 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Probiotic products have been shown to have beneficial effects on human hosts, but what happens in the gastrointestinal tract after its ingestion remains unclear. Our aim was to investigate the changes within the small intestines after a single intake of a fermented milk product containing a probiotic. We have periodically collected the small-intestinal fluids from the terminal ileum of seven healthy subjects for up to 7 h after ingestion by small-intestinal fluid perfusion using an endoscopic retrograde bowel insertion technique. The bacterial composition of the terminal ileum clearly revealed that the ingested probiotics (Lactobacillus casei strain Shirota: LcS and Bifidobacterium breve strain Yakult: BbrY) occupied the ileal microbiota for several hours, temporarily representing over 90% of the ileal microbiota in several subjects. Cultivation of ileal fluids showed that under a dramatic pH changes before reaching the terminal ileum, a certain number of the ingested bacteria survived (8.2 ± 6.4% of LcS, 7.8 ± 11.0% of BbrY). This means that more than 1 billion LcS and BbrY cells reached the terminal ileum with their colony-forming ability intact. These results indicate that there is adequate opportunity for the ingested probiotics to continuously stimulate the host cells in the small intestines. Our data suggest that probiotic fermented milk intake affects intestinal microbes and the host, explaining part of the process from the intake of probiotics to the exertion of their beneficial effects on the host.
Introduction
Probiotics, which include lactic acid bacteria and bifidobacteria, have been marketed in various forms, such as fermented milk, tablets, biscuits, and chocolates. The Food and Agriculture Organization and World Health Organization defined probiotics as “live microorganisms which when administered in adequate amounts confer a health benefit on the host.”Citation1 The International Life Science Institute has a similar definition for a probiotic: “a live microbial food ingredient that, when ingested in sufficient quantities, confers health benefits on the consumer.”Citation2 As is clear from these definitions, probiotics must be alive and have beneficial effects on the host.
In terms of benefit to the host, probiotics have been shown in numerous human and animal studies to improve gastrointestinal symptoms,Citation3,Citation4 bowel habits,Citation5 constipation,Citation6 host defense against infection,Citation7-Citation10 immunological control,Citation11,Citation12 cancer prevention,Citation13,Citation14 stress-associated symptoms,Citation15-Citation17 and sleep.Citation18 The recovery of orally ingested strains in fecal samples has been investigated in several studiesCitation19,Citation20 to examine the ability of probiotics to reach the intestines in a viable state. However, reports showing the behavior of ingested probiotics within the intestinal tract have been limited.Citation21,Citation22 When fermented milk containing lactic acid bacteria and/or bifidobacteria is ingested, these probiotics are exposed to bactericidal gastric juice and bile and interact closely with various resident bacteria during the transit through the gastrointestinal tract from the oral cavity to the colon.
Small-intestinal fluid perfusion using an endoscopic retrograde bowel insertion (ERBI) technique is a powerful tool which allows to collect ileal perfusion fluid periodically.Citation23 A double-lumen tube, featuring an occluding balloon, is inserted retrograde through the colon and placed at the terminal ileum. This makes it possible to collect the ileal fluids over time under physiological conditions without affecting secretion of gastric juice and bile. This ERBI technique has been used for analyses of the dynamics and metabolism of indigestible polysaccharides such as resistant starchCitation24 and raffinose,Citation25 and of dietary fibers such as pectinCitation26 and cellulose.Citation27
Our aim was to elucidate the environmental change in the gastrointestinal tract after ingestion of a probiotic product. For this purpose, we periodically collected fluids from the terminal ileum of healthy subjects for up to 7 h after intake of probiotic (Lactobacillus casei strain Shirota: LcS; Bifidobacterium breve strain Yakult: BbrY)-containing fermented milk using ERBI technique. We focused on the questions to clarify the following: the duration of the ingested strain to pass through the digestive tract; whether a single ingested strain become dominant within the digestive tract under coexisting with various resident microbial species; the proportion of an ingested strain maintains metabolic activity and the ability to grow (i.e., colony-forming ability) after reaching the colon; whether there is a negative correlation between the viability rate of the ingested strain and the concentration of bile acids.
Results
Recovery rate of ingested strains
The intestinal transit time and the total bacterial counts of the ingested strains were investigated. The sum of viable and dead cells of LcS, BbrY, or both in each ileal fluid sample collected at different time points was counted by using a fluorescent antibody technique. Then, the recovery rate of the ingested probiotic strains over time within the terminal ileum was calculated (). Ingested strains started to increase within the terminal ileum around 2 h after intake of fermented milk containing LcS (LFM), BbrY (BFM), or both LFM and BFM. Each strain increased with time, and reached a peak around 4 h after the ingestion. These results indicate that probiotics reaches the terminal ileum within hours of ingestion. When both LFM and BFM were ingested simultaneously, the recovery rates of LcS and BbrY transitioned in the same manner.
Figure 1. Changes in the cumulative recovery rate of (a) ingested probiotic strains, (b) PEG, and (c) lactose.
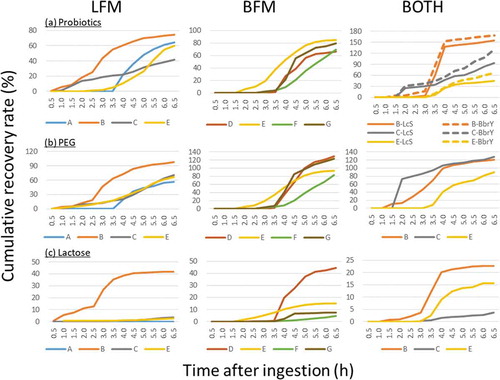
The non-absorbable marker polyethylene glycol (PEG) was ingested at the same time as the fermented milk in all cases, and it transitioned with a time similar to that of the bacterial strains in all trials (). The correlation coefficient between the recovery rate of PEG and the ingested strain at each collection time for each trail, except the case of subject C ingested LFM alone, was 0.73 to 0.98 (n = 10, p < .01).
