistance to gastric and pancreatic juices and bile salts, antibiotic resistance and antipathogenicity, were tested. Of the 14 isolated Lactobacillus strains, 5 could tolerate gastric stress. These 5 potential probiotic strains also had clindamycin resistance and significant antimicrobial capacity in inhibiting pathogenic strains. Morphological and molecular characterization identified that 3 of these strains were Lactobacillus reuteri, and the other two were Lactobacillus johnsonii. Identification of Lactobacillus reuteri in vitro confirmed its ability to ferment sugar and adhere to HT29 epithelial cells; thus, Lactobacillus reuteri may be a potential probiotic.Citation65
Isolating potential new probiotic strains is crucial for probiotic therapy. Culturing new bacterial isolates is a research frontier in the human gut microbiota, and probiotic strains isolated from the human gut have played important roles in modulating the gut microbiome balance. Probiotic isolation and therapy could become a new method of treating canine GI diseases.
Similarities between the canine and human gut microbiotas
A recent metagenomic study reported that in dogs who randomly received either an HP or HC diet, the canine microbiome diversity was closer to that of the human microbiome than to the microbiomes of either pigs or mice.Citation66 Factors including breed and sex had little influence on the gut microbiome changes. As the results were consistent with those of previous human studies, canine studies may predict results in humans. Other studies on canines validate this conclusion.
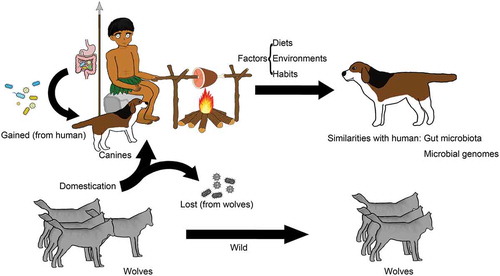
Dogs are the oldest known example of domestication. Domestication of canines from wolves is a key factor influencing the microbiota diversity between domesticated canines and wolves. Of six bacterial genera isolated from canine fecal samples, Fusobacterium and Ruminococcus gnavus are typical inhabitants of canine guts, whereas these two genera are not represented in the healthy human gut microbiota. Compared with wild wolves, canine domestication seems to have caused the loss of these six bacterial genera in canines.Citation67 Interestingly, domesticated canines acquired five bacterial taxa that exist in the healthy human core gut microbiota. The Bifidobacterium population in the domesticated canine gut microbiota showed a higher average relative abundance and diversity in species numbers than those of nondomesticated canines. Hence, these results revealed that under the circumstances of cohabitation with humans, the canine gut microbiota coevolved with that of its host. Canines can adapt and gain resilience against dietary changes, and this resilience is also seen in the human gut microbiota.Citation68 Domestic canines had several microbial taxa related to cellulose and starch digestion. Metagenomics showed that the metabolic pathways of domestic canines and wolves also differed in the abundances of encoding genes, and significant differences existed in genes encoding glycosyltransferase family 34 (GT34), carbohydrate-binding module family 25 (CBM25), and glycoside hydrolase family 13 (GH13) between domestic canines and wolves. Such differences also occurred in carbohydrate enzymes, especially amylose, sucrose, and maltose. Compared with other mammals, domestic canines are more similar to humans in their gut microbiota, and consuming similar diets to those of humans played an important role in canine domestication.Citation69
Further research on canine domestication provides additional evidence of the similarities in the gut microbiota between canines and humans. Population genetics analysis shows that humans and canines both experienced similar environments during canine domestication (). Natural selection, driven by convergent environmental pressures, may have played an important role in modulating a similar set of genes between the two genomes of humans and canines. Environmental factors might have driven positive selection of some genes. A group of genes related to digestion and metabolism in both species appear to have been under positive selection during domestication. Many positive selections in both species show parallel evolution. Another interesting conclusion is that positive selection acted on genes involved in neurological processes in both humans and canines. The genes involved in canine neurological processes overlap extensively with these genes in humans.Citation70 The close interactions between humans and canines as well as drastic changes in the canines’ living environments may have caused some of the striking parallelism between human and canine genetic evolution. These interesting conclusions may provide potential approaches in using canine models to investigate neurological diseases.
Figure 1. Domestication played an important role in altering the canine gut microbiota. Canine domestication led to the loss of some gut bacteria compared with those of nondomesticated wolves, while interaction with humans resulted in new gastrointestinal bacteria in canines.
Research on the gut-brain axis has shown that the gut microbiota is significantly correlated with neurological diseases. The gut-brain axis presents new opportunities for medical and probiotic therapies in neurogastroenterology.Citation71 However, few articles to date have reported the impacts of the canine gut microbiota on the gut-brain axis and neurological diseases in canine models.