Microbiota composition
DNA was extracted from the collected ileal fluids, and the microbiota composition was analyzed using 16 S rRNA gene profiling ().
Figure 2. Changes in relative abundance of ileal microbiota.
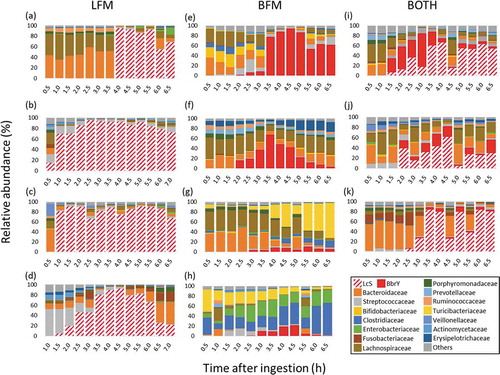
LFM single ingestion: After the ingestion of LFM alone, LcS was first detected in ileal fluid anywhere from 0.5 h (subject B) to 4 h (subject A) ). In subject E (), Streptococcaceae occupied about half of the ileal microbiota until 2 h after LFM ingestion, after which LcS predominated. The relative abundance of LcS in the ileal microbiota peaked above 90%, and for subjects A (), B (), and C () remained high in the last sample obtained.
BFM single ingestion: BbrY transited the gastrointestinal tract more slowly than LcS and, except in subject D (), did not persist as long (). In addition, its relative abundance was lower than that of LcS.
Both LFM and BFM: In subjects who ingested both probiotics, LcS and BbrY were detected in ileal fluid at similar times (). The combined relative abundance varied among the subjects and over time within the subjects. In all subjects, the relative abundance of LcS was much higher than that of BbrY, except for subject B (), in whom the relative abundance of BbrY was higher than that of LcS until 3.5 h after ingestion, after which LcS was more abundant.
Subjects B (), C (), and E () each underwent an additional LFM single-ingestion trial on a separate day. These subjects showed differences between the two trials such as the time until the first detection and the occupancy degree of ingested strain as well as the presence or absence of Streptococcaceae.
Viability rate of ingested strains
Viable LcS and BbrY were detected from all ileal fluid samples that were determined to be positive by 16 S rRNA gene analysis (). The viable LcS and BbrY in each ileal fluid sample were counted, and the changes in the viability rate over time in each subject were plotted (Supplementary Figure 1). The viability rate for LcS was maintained at 10% or more from 1 h to 6.5 h after LFM ingestion in subject C, with a maximum of about 55%, but was only a few percent throughout the trial in subjects A and E. The viability rate of BbrY also differed substantially among the subjects, from <2% in subjects F and G to peaks above 60% in subject D. When LFM and BFM were ingested simultaneously, the viability rate of LcS remained higher than that of BbrY throughout each trial, although live bacteria of both strains were detected. The viability rates for both LcS and BbrY were higher in the early phase after ingestion and then decreased with time. Subject E underwent trials of all three conditions (LFM, BFM, and both), and the pattern of viability over time for this subject differed between conditions.
Survival rate of ingested strains
The total cell numbers of the two probiotics were determined by fluorescent antibody technique, using monoclonal antibodies for LcSCitation19 or BbrY.Citation28 Subject numbers of LFM and BFM ingestion were calculated by adding the single ingestion (n = 4 each) and the simultaneous ingestion (n = 3). Each bottle of fermented milk contained 10.9 ± 0.1 log10 LcS cells, 10.8 ± 0.1 log10 BbrY cells. The average number of recovered cells (recovery rate) at the terminal ileum was 10.8 ± 0.2 log10 cells (78 ± 39%) after ingestion of LFM (n = 7) and 10.8 ± 0.2 log10 cells (104 ± 39%) after ingestion of BFM (n = 7). The recovery rates of LcS and BbrY positively correlated with that of PEG (r = 0.69, p = .09 for LcS and r = 0.60, p = .16 for BbrY).
The ileal fluids were cultured on a rich nutrient agar plate medium, and cells with colony-forming ability were counted as viable cells. Before ingestion, the viable LcS in LFM and BbrY in BFM were 10.9 ± 0.1 log10 cfu/bottle and 10.8 ± 0.0 log10 cfu/bottle, respectively, suggesting that almost all of the cells contained in each type of fermented milk were viable. The viable cell numbers (survival rates) of LcS (n = 7) and BbrY (n = 7) at the terminal ileum after ingestion of LFM and BFM were 9.6 ± 0.7 log10 cfu (range: 0.2%-19.4%, mean: 8.2 ± 6.4%) and 9.2 ± 0.9 log10 cfu (range: 0.08%-31.6%, mean: 7.8 ± 11.0%), respectively.
The advantage of the combination intake trial is that the survival rate of both strains can be evaluated under the same condition because the strains are taken simultaneously within a single subject. In all three subjects who simultaneously ingested both LFM and BFM, the survival rate of LcS was higher than that of BbrY (Subject B: 19.4% [LcS] >4.0% [BbrY], Subject C: 12.4% [LcS] >6.6% [BbrY], Subject E: 9.3% [LcS] >2.6% [BbrY]).
Bile acid
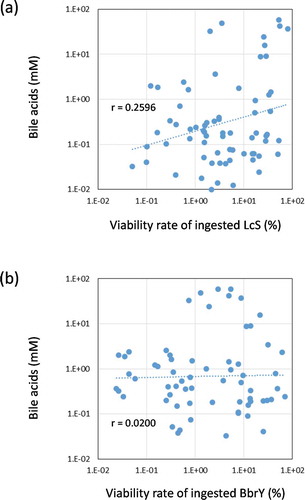
The total bile acid concentration in the ileal fluids varied among individuals and even within the individual, ranging from 0.7 to 2,383 mM, with an average of 251 ± 714 mM (Supplementary Figure 2a). The mean ± SD of the deconjugation ratio (Supplementary Figure 2b) and 7α-dehydroxylation ratio (Supplementary Figure 2 c) was 19% ± 15% and 12% ± 12%, respectively, showing that both reactions occurred rarely in the upper digestive tract. Although previous studiesCitation21,Citation22 have made clear that gastric acid and bile strongly influence the survival of ingested probiotics, no negative correlation was found between the bile acid concentration and the viability rate of LcS () or BbrY () in the ileal fluids collected in the present study.
Figure 3. Relationships between viability rate of ingested strains and bile acid concentration in ileal fluids.
Lactose
LFM and BFM contain 1.4 g and 4.0 g of lactose, respectively. To determine whether the lactose reached the terminal ileum without being digested and absorbed in the upper gastrointestinal tract, we measured the amount of lactose in the ileal fluids collected at each time point. The percentage of the ingested amount is shown as the cumulative recovery rate (). Although the transit time to reach the terminal ileum was different for each trial, lactose was detected in all subjects, and the recovery rate generally reached a peak at around 3.5 h after ingestion. The recovery rate of lactose in subject B was more than four times that in subject C after LFM single ingestion and after simultaneous ingestion of both. With some exceptions (LFM single ingestion by subjects A and E, and ingestion of both LFM and BFM by subject C), the time course of lactose recovery was similar to that of PEG (). The correlation coefficient of the eight cases excluding the three exceptions noted above was 0.87 to 0.97.
Streptococcaceae
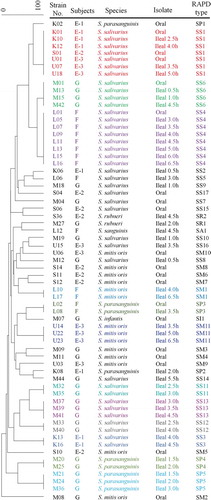
Microbiomics of the ileal fluids showed that the relative abundance of Streptococcaceae increased along with the ingested strains in some cases (Supplementary Figure 3). The Feature ID analysis indicated that most of the Streptococcaceae were oral-predominant bacterial species such as the Streptococcus mitis/oralis group or S. salivarius, leading us to speculate that these originated in the oral cavity. To verify this hypothesis, Streptococcaceae strains were isolated from both the oral fluid and ileal fluid of subjects E, F, and G, and RAPD-PCR typing was performed (). Three RAPD-types of S. salivarius (SS1, SS4, and SS6) and 1 type of S. parasanguinis (SP3) were isolated from both the oral fluid and ileal fluid of each subject. In subject E, SS1 was isolated from the oral fluid in all three trials and also from the ileal fluid in two out of the three trials.
Figure 4. Dendrogram derived from a comparison of RAPD-PCR profiles of the 65 individual Streptococcaceae strains found in subjects.
Discussion
A large number of probiotics have been proposed, and some have been shown to reach the intestines in a viable stateCitation19,Citation20 and to beneficially affect the host.Citation3-Citation18 However, what happens to the ingested probiotics between ingestion and excretion in feces and how they exert their physiological effects has been unclear. Here we have shed light on some of these processes that were previously considered to be a “black box.”
The recovery rates of the ingested LcS, BbrY, and the non-absorbable marker PEG showed the same pattern (), suggesting that LcS and BbrY passed through the upper digestive tract without colonizing it or stagnating in that region. LcS has been thought to pass out of the intestines without colonizing them because it becomes undetectable about a week after stopping the continuous intake of LFM.Citation29 Our results support this hypothesis.
The bacterial composition of the terminal ileum () clearly showed that the ingested LcS and BbrY occupied the ileal microbiota for several hours, temporarily representing over 90% of the ileal microbiota in several subjects. On the other hand, major indigenous members of the intestinal microbiome such as Bacteroidaceae and Lachnospiraceae have relatively decreased. Previous reports indicated that LcS is transported by M cells in the follicle-associated epithelium and is then recognized by macrophages and dendritic cells in Peyer’s patches, inducing interleukin (IL)-12 production and Th1 enhancement.Citation11 Continuous ingestion of 100 ml of fermented milk containing 100 billion live LcS for 8 weeks relieves temporary stressCitation15-Citation17 and improves sleep quality.Citation18 These effects are induced through the vagus nerve,Citation16,Citation30 which is spread throughout the gastrointestinal tract. Because the intestinal microbiota is occupied by the ingested LcS for a period of time, there is adequate opportunity for the ingested LcS to continuously stimulate the immune cells, nerve cells, and glia cells in the upper gastrointestinal tract. We believe this discovery is a major step toward elucidating the mechanisms of the physiological effects on probiotics.
After oral ingestion of fermented milk, LcS and BbrY are first exposed to strongly acidic gastric juice, and then placed in a neutral-to-alkaline environment when mixed with bile and pancreatic juice. Thus, these probiotics experience a dramatic pH change before reaching the terminal ileum. Cultivation of ileal fluids showed that under such severe environmental changes, a certain number of the ingested bacteria survived (8.2 ± 6.4% of LcS, 7.8 ± 11.0% of BbrY); this means that more than 1 billion LcS and BbrY cells reached the terminal ileum with their colony-forming ability intact, even after exposure to gastric acid and bile during the transit through the upper digestive tract. This is consistent with previous studies that have reported survival rates of LcS in human feces after ingestion of LFM of 1.1% to 10%.Citation31
Although clarification of the growth and death of LcS and BbrY while passing through the colon are needed to draw firm conclusions, the results of this study indicate that the environment in the upper digestive tract from the oral cavity to the terminal ileum largely determines the survival rate of LcS and BbrY. Although it cannot be determined how much gastric juice and bile influenced the ingested probiotics during passage through the upper digestive tract, no correlation between the transit time to reach the terminal ileum and the viability rate of LcS and BbrY was observed (Supplementary Figure 1). Bile acid is an important biophylactic factor that, together with gastric acid, prevents the invasion of pathogenic microorganisms. For example, bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats.Citation32 Bile acids damage cell membranes because of their surfactant characteristics, and hydrophobic deconjugated and secondary bile acids have stronger activity than hydrophilic conjugated and primary bile acids.Citation33 Bile acids undergo deconjugation and dehydration by intestinal bacteria.Citation34 Our present study showed that deconjugation (Supplementary Figure 2b) and dehydration (Supplementary Figure 2c) were not complete in the terminal ileum, and no negative correlation was found between total bile acid concentration and the viability ratio of LcS and BbrY (). Additionally, there were no correlations between the concentration of each molecular species of bile acid and the viability rate of LcS and BbrY (data not shown). These results indicate that analyzing only the bile acids is not enough to explain what biological factors are involved in the survival through the gastrointestinal tract. Analyzing not only bile acids but other factors, such as gastric acid, antimicrobial defensin, etc., and clarifying the relationship between bacterial survival rates will be necessary to elucidate the mechanism of the gastrointestinal survival ability.