Some research on human GI diseases has been based on rodent models, which have provided meaningful results regarding the gut microbiota.Citation72 However, some researchers have questioned how informative mouse models are for research on human GI diseases.Citation73 Discrepancies in researching GI diseases, as well as whether these inferences are reliable, should be considered when translating the results from rodent models to humans.
Studies regarding taxonomic classification of the gut microbiota have found that approximately 85% of bacterial genera in the mouse gut microbiota are not present in the healthy human gut.Citation74 While the latest results show that the human gut microbiota is similar to that of both canines and mice, the canine gut microbiota is more similar to that of humans.Citation66,Citation75 Rodent models remain the predominant animal models in gastrointestinal microbiome research, and the research on mouse gastroenterology, neurology, immunology and genetics far surpasses that of other animal models.Citation76 Here, we discuss the similarities and biases in some representative cases related to GI diseases: obesity and IBD among humans, canines and mice.
In obesity:
Regarding the relevance of canine models in studying human obesity and its relation of canine models to mouse models, several factors should be considered. Obesity is usually accompanied by an altered gut microbiota.Citation77–Citation79 Some common trends exist between obese humans and mice regardless of dietary intake. Gut microbiota diversity is lower in obese canines, and this lower diversity of the gut bacterial community is also observed in obese mice and humans.Citation46,Citation79–Citation83 Combined with obesity and diet, canines who consume both high-fat and high-protein diets have lower relative abundances of Prevotella, whereas the Prevotella-dominated enterotype in the human gut is associated with high carbohydrate consumption.Citation84 Additionally, all 3 species present an increased concentration of SCFAs.Citation15,Citation85,Citation86
Increased bacterial Firmicutes/Bacteroidetes ratios have been shown in obese mouse gut microbiotas;Citation74,Citation78 however, contradictory results showing decreased Firmicutes/Bacteroidetes ratios have also been reported.Citation85,Citation87 In studies of the gut microbiota in obese humans, several conflicting reports also showed different Firmicutes/Bacteroidetes ratios, including a higher ratio,Citation82 lower ratioCitation85 and no difference.Citation88 However, the Firmicutes/Bacteroidetes ratio is higher in lean canines.Citation46 These conflicting results indicate that the Firmicutes/Bacteroidetes ratio in canines and mice should not be translated to obese humans, and further studies are needed.
Genome-wide association has shown that some genes associated with human obesity overlap with some identified obesity genes in mice.Citation89 Limited studies have investigated the genome-wide association in obesity between humans and canines, but recent research showed that canines share a significantly higher fraction of genes with humans than with mice;Citation66 therefore, obese canines may have more genes that overlap with those of obese humans.
In IBD:
Gut microbiome dysbiosis is usually observed with IBD and has been proposed as an inducing factor in IBD pathogenesis.Citation90,Citation91 IBD has been widely reported in human patients, rodent models and canine models. Studies have shown that compared with the control group, the most significant phenomenon in IBD is microbiota dysbiosis in parallel with metabolic disorders in humans, mice and canines.Citation37,Citation92–Citation95 For example, Enterobacteriaceae are increased in all 3 species with IBD,Citation96–Citation99 and Enterobacteriaceae acts in conjunction with the gut microbiota to cause IBD.Citation100 Faecalibacterium prausnitzii has anti-inflammatory effects, and research on human IBD has shown decreased Faecalibacterium prausnitzii.Citation101–Citation103 Faecalibacterium spp. are also decreased in canine IBD,Citation39 while in mice with dextran sulfate sodium (DSS)-induced and 2,4,6-trinitrobenzene sulfonic acid (TNBS)-induced colitis, Faecalibacterium spp. showed no significant changes.Citation104,Citation105 The decrease in Faecalibacterium prausnitzii may cause a research bias in IBD because Faecalibacterium prausnitzii can secrete anti-inflammatory peptides and play an important role in IBD.Citation47,Citation106 Another important bacterium, Akkermansia, is beneficial for gastrointestinal health, and Akkermansia is decreased in human IBD but increased in DSS-induced mouse models.Citation103,Citation104,Citation107 No published studies on canine IBD have discussed Akkermansia.