As described above, cultivation of ileal fluids showed that more than 1 billion cells of LcS and BbrY reached the terminal ileum with maintained colony-forming ability. However, the bacterial counts were not directly measured from the ileal fluid samples but by culturing the samples on an enriched culture medium, which is not sufficient for determining whether the probiotics were actually metabolizing or growing in the small intestines. Lactose is a component of fermented milk and can be used by LcS and BbrY as an energy source. Our results showed that some lactose escaped digestion and absorption, and reached the terminal ileum (). A previous clinical studyCitation35 reported that intake of LcS in dairy increased indigenous bifidobacteria, suggesting that the probiotics produce an acidic environment that is favorable for bifidobacteria by producing lactate in the gastrointestinal tract. Further investigations are needed to clarify whether LcS produces lactate in the colon. Lactate is an antibacterial substanceCitation36 as well as an effector for proliferation of epithelial cells,Citation37 and it is a substrate of butyrate productionCitation38 by butyric acid bacteria. Butyrate is a short-chain fatty acid that is an energy source for colonic epithelial cellsCitation39,Citation40 and stimulates cell proliferationCitation41 and mucus secretion.Citation42
In this study, it was revealed that as the ingested LFM reach the terminal ileum, not only LcS but also the abundance of Streptococcaceae relatively increased (Supplementary Figure 3). Streptococcaceae strains isolated from the oral fluids before the fermented milk intake and the ileal fluids collected after the intake were identified as monophyletic (). This result clearly reveals that Streptococcaceae strains living in the oral cavity reached the terminal ileum in association with the ingested fermented milk without losing their colony-forming ability. This is a new finding that has not been reported in the past. The SS1 strain was isolated from oral fluids collected in three independent studies of subject E (), showing that the identical strain colonized the oral cavity for a year. The oral cavity has a unique microbiota, which is different from the intestinal microbiota and is composed of approximately 700 species of bacteria.Citation43 The bacteria that cause periodontitis, bacterial endocarditis, and aspiration pneumonia are included among the oral microbiota, and oral bacteria are causally related to inflammatory bowel disease.Citation43-Citation46 Our result shows that non-negligible amount of bacteria living in the oral cavity pass through the upper gastrointestinal tract with food and drink while being exposed to gastric acid and bile acid, and some of them can reach the terminal ileum alive. Thus, intraoral care is important from the perspective of preventing gastrointestinal diseases as well as oral diseases.
We used the small intestinal fluid perfusion by ERBI technique to periodically examine the state of the terminal ileum after intake of fermented milk. The ERBI method has been used for studies on digestion and absorption of indigestible polysaccharides and water-soluble and water-insoluble dietary fibers in the small intestine.Citation24-Citation27 Our results have confirmed, for the first time, that this method is also useful for the analysis of bacteria and their metabolites. The small intestinal fluid perfusion using ERBI technique entails placing a balloon in the intestinal tract to stop the downstream movement of contents, introducing a perfusion solution, and collecting the contents coming down from higher in the gastrointestinal tract. In principle, if the indwelling position of the balloon moves downstream during the experiment, the newly exposed endogenous intestinal bacteria will also be collected simultaneously. In this case, the evaluation for the recovery of ingested probiotics is not affected, but the relative abundance of ingested probiotics in the ileal microbiota cannot be accurately estimated. Moreover, if the balloon moves beyond the ileocecal valve into the colon, the ingested probiotics will be buried in a large population of resident bacteria. This could explain why the BbrY occupancy rate was extremely low after the single BFM ingestion for subjects F and G (). Indeed, in subject G, X-ray imaging confirmed that the balloon, which was placed at the terminal ileum at the beginning of the trial, had moved to the colon by the end of the trial, 6.5 h after the ingestion of BFM. Fixing the indwelling position of the balloon is therefore an important point to be noted for the use of this method. By carefully adjusting so that the indwelling site of the balloon does not move during the trial, the ERBI method can be applied to the dynamic analysis of ingested bacteria in not only the terminal ileum but also the ascending, transverse, and descending colon.
As mentioned in the introduction, one of our main objective was to clarify several questions regarding the environmental change in the gastrointestinal tract after ingestion of a probiotic product. Results from this study obtained the following answers: the ingested LcS and BbrY generally reached the terminal ileum around 2 h after intake of probiotic product; the ingested LcS and BbrY occupied the ileal microbiota for several hours; more than 1 billion of the ingested LcS and BbrY maintained their colony-forming ability after reaching the terminal ileum; no negative correlation was found between the bile acid concentration and the viability rate of the ingested LcS or BbrY in the ileal fluids.
These findings indicate the behavior of probiotic strains and ingredients in the intestinal tract after intake of probiotic products, and are expected to be a step toward to understand the significance of probiotics intake.
It has been noted that the effects of probiotics on their hosts and/or their gut microbiomes might be influenced by the highly heterogeneous in terms of diet, age range, genetic background, gut microbiome configuration, and the degree of gut colonization by probiotics.Citation47 From our present study, it became clear that changes in the intestinal environment after ingestion of probiotics products, e.g., the duration and the degree of the ingested strain to occupy the small intestines; the survival rate of ingested strain in the small intestines; the bile acid concentration in the ileal fluids, differed not only between product/strain types but also between individuals and within individuals. These heterogeneities may account for the differential effects of probiotics observed between individuals.
Since this research area had remained as a complete “black box,” our priority was to reliably acquire data over time, regardless of the number of cases. The trial numbers of this study were limited to discuss the statistical interpretation among each ingestion group. However, our results have shown high potential for future use in studies elucidating the behavior of probiotics in the digestive tract and the mechanisms of their various physiological effects, and performing a large scale trail is a next step.
In summary, this study has clearly shown that the ingested probiotics will occupy the ileal microbiota for several hours. Understanding the process of the ingested probiotics between ingestion and excretion in feces and how they affect the intestinal microbes and host, is a major step toward elucidating the mechanisms of the physiological effects on probiotics.
Materials and methods
Subjects
Seven healthy men (mean age 37.6 ± 13.2 years) were recruited by the department of Gastroenterology and Hematology, Hirosaki University Graduate School of Medicine. All subjects provided written informed consent. This study was approved by the Ethics Committee of the Graduate School of Medicine at Hirosaki University (UMIN ID: UMIN000018128) and conducted in compliance with ethical principles from the Declaration of Helsinki, guidelines of good clinical practice, and applicable regulations.
Test meal
Commercially available fermented daily products, LFM and BFM, were used as the test beverages. The total (viable and dead) and viable LcS numbers contained in LFM (80 mL per bottle) were 10.9 ± 0.1 log10 cells and 10.9 ± 0.1 log10 cfu, respectively. The total and viable BbrY numbers contained in BFM (100 mL per bottle) were 10.8 ± 0.1 log10 cells and 10.8 ± 0.0 log10 cfu, respectively. At the time of the trial, a meal-replacement drink (Calorie Mate, Otsuka Pharmaceutical) was used as breakfast. In addition, the test beverage (1 bottle) was taken with a solution prepared by dissolving 5 g of the non-absorbable marker polyethylene glycol-4000 (PEG: Sigma-Aldrich [cat. no. 81240]) in 100 ml of water. During the trial, the subjects could freely ingest water.
Trial and sample collections
Eleven trials were performed in three test groups: LFM single ingestion (n = 4; subjects A, B, C, and E) for the first test, BFM single ingestion (n = 4; subjects D, E, F, and G) for the second test, and LFM and BFM simultaneous ingestion (n = 3; subject B, C, and E) for the third test. The trial interval for the subjects participated in more than one test (subjects B, C, and E), was set as more than 4 months. The day before each trial, all subjects collected 0.5 g of fecal sample into a fecal collection tube (Sarstedt AG & Co. KG [cat. no. 80.734.001]) containing 3 ml of RNAlater® (Thermo Fisher Scientific [cat. no. AM7021]). Oral fluids were collected at the time of awakening on the day of the trial for five trials: LFM single ingestion (subject E), BFM single ingestion (subjects E, F, and G), and LFM and BFM simultaneous ingestion (subject E). The subjects were instructed to gargle 2 to 3 times with 20 ml of saline, and expectorate the oral fluid into a specimen tube. During the trial, ileal fluids were collected from all subjects by small-intestinal fluid perfusion using the ERBI technique (described below). All samples were kept at 4°C immediately after the collection.
Small-intestinal fluid perfusion
On the day before the trial, all subjects were requested to consume a low-residual food “Darm Space Rich III” (House Foods Group) instead of a normal meal. Also, they were requested to consume more than 200 ml of fluids at 7 pm, and after that, were instructed not to eat until the start of the trial. Subjects were not permitted to drink alcohol or smoke from the day before the trial until the trial end. Suppositories were administered at the night before and in the morning of the trial, and carbon dioxide-releasing suppositories were used for the subjects before the colonoscopic insertion. Intestinal lavage was not performed. Small-intestinal fluid perfusion using the ERBI technique was performed as previously reported,Citation23 with slight modification. A colonoscope (PCF-PQ260 L, Olympus) covered with a single-balloon endoscopic overtube (Disposable sliding tube ST-SB1, Olympus) was inserted into the terminal ileum. The tip of the tube was fixed in place by inflating the balloon using the overtube balloon control unit (Olympus Balloon Control Unit, Olympus). After the guide wire was passed through the forceps channel of the endoscope, only the endoscope was removed. Then, the end of the double-lumen tube with the balloon was inserted through the guide wire to a position approximately 10 cm from the ileocecal valve. After inflating the balloon with 15 to 20 ml of sterile-distilled water using a syringe, the overtube balloon was deflated. After the start of perfusion, subjects were placed supine on a bed, and ingested test meal and breakfast within 5 minutes. 0.03% phenolsulfonphthalein (PSP) was continuously infused through the lumen whose end was positioned 10 cm proximally to the second lumen at a rate of 1.0 ml/min to calculate ileal flow. Ileal fluid was continuously aspirated at – 20 to – 100 mmH2O from the second lumen and collected every 30 min until a maximum of 7 h after ingestion of the test beverage. The volume of each ileal fluid sample was measured as soon as it was collected, and it was immediately subjected to various other measurements. Some of each ileal fluid was suspended in 3 times its volume of RNAlater® and was stored at – 80°C for microbiomics.