Metabolic disorders in IBD have been widely researched in human patients and mouse colitis models and have surpassed canine models. Fecal SCFAs are decreased by nearly the same amount in both humans and mice with IBD.Citation108–Citation110 Canines with IBD have a lower diversity of several bacteria, which are thought to produce SCFAs.Citation41,Citation111,Citation112 Fecal SCFAs are also decreased in other canine GI diseases such as chronic enteropathy.Citation42 Serum metabolites differ in humans, mice and canines. Decreased serum alanine and valine have been observed in human IBD patients as well as mice, while both of these serum metabolites are increased in canines with IBD.Citation40,Citation59,Citation113,Citation114 Alanine is an important transport metabolite for amino groups in animals; altered concentrations of alanine may indicate an altered amino acid turnover ability in IBD.Citation40 The difference in valine in IBD remains unknown. One possibility is the factor of different nutrient intakes among humans, mice and canines. However, no articles have been published to date regarding abnormal concentrations of serum metabolites in canine IBD. Deep analysis of the bacteria in both the canine and human gut provide opportunities for translating IBD research from canines to humans. A limitation of the studies on canine IBD is that few canines have been evaluated, and further research is essential to explain the similarities and biases of serum metabolites in humans and canines.
Future directions for canine gut microbiota research
While interesting similarities between the gut microbiotas in humans and canines have recently been addressed, comprehensive studies of these similarities and their mechanisms are unresolved. Research on the canine gut microbiota and GI diseases may predict results in humans. Symptoms of GI diseases in humans and canines have not been fully compared. Relationships between the canine gut microbiota and its role in the pathogenesis of canine GI diseases should be clarified. Culturomics has been widely used in studying the human gut microbiota for several years.Citation115–Citation117 Culturomics involves isolating uncultured gut bacteria to the greatest extent possible under many culture conditions. Research on probiotic therapies for canine GI diseases and beneficial functions remains limited. Potential new probiotics must be isolated to treat canine GI diseases in compliance with ethical standards.
Additional information
Funding
References
- Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall L-I, McDonald D, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16:410–422. doi:10.1038/s41579-018-0029-9.
- Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31:69–75. doi:10.1097/MOG.0000000000000139.
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi:10.1038/nrg3182.
- Davis CP, Cleven D, Balish E, Yale CE. Bacterial association in the gastrointestinal tract of beagle dogs. Appl Environ Microbiol. 1977;34:194–206.
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700.
- Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek. 1998;73:127–141. doi:10.1023/A:1000669317571.
- Suchodolski JS, Ruaux CG, Steiner JM, Fetz K, Williams DA. Application of molecular fingerprinting for qualitative assessment of small-intestinal bacterial diversity in dogs. J Clin Microbiol. 2004;42:4702–4708. doi:10.1128/JCM.42.10.4702-4708.2004.
- Suchodolski JS, Ruaux CG, Steiner JM, Fetz K, Williams DA. Assessment of the qualitative variation in bacterial microflora among compartments of the intestinal tract of dogs by use of a molecular fingerprinting technique. Am J Vet Res. 2005;66:1556–1562. doi:10.2460/ajvr.2005.66.1556.
- Apajalahti JH, Kettunen A, Bedford MR, Holben WE. Percent G+C profiling accurately reveals diet-related differences in the gastrointestinal microbial community of broiler chickens. Appl Environ Microbiol. 2001;67:5656–5667. doi:10.1128/AEM.67.12.5656-5667.2001.
- Hang I, Rinttila T, Zentek J, Kettunen A, Alaja S, Apajalahti J, Harmoinen J, de Vos WM, Spillmann T. Effect of high contents of dietary animal-derived protein or carbohydrates on canine faecal microbiota. BMC Vet Res. 2012;8:90.
- Holben WE, Feris KP, Kettunen A, Apajalahti JH. GC fractionation enhances microbial community diversity assessment and detection of minority populations of bacteria by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2004;70:2263–2270. doi:10.1128/AEM.70.4.2263-2270.2004.
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi:10.1093/dnares/dsm018.
- Shoaib M, Shehzad A, Omar M, Rakha A, Raza H, Sharif HR, Shakeel A, Ansari A, Niazi S. Inulin: properties, health benefits and food applications. Carbohydr Polym. 2016;147:444–454.
- Middelbos IS, Vester Boler BM, Qu A, White BA, Swanson KS, Fahey GC Jr. Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS One. 2010;5:e9768.
- Alexander C, Cross TL, Devendran S, Neumer F, Theis S, Ridlon JM, Suchodolski JS, de Godoy MR, Swanson KS. Effects of prebiotic inulin-type fructans on blood metabolite and hormone concentrations and faecal microbiota and metabolites in overweight dogs. Br J Nutr. 2018;120:711–720.