Microbiomics
The RNAlater®-fixed fecal samples and ileal samples were washed twice with phosphate-buffered saline (PBS) solution before DNA extraction. DNA was extracted by using glass beads and buffer-saturated phenol (NIPPON GENE Co. Ltd. [code no. 319–90093]).Citation48 The V1-V2 region of the 16 S rRNA gene was amplified from the extracted DNA by using the primers 27F mod2-MiSeq and 338R-MiSeq.Citation49 The PCR cycle was as follows: 50°C for 2 min; 95°C for 10 min; and a repeated cycle of 95°C for 30 s, 55°C for 30 s, 72°C for 90 s. The PCR was stopped before signal saturation. The MiSeq library was prepared with a MiSeq Reagent Kit v2 (Illumina [cat. no. MS-102-2001]). Sequencing reads were pre-processed using QIIME2Citation50 and its plugins. Noise, chimeras, and trimming of the sequence were removed by using the DADA2Citation51 plug-in (using the settings denoise-paired --p-trim-left-f 20 --p-trim-left-r 17 --p-trunc-len-f 220 --p-trunc-len-r 200). The processing by DADA2 was performed separately for each MiSeq run with the same parameters, and the feature tables and feature sequences were merged. Subsequently, the feature sequence was classified using the feature-classifier plug-in classify-sklearn. Classification of bacterial 16S rRNA gene sequences was made using the Greengenes (13_8 release) (99% identity clusters) database using the feature-classifier classify-sklearn function. Taxonomic names obtained from the feature table after merging were normalized so that each family would be assigned to the same lineage.
Viable bacterial count
A series of 10-fold dilutions of the test beverages or ileal fluids was prepared with sterilized PBS and spread on lactitol-LBS vancomycin (LLV) agar mediumCitation19 (for isolation of LcS) or TOS-MUP agar medium (for isolation of BbrY; TOS-Propionate agar base (Yakult Pharmaceutical Ind. [cat. no. 8-MJ54]) supplemented with lithium mupirocin (Merck KGaA [cat. no. 1. 00045.0010]) solution [50 mg/l]), respectively. The agar plates were incubated aerobically (for LLV agar plate) or anaerobically (for TOS-MUP agar plate) at 37°C for 72 h. Each type of colony was picked up and identified as LcS or BbrY by using strain-specific PCR-based assays.Citation29,Citation52 The number of LcS or BbrY per milliliter of ileal fluid was estimated from the number of colonies that were identified as LcS or BbrY. The survival rate of ingested strain from ileal fluid was calculated by using the following equation:
PEG and PSP analysis
The concentration of PEG was measured with Hyden’s methodCitation53 and that of PSP with spectrophotometry.Citation54 Dilution of the constantly perfused PSP marker was used to calculate the proportion of the ileal contents recovered by aspiration.Citation55 The ileal flow volume over 30 min was determined with the following formula:
where 1.0 was the PSP infusion rate in ml/min.
The bacterial count and amount of PEG and bile acids in each sample were determined from the calculated ileal flow volume over 30 min.
Fluorescent antibody technique
Some ileal fluids were suspended in 3 times its volume of 4% paraformaldehyde (Merck [cat no. 30525–89-4])-PBS solution and left overnight at 4°C. After the samples were centrifuged at 20,400 x g for 5 min, the supernatant was removed and the residue was washed and suspended in PBS. Then, the fixed ileal fluids were dropped onto a MAS-coated slide glass (Matsunami Glass [cat. no. SF17279]), air dried, and treated with 96% ethanol solution. A solution of 0.2% TRITON-X (Sigma-Aldrich [cat. no. 9002–93-1]), 1% Bovine Serum Albumin (Roche Diagnostics GmbH [ref. 10 735 086 001])-PBS (PBS-TB) was dropped onto the slide glass, which was incubated for 30 min under wet conditions. Monoclonal antibodiesCitation19,Citation28 for LcS or BbrY were diluted 100-fold in PBS-TB, dropped onto the slide glass, and incubated for 30 min under wet conditions. After brief washes with PBS, 100-fold dilution of Alexa Fluor 568 labelled IgM antibody (Abcam [cat. no. ab175702]) was dropped onto the slide glass and incubated for 30 min in the dark under wet conditions. After the slide glass was briefly washed with PBS and air dried, it was mounted with VECTASHIELD (Vector laboratories [cat. no. H-1200]). Fluorescent images of the slide glass were obtained using a Leica fluorescent microscopy system, and LcS- or BbrY-positive bacterial cells in the fluorescent images were enumerated. The probiotics recovery rate of ingested strain from ileal fluid was calculated by using the following equation:
The probiotics viability rate of ingested strain from ileal fluid was calculated by using the following equation:
Bile acid analysis
A 10-fold dilution of fecal samples was prepared with sterilized PBS, and 1 ml of the diluted fecal solutions and ileal fluids were separately freeze dried with 100 μl of 1 mM 5β-pregnan-3α, 17α, 20α-triol (pregnanetriol: M.W. 336.5, Sigma [cat. no. P8629-100 MG])-methanol added as an internal standard. Then, 5 ml of 99.5% ethanol was added and thermally extracted at 70°C for 2 h. The supernatants after centrifugation at 20,400 x g for 5 min were evaporated to dryness, dissolved with 1 ml of methanol, and these samples were filtered and used to measure bile salts. Bile acid standards were obtained from JASCO Corp. High-performance liquid chromatography (HPLC) was performed using an LC-2000 Plus HPLC system (JASCO corp.).Citation56 The bile acids were separated on a Bilepak II separation column (4.6 mm × 125 mm, JASCO [code no. D511]), and the eluted bile acids were selectively detected by an enzymatic reaction using immobilized 3α-hydroxysteroid dehydrogenase (HSDH) on an Enzymepak 3α-HSD column (4 mm × 20 mm; JASCO [code no. D512]). Total bile acid concentration was calculated as the sum of individual bile acids. The conjugated/deconjugated and primary/secondary bile acid ratios were calculated for each of the ileal fluids after PEG reached the terminal ileum.