- Salas-Mani A, Jeusette I, Castillo I, Manuelian CL, Lionnet C, Iraculis N, Sanchez N, Fernández S, Vilaseca L, Torre C, et al. Fecal microbiota composition changes after a BW loss diet in Beagle dogs. J Anim Sci. 2018;96:3102–3111. doi:10.1093/jas/sky193.
- Diez M, Nguyen P, Jeusette I, Devois C, Istasse L, Biourge V. Weight loss in obese dogs: evaluation of a high-protein, low-carbohydrate diet. J Nutr. 2002;132:1685S–7S. doi:10.1093/jn/132.6.1685S.
- Li Q, Lauber CL, Czarnecki-Maulden G, Pan Y, Hannah SS. Effects of the dietary protein and carbohydrate ratio on gut microbiomes in dogs of different body conditions. MBio. 2017;8. doi:10.1128/mBio.01703-16
- Birkett A, Muir J, Phillips J, Jones G, O’Dea K. Resistant starch lowers fecal concentrations of ammonia and phenols in humans. Am J Clin Nutr. 1996;63:766–772. doi:10.1093/ajcn/63.5.766.
- Zhang LS, Davies SS. Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome Med. 2016;8:46. doi:10.1186/s13073-016-0296-x.
- Bridgman SL, Azad MB, Field CJ, Haqq AM, Becker AB, Mandhane PJ, Subbarao P, Turvey SE, Sears MR, Scott JA, et al. Fecal short-chain fatty acid variations by breastfeeding status in infants at 4 months: differences in relative versus absolute concentrations. Front Nutr. 2017;4:11. doi:10.3389/fnut.2017.00011.
- Xu J, Verbrugghe A, Lourenco M, Cools A, Liu DJX, Van de Wiele T, Marzorati M, Eeckhaut V, Van Immerseel F, Vanhaecke L, et al. The response of canine faecal microbiota to increased dietary protein is influenced by body condition. BMC Vet Res. 2017;13:374. doi:10.1186/s12917-017-1276-0.
- Schauf S, de la Fuente G, Newbold CJ, Salas-Mani A, Torre C, Abecia L, Castrillo C. Effect of dietary fat to starch content on fecal microbiota composition and activity in dogs. J Anim Sci. 2018. doi:10.1093/jas/sky264
- Kerr KR, Forster G, Dowd SE, Ryan EP, Swanson KS. Effects of dietary cooked navy bean on the fecal microbiome of healthy companion dogs. PLoS One. 2013;8:e74998.
- Kim J, An JU, Kim W, Lee S, Cho S. Differences in the gut microbiota of dogs (Canis lupus familiaris) fed a natural diet or a commercial feed revealed by the Illumina MiSeq platform. Gut Pathog. 2017;9:68. doi:10.1186/s13099-017-0218-5.
- Sandri M, Dal Monego S, Conte G, Sgorlon S, Stefanon B. Raw meat based diet influences faecal microbiome and end products of fermentation in healthy dogs. BMC Vet Res. 2017;13:65. doi:10.1186/s12917-017-0981-z.
- Herstad KMV, Gajardo K, Bakke AM, Moe L, Ludvigsen J, Rudi K, Rud I, Sekelja M, Skancke E. A diet change from dry food to beef induces reversible changes on the faecal microbiota in healthy, adult client-owned dogs. BMC Vet Res. 2017;13:147.
- Bermingham EN, Maclean P, Thomas DG, Cave NJ, Young W. Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ. 2017;5:e3019. doi:10.7717/peerj.3019.
- Szczesniak O, Hestad KA, Hanssen JF, Rudi K. Isovaleric acid in stool correlates with human depression. Nutr Neurosci. 2016;19:279–283. doi:10.1179/1476830515Y.0000000007.
- Henderson G, Cox F, Ganesh S, Jonker A, Young W, Collaborators GR, Abecia L, Angarita E, Aravena P, Arenas GN, Ariza C. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. 2015;5:14567. doi:10.1038/srep14567.
- He C, Shan Y, Song W. Targeting gut microbiota as a possible therapy for diabetes. Nutr Res. 2015;35:361–367. doi:10.1016/j.nutres.2015.03.002.
- Hartstra AV, Bouter KE, Backhed F, Nieuwdorp M. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care. 2015;38:159–165. doi:10.2337/dc14-0769.
- Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. doi:10.1016/j.cell.2016.05.041.
- Kimura I, Inoue D, Hirano K, Tsujimoto G. The SCFA Receptor GPR43 and Energy Metabolism. Front Endocrinol (Lausanne). 2014;5:85. doi:10.3389/fendo.2014.00085.
- Ang Z, Ding JL. GPR41 and GPR43 in Obesity and Inflammation - Protective or Causative? Front Immunol. 2016;7:28. doi:10.3389/fimmu.2016.00028.
- Jergens AE. Inflammatory bowel disease in veterinary medicine. Frontiers in Bioscience. 2012;E4:1404–1419. doi:10.2741/e470.
- Vazquez-Baeza Y, Hyde ER, Suchodolski JS, Knight R. Dog and human inflammatory bowel disease rely on overlapping yet distinct dysbiosis networks. Nat Microbiol. 2016;1:16177. doi:10.1038/nmicrobiol.2016.177.
- Blake AB, Suchodolski JS. Importance of gut microbiota for the health and disease of dogs and cats. Animal Frontiers. 2016;6:37–42. doi:10.2527/af.2016-0032.
- Suchodolski JS, Markel ME, Garcia-Mazcorro JF, Unterer S, Heilmann RM, Dowd SE, Kachroo P, Ivanov I, Minamoto Y, Dillman EM, et al. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One. 2012;7:e51907. doi:10.1371/journal.pone.0051907.
- Xu J, Verbrugghe A, Lourenco M, Janssens GP, Liu DJ, Van de Wiele T, Eeckhaut V, Van Immerseel F, Van de Maele I, Niu Y, et al. Does canine inflammatory bowel disease influence gut microbial profile and host metabolism? BMC Vet Res. 2016;12:114. doi:10.1186/s12917-016-0736-2.
- Minamoto Y, Otoni CC, Steelman SM, Buyukleblebici O, Steiner JM, Jergens AE, Suchodolski JS. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes. 2015;6:33–47.
- Minamoto Y, Minamoto T, Isaiah A, Sattasathuchana P, Buono A, Rangachari VR, McNeely IH, Lidbury J, Steiner JM, Suchodolski JS, et al. Fecal short-chain fatty acid concentrations and dysbiosis in dogs with chronic enteropathy. J Vet Intern Med. 2019. doi:10.1111/jvim.2019.33.issue-4.
- AlShawaqfeh MK, Wajid B, Minamoto Y, Markel M, Lidbury JA, Steiner JM, Serpedin E, Suchodolski JS. A dysbiosis index to assess microbial changes in fecal samples of dogs with chronic inflammatory enteropathy. FEMS Microbiol Ecol. 2017;93. doi:10.1016/j.celrep.2016.05.092
- Heilmann RM, Guard MM, Steiner JM, Suchodolski JS, Unterer S. Fecal markers of inflammation, protein loss, and microbial changes in dogs with the acute hemorrhagic diarrhea syndrome (AHDS). J Vet Emerg Crit Care (San Antonio). 2017;27:586–589. doi:10.1111/vec.12636.
- Guard BC, Barr JW, Reddivari L, Klemashevich C, Jayaraman A, Steiner JM, Vanamala J, Suchodolski JS. Characterization of microbial dysbiosis and metabolomic changes in dogs with acute diarrhea. PLoS One. 2015;10:e0127259.
- Park HJ, Lee SE, Kim HB, Isaacson RE, Seo KW, Song KH. Association of obesity with serum leptin, adiponectin, and serotonin and gut microflora in beagle dogs. J Vet Intern Med. 2015;29:43–50. doi:10.1111/jvim.12455.
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux -J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi:10.1073/pnas.0804812105.
- Fontana L, Cummings NE, Arriola Apelo SI, Neuman JC, Kasza I, Schmidt BA, Cava E, Spelta F, Tosti V, Syed FA, et al. Decreased Consumption of Branched-Chain Amino Acids Improves Metabolic Health. Cell Rep. 2016;16:520–530. doi:10.1016/j.celrep.2016.05.092.
- Jang C, Oh SF, Wada S, Rowe GC, Liu L, Chan MC, Rhee J, Hoshino A, Kim B, Ibrahim A, et al. A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat Med. 2016;22:421–426. doi:10.1038/nm.4057.
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. doi:10.1038/nm.3145.
- Mihalik SJ, Goodpaster BH, Kelley DE, Chace DH, Vockley J, Toledo FG, DeLany JP. Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity (Silver Spring). 2010;18:1695–1700.
- Soeters MR, Sauerwein HP, Duran M, Wanders RJ, Ackermans MT, Fliers E, Houten SM, Serlie MJ. Muscle acylcarnitines during short-term fasting in lean healthy men. Clin Sci (Lond). 2009;116:585–592.
- Ghonimy A, Zhang DM, Farouk MH, Wang Q. The impact of carnitine on dietary fiber and gut bacteria metabolism and their mutual interaction in monogastrics. Int J Mol Sci. 2018;19:1008. doi:10.3390/ijms19041008.