Randomly amplified polymorphic DNA (RAPD) analysis
A series of 10-fold dilutions of oral fluids and ileal fluids wereas prepared with sterilized PBS, and the diluted solutions were spread on DifcoTM Mitis Salivarius agar (Becton Dickinson and Company [cat. no. 229810]) supplemented with 0.001% sodium tellurite. The agar plates were incubated at 37°C for 72 h, aerobically. Each colony with different morphologies were collected, and DNA was extracted as described above.Citation48 RAPD analysis was performed using the random primer 1252 (5ʹ-CCGCAGCCAA-3ʹ). PCR amplifications were performed with initial heating at 94°C for 2 min; 6 cycles of 94°C for 30 s, 36°C for 1 min, and 72°C for 90 s; 30 cycles of 94°C for 20 s, 36°C for 30 min, and 72°C for 90 s; and a final extension at 72°C for 3 min. Fingerprint profiles were separated by gel electrophoresis at 100 V for approximately 30 min in 1.0% agarose gels. The RAPD fingerprint profiles were compared by using BioNumerics software (Applied Math). The similarity index was calculated using the Jaccard coefficient, and the unweighted pair-group method using average linkages was used to construct a dendrogram.
16 S rRNA gene sequencing
A single strain was selected from each unique RAPD fingerprint, and identification of the selected strains was carried out by sequencing the 16 S rRNA gene as previously described.Citation48 Bacterial universal primers 8 F (5ʹ-AGAGTTTGATCMTGGCTCAG-3ʹ) and 15 R (5ʹ-AAGGAGGTGATCCARCCGCA-3ʹ) were employed for PCR amplifications.Citation57 Each 30-µl reaction mixture contained 0.2 mM dNTPs, 1.5 mM MgCl2, 1.5 U of rTaq polymerase (Takara Bio [cat. no. R001AM]), primers 8 F and 15 R (1 µM each), and 1 ng of template DNA. The PCR amplification program consisted of initial heating at 95°C for 5 min; 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and a final extension at 72°C for 5 min. The amplicons were purified by using ExoSAP-IT (Thermo Fisher Scientific [cat. no. 78200.200.UL]) and were sequenced with a BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific [cat. no. 4337454]) on a 3500xL Genetic Analyzer (Thermo Fisher Scientific Inc.). Closely related sequences were retrieved from the DDBJ (http://www.ddbj.nig.ac.jp) using the BLAST program.Citation58
Lactose analysis
Lactose content in the test beverages and ileal fluids was determined with the lactose/D-glucose kit (J. K. International [cat. no. 986 119]). The lactose recovery rate from ileal fluid was calculated by using the following equation:
The ingested amount was 950 mg for LFM and 4,120 mg for BFM.
Statistical analysis
Measured values are presented as mean ± standard deviation. Pearson’s correlation coefficient was calculated to investigate correlations between recovered bacterial amounts and PEG concentrations in each individual. The level of statistical significance was set at p < .05.
Disclosure of potential conflicts of interest
T. Takada, K. Shimizu, K. Oana, M. Katto, Y. Nagara, H. Makino, A. Kushiro, & K. Oishi are employee of Yakult Honsha Co., Ltd, Tokyo, Japan. Test beverages used in this study were produced by Yakult Honsha Co., Ltd, Tokyo, Japan.
Author contribution
K.Oi, A.K., and S.F. directed the study. T.T., A.K., K.Oi, D.C., T.M., and S.F. designed the study. T.T, K.S., K.Oa, M.K, Y.N., K.Oi, D.C., T.M., T.A., K.M., and S.H. conducted the experiments. T.T., K.S., Y.N., H.M, and D.C. analyzed the data. T.T., K.Oi, K.S., Y.N., H.M., K.Oa, D.C., A.K., and S.F. reviewed the manuscript.
Ethics approval
This study was approved by the Ethics Committee of the Graduate School of Medicine at Hirosaki University (UMIN ID: UMIN000018128) and conducted in compliance with ethical principles from the Declaration of Helsinki, guidelines of good clinical practice, and applicable regulations.
Supplemental Material
Download MS Word (270.3 KB)Supplementary data
Supplemental material for this article can be accessed on the Publisher’s website.
References
- Food and Agricultural Organization of the United Nations and World Health Organization. Guidelines for the evaluation of probiotics in food. Joint FAO/WHO working group report on drafting guidelines for the evaluation of probiotics in food. London Ontario (CA); 2002 Apr 30 and May 1. Accessed 2018 December 10. https://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf.
- Ashwell M Concepts of functional foods. Brussels (BE): International Life Sciences Institute (ILSI) Europe; 2002 Jan 1. Accessed 2018 December 10. https://ilsi.eu/publication/concepts-of-functional-foods/.
- Koebnick C, Wagner I, Leitzmann P, Stern U, Zunft HJ. Probiotic beverage containing Lactobacillus casei Shirota improves gastrointestinal symptoms in patients with chronic constipation. Can J Gastroenterol. 2003;17(11):655–659. doi:10.1155/2003/654907. PMID: 14631461.
- Aoki T, Asahara T, Matsumoto K, Takada T, Chonan O, Nakamori K, Nonaka C, Yamaji I, Hisamoto T, Sato M, et al. Effects of the continuous intake of a milk drink containing Lactobacillus casei strain Shirota on abdominal symptoms, fecal microbiota, and metabolites in gastrectomized subjects. Scand J Gastroenterol. 2014;49(5):552–563. doi:10.3109/00365521.2013.848469. PMID: 24621348.
- Kawai M, Setoyama H, Takada T, Shimizu K, Satoh M, Manabe K, Makino T, Watanabe O, Yoshioka M, Nonaka C, et al. Effect of fermented milk containing Bifidobacterium on bowel habits of healthy volunteers with mild constipation. J Intestinal Microbiol. 2011;25:181–187. doi:10.11209/jim.25.181.
- Tabbers MM, de Milliano I, Roseboom MG, Benninga MA. Is Bifidobacterium breve effective in the treatment of childhood constipation? Results from a pilot study. Nutr J. 2011;10(19). doi:10.1186/1475-2891-10-19. PMID: 21345213.
- Nagata S, Asahara T, Ohta T, Yamada T, Kondo S, Bian L, Wang C, Yamashiro T, Nomoto K. Effect of the continuous intake of probiotic-fermented milk containing Lactobacillus casei strain Shirota on fever in a mass outbreak of norovirus gastroenteritis and the faecal microflora in a health service facility for the aged. Br J Nutr. 2011;106:549–556. doi:10.1017/S000711451100064X. PMID: 21521545.