- Ouaknine Krief J, Helly de Tauriers P, Dumenil C, Neveux N, Dumoulin J, Giraud V, Labrune S, Tisserand J, Julie C, Emile J-F, et al. Role of antibiotic use, plasma citrulline and blood microbiome in advanced non-small cell lung cancer patients treated with nivolumab. J Immunother Cancer. 2019;7:176. doi:10.1186/s40425-019-0658-1.
- Vockley J, Ensenauer R. Isovaleric acidemia: new aspects of genetic and phenotypic heterogeneity. Am J Med Genet C Semin Med Genet. 2006;142C:95–103. doi:10.1002/(ISSN)1552-4876.
- Chinen Y, Nakamura S, Tamashiro K, Sakamoto O, Tashiro K, Inokuchi T, Nakanishi K. Isovaleric acidemia: therapeutic response to supplementation with glycine, l-carnitine, or both in combination and a 10-year follow-up case study. Mol Genet Metab Rep. 2017;11:2–5.
- van Hove JL, Rutledge SL, Nada MA, Kahler SG, Millington DS. 3-Hydroxyisovalerylcarnitine in 3-methylcrotonyl-CoA carboxylase deficiency. J Inherit Metab Dis. 1995;18:592–601. doi:10.1007/BF02436004.
- Visentin M, Diop-Bove N, Zhao R, Goldman ID. The intestinal absorption of folates. Annu Rev Physiol. 2014;76:251–274. doi:10.1146/annurev-physiol-020911-153251.
- Schicho R, Nazyrova A, Shaykhutdinov R, Duggan G, Vogel HJ, Storr M. Quantitative metabolomic profiling of serum and urine in DSS-induced ulcerative colitis of mice by (1)H NMR spectroscopy. J Proteome Res. 2010;9:6265–6273. doi:10.1021/pr100547y.
- Burzle M, Hediger MA. Functional and physiological role of vitamin C transporters. Curr Top Membr. 2012;70:357–375.
- Verna EC, Lucak S. Use of probiotics in gastrointestinal disorders: what to recommend? Therap Adv Gastroenterol. 2010;3:307–319. doi:10.1177/1756283X10373814.
- Strompfova V, Kubasova I, Laukova A. Health benefits observed after probiotic Lactobacillus fermentum CCM 7421 application in dogs. Appl Microbiol Biotechnol. 2017;101:6309–6319. doi:10.1007/s00253-017-8425-z.
- White R, Atherly T, Guard B, Rossi G, Wang C, Mosher C, Webb C, Hill S, Ackermann M, Sciabarra P, et al. Randomized, controlled trial evaluating the effect of multi-strain probiotic on the mucosal microbiota in canine idiopathic inflammatory bowel disease. Gut Microbes. 2017;8:451–466. doi:10.1080/19490976.2017.1334754.
- Xu H, Zhao F, Hou Q, Huang W, Liu Y, Zhang H, Agus A, Denizot J, Thévenot J, Martinez-Medina M, et al. Metagenomic analysis revealed beneficial effects of probiotics in improving the composition and function of the gut microbiota in dogs with diarrhoea. Food Funct. 2019;10:2618–2629. doi:10.1039/C9FO00087A.
- Coman MM, Verdenelli MC, Cecchini C, Bela B, Gramenzi A, Orpianesi C, Cresci A, Silvi S. Probiotic characterization of Lactobacillus isolates from canine faeces. J Appl Microbiol. 2019;126:1245–1256.
- Coelho LP, Kultima JR, Costea PI, Fournier C, Pan Y, Czarnecki-Maulden G, Hayward MR, Forslund SK, Schmidt TSB, Descombes P, et al. Similarity of the dog and human gut microbiomes in gene content and response to diet. Microbiome. 2018;6:72. doi:10.1186/s40168-018-0450-3.
- Alessandri G, Milani C, Mancabelli L, Mangifesta M, Lugli GA, Viappiani A, Duranti S, Turroni F, Ossiprandi MC, van Sinderen D., et al. Metagenomic dissection of the canine gut microbiota: insights into taxonomic, metabolic and nutritional features. Environ Microbiol. 2019;21:1331–1343. doi:10.1111/1462-2920.14540.
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi:10.1038/nature11550.
- Lyu T, Liu G, Zhang H, Wang L, Zhou S, Dou H, Pang B, Sha W, Zhang H. Changes in feeding habits promoted the differentiation of the composition and function of gut microbiotas between domestic dogs (Canis lupus familiaris) and gray wolves (Canis lupus). AMB Express. 2018;8:123.
- Wang GD, Zhai W, Yang HC, Fan RX, Cao X, Zhong L, Wang L, Liu F, Wu H, Cheng L-G, et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun. 2013;4:1860. doi:10.1038/ncomms2814.
- Saulnier DM, Ringel Y, Heyman MB, Foster JA, Bercik P, Shulman RJ, Versalovic J, Verdu EF, Dinan TG, Hecht G, et al. The intestinal microbiome, probiotics and prebiotics in neurogastroenterology. Gut Microbes. 2013;4:17–27. doi:10.4161/gmic.22973.
- Hugenholtz F, de Vos WM. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci. 2018;75:149–160. doi:10.1007/s00018-017-2693-8.
- Nguyen TL, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8:1–16. doi:10.1242/dmm.017400.
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi:10.1073/pnas.0504978102.
- Swanson KS, Dowd SE, Suchodolski JS, Middelbos IS, Vester BM, Barry KA, Nelson KE, Torralba M, Henrissat B, Coutinho PM, et al. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. Isme J. 2011;5:639–649. doi:10.1038/ismej.2010.162.
- Ma Q, Xing C, Long W, Wang HY, Liu Q, Wang RF. Impact of microbiota on central nervous system and neurological diseases: the gut-brain axis. J Neuroinflammation. 2019;16:53. doi:10.1186/s12974-019-1434-3.
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi:10.1126/science.1241214.
- Lu Y, Fan C, Li P, Lu Y, Chang X, Qi K. Short chain fatty acids prevent high-fat-diet-induced obesity in mice by regulating G protein-coupled receptors and gut microbiota. Sci Rep. 2016;6:37589. doi:10.1038/srep37589.
- Kiilerich P, Myrmel LS, Fjaere E, Hao Q, Hugenholtz F, Sonne SB, Derrien M, Pedersen LM, Petersen RK, Mortensen A, et al. Effect of a long-term high-protein diet on survival, obesity development, and gut microbiota in mice. Am J Physiol Endocrinol Metab. 2016;310:E886–99. doi:10.1152/ajpendo.00363.2015.
- Bouter KE, van Raalte DH, Groen AK, Nieuwdorp M. Role of the gut microbiome in the pathogenesis of obesity and obesity-related metabolic dysfunction. Gastroenterology. 2017;152:1671–1678. doi:10.1053/j.gastro.2016.12.048.
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi:10.1038/nature07540.
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi:10.1038/4441022a.
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J-M, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi:10.1038/nature12506.
- Deng P, Swanson KS. Gut microbiota of humans, dogs and cats: current knowledge and future opportunities and challenges. Br J Nutr. 2015;113(Suppl):S6–17. doi:10.1017/S0007114514002943.
- Schwiertz A, Taras D, Schafer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring). 2010;18:190–195.
- Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, Clarke SF, O’Toole PW, Quigley EM, Stanton C, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635–1642. doi:10.1136/gut.2010.215665.
- Serino M, Luche E, Gres S, Baylac A, Berge M, Cenac C, Waget A, Klopp P, Iacovoni J, Klopp C, et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut. 2012;61:543–553. doi:10.1136/gutjnl-2011-301012.
- Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, Flint HJ. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond). 2008;32:1720–1724.
- Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau C, Bennett B, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17:141–152. doi:10.1016/j.cmet.2012.12.007.
- Duboc H, Rajca S, Rainteau D, Benarous D, Maubert M-A, Quervain E, Thomas G, Barbu V, Humbert L, Despras G, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi:10.1136/gutjnl-2012-302578.
- Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis in inflammatory bowel disease. Gut. 2004;53:1–4. doi:10.1136/gut.53.1.1.
- Jergens AE, Simpson KW. Inflammatory bowel disease in veterinary medicine. Front Biosci (Elite Ed). 2012;4:1404–1419. doi:10.2741/e470.
- Khan I, Ullah N, Zha L, Bai Y, Khan A, Zhao T, Che T, Zhang C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): cause or consequence? IBD treatment targeting the gut microbiome. Pathogens. 2019;8. doi:10.3390/pathogens8030126
- Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap Adv Gastroenterol. 2013;6:295–308. doi:10.1177/1756283X13482996.
- Nguyen GC. Editorial: bugs and drugs: insights into the pathogenesis of inflammatory bowel disease. Am J Gastroenterol. 2011;106:2143–2145. doi:10.1038/ajg.2011.308.
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129.
- Berry D, Schwab C, Milinovich G, Reichert J, Ben Mahfoudh K, Decker T, Engel M, Hai B, Hainzl E, Heider S, et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. Isme J. 2012;6:2091–2106. doi:10.1038/ismej.2012.39.
- Cassmann E, White R, Atherly T, Wang C, Sun Y, Khoda S, Mosher C, Ackermann M, Jergens A. Alterations of the ileal and colonic mucosal microbiota in canine chronic enteropathies. PLoS One. 2016;11:e0147321. doi:10.1371/journal.pone.0147321.
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi:10.1186/gb-2012-13-9-r79.
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi:10.1016/j.chom.2010.08.004.
- Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637.
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Doré J, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi:10.1002/ibd.20903.
- Lopez-Siles M, Enrich-Capo N, Aldeguer X, Sabat-Mir M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Alterations in the Abundance and Co-occurrence of Akkermansia muciniphila and Faecalibacterium prausnitzii in the Colonic Mucosa of Inflammatory Bowel Disease Subjects. Front Cell Infect Microbiol. 2018;8:281.
- Hakansson Å, Tormo-Badia N, Baridi A, Xu J, Molin G, Hagslatt M-L, Karlsson C, Jeppsson B, Cilio CM, Ahrné S, et al. Immunological alteration and changes of gut microbiota after dextran sulfate sodium (DSS) administration in mice. Clin Exp Med. 2015;15:107–120. doi:10.1007/s10238-013-0270-5.
- Ettreiki C, Gadonna-Widehem P, Mangin I, Coeffier M, Delayre-Orthez C, Anton PM. Juvenile ferric iron prevents microbiota dysbiosis and colitis in adult rodents. World J Gastroenterol. 2012;18:2619–2629. doi:10.3748/wjg.v18.i21.2619.
- Quevrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermúdez-Humarán LG, Pigneur B, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut. 2016;65:415–425. doi:10.1136/gutjnl-2014-307649.
- Lopetuso LR, Petito V, Graziani C, Schiavoni E, Paroni Sterbini F, Poscia A, Gaetani E, Franceschi F, Cammarota G, Sanguinetti M, et al. Gut microbiota in health, diverticular disease, irritable bowel syndrome, and inflammatory bowel diseases: time for microbial marker of gastrointestinal disorders. Dig Dis. 2018;36:56–65. doi:10.1159/000477205.
- Huda-Faujan N, Abdulamir AS, Fatimah AB, Anas OM, Shuhaimi M, Yazid AM, Loong YY. The impact of the level of the intestinal short chain Fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem J. 2010;4:53–58.
- Parada Venegas D, De la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, Harmsen HJ, Faber KN. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019;10:277.
- Agus A, Denizot J, Thevenot J, Martinez-Medina M, Massier S, Sauvanet P, Bernalier-Donadille A, Denis S, Hofman P, Bonnet R, et al. Western diet induces a shift in microbiota composition enhancing susceptibility to adherent-invasive E. coli infection and intestinal inflammation. Sci Rep. 2016;6:19032. doi:10.1038/srep19032.
- Guard BC, Suchodolski JS. HORSE SPECIES SYMPOSIUM: canine intestinal microbiology and metagenomics: from phylogeny to function. J Anim Sci. 2016;94:2247–2261. doi:10.2527/jas.2015-0029.
- Omori M, Maeda S, Igarashi H, Ohno K, Sakai K, Yonezawa T, Horigome A, Odamaki T, Matsuki N. Fecal microbiome in dogs with inflammatory bowel disease and intestinal lymphoma. J Vet Med Sci. 2017;79:1840–1847.
- Schicho R, Shaykhutdinov R, Ngo J, Nazyrova A, Schneider C, Panaccione R, Kaplan GG, Vogel HJ, Storr M. Quantitative metabolomic profiling of serum, plasma, and urine by (1)H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J Proteome Res. 2012;11:3344–3357.
- Dawiskiba T, Deja S, Mulak A, Zabek A, Jawien E, Pawelka D, Banasik M, Mastalerz-Migas A, Balcerzak W, Kaliszewski K, et al. Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J Gastroenterol. 2014;20:163–174. doi:10.3748/wjg.v20.i1.163.
- Lagier J-C, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Seck EH, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi:10.1038/nmicrobiol.2016.203.
- Greub G. Culturomics: a new approach to study the human microbiome. Clin Microbiol Infect. 2012;18:1157–1159. doi:10.1111/1469-0691.12032.
- Lagier JC, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, Levasseur A, Rolain J-M, Fournier P-E, Raoult D, et al. Culturing the human microbiota and culturomics. Nat Rev Microbiol. 2018;540–550. doi:10.1038/s41579-018-0041-0