- Nagata S, Asahara T, Wang C, Suyama Y, Chonan O, Takano K, Daibou M, Takahashi T, Nomoto K, Yamashiro Y. The effectiveness of Lactobacillus beverages in controlling infections among the residents of an aged care facility: a randomized placebo-controlled double-blind trial. Ann Nutr Metab. 2016;68(1):51–59. doi:10.1159/000442305. PMID: 26599038.
- Shimizu K, Ogura H, Asahara T, Nomoto K, Morotomi M, Tasaki O, Matsushima A, Kuwagata Y, Shimazu T, Sugimoto H. Probiotic/synbiotic therapy for treating critically III patients from a gut microbiota perspective. Dig Dis Sci. 2013;58(1):23–32. doi:10.1007/s10620-012-2334-x. PMID: 22903218.
- Okazaki T, Yamataka A, Asahara T, Nomoto K, Yamashiro Y. The high incidence of bacteremia in children undergoing surgery can be prevented by Bifidobacterium supplementation. Ann Nutr Metab. 2017;71(1):31–36. doi:10.1159/000479921. PMID: 28950282.
- Shida K, Nanno M, Nagata S. Flexible cytokine production by macrophages and T cells in response to probiotic bacteria. Gut Microbes. 2011;2:109–114. doi:10.4161/gmic.2.2.15661. PMID: 21637028.
- Murosaki S, Yamamoto Y, Ito K, Inokuchi T, Kusaka H, Ikeda H, Yoshikai Y. Heat-killed Lactobacillus plantarum L-137 suppresses naturally fed antigen–specific IgE production by stimulation of IL-12 production in mice. J Allergy Clin Immunol. 1998;102(1):57–64. doi:10.1016/s0091-6749(98)70055-7. PMID: 9679848.
- Ohashi Y, Nakai S, Tsukamoto T, Masumori N, Akaza H, Miyanaga N, Kitamura T, Kawabe K, Kotake T, Kuroda M, et al. Habitual intake of lactic acid bacteria and risk reduction of bladder cancer. Urol Int. 2002;68(4):273–280. doi:10.1159/000058450. PMID: 12053032.
- Ishikawa H, Akedo I, Otani T, Suzuki T, Nakamura T, Takeyama I, Ishiguro S, Miyaoka E, Sobue T, Kakizoe T. Randomized trial of dietary fiber and Lactobacillus casei administration for prevention of colorectal tumors. Int J Cancer. 2005;116(5):762–767. doi:10.1002/ijc.21115. PMID: 15828052.
- Kato-Kataoka A, Nishida K, Takada M, Suda K, Kawai M, Shimizu K, Kushiro A, Hoshi R, Watanabe O, Igarashi T, et al. Fermented milk containing Lactobacillus casei strain Shirota prevents the onset of physical symptoms in medical students under academic examination stress. Benef Microbes. 2016;7(2):153–156. doi:10.3920/BM2015.0100. PMID: 26689231.
- Takada M, Nishida K, Kataoka-Kato A, Gondo Y, Ishikawa H, Suda K, Kawai M, Hoshi R, Watanabe O, Igarashi T, et al. Probiotic Lactobacillus casei strain Shirota relieves stress-associated symptoms by modulating the gut-brain interaction in human and animal models. Neurogastroenterol Motil. 2016;28(7):1027–1036. doi:10.1111/nmo.12804. PMID: 26896291.
- Kato-Kataoka A, Nishida K, Takada M, Kawai M, Kikuchi-Hayakawa H, Suda K, Ishikawa H, Gondo Y, Shimizu K, Matsuki T, et al. Fermented milk containing Lactobacillus casei strain Shirota preserves the diversity of the gut microbiota and relieves abdominal dysfunction in healthy medical students exposed to academic stress. Appl Environ Microbiol. 2016;82(12):3649–3658. doi:10.1128/AEM.04134-15. PMID: 27208120.
- Takada M, Nishida K, Gondo Y, Kikuchi-Hayakawa H, Ishikawa H, Suda K, Kawai M, Hoshi R, Kuwano Y, Miyazaki K, et al. Beneficial effects of Lactobacillus casei strain Shirota on academic stress-induced sleep disturbance in healthy adults: a double-blind, randomised, placebo-controlled trial. Benef Microbes. 2017;8(2):153–162. doi:10.3920/BM2016.0150. PMID: 28443383.
- Yuki N, Watanabe K, Mike A, Tagami Y, Tanaka R, Ohwaki M, Morotomi M. Survival of a probiotic, Lactobacillus casei strain Shirota, in the gastrointestinal tract: selective isolation from faeces and identification using monoclonal antibodies. Int J Food Microbiol. 1999;48(1):51–57. doi:10.1016/s0168-1605(99)00029-x. PMID: 10375134.
- Kado Y, Yuki N, Kushiro A, Watanabe K, Morotomi M. Survival of a probiotic, Bifidobacterium breve strain Yakult, in the human gastrointestinal tract: selective isolation from feces and identification using randomly amplified polymorphic. J Intest Microbiol. 2001;15:9–14. doi:10.11209/jim1997.15.9.
- Oozeer R, Leplingard A, Mater DD, Mogenet A, Michelin R, Seksek I, Marteau P, Doré J, Bresson JL, Corthier G. Survival of Lactobacillus casei in the human digestive tract after consumption of fermented milk. Appl Environ Microbiol. 2006;72(8):5615–5617. doi:10.1128/AEM.00722-06. PMID: 16885316.
- Pochart P, Marteau P, Bouhnik Y, Goderel I, Bourlioux P, Rambaud JC. Survival of bifidobacteria ingested via fermented milk during their passage through the human small intestine: an in vivo study using intestinal perfusion. Am J Clin Nutr. 1992;55(1):78–80. doi:10.1093/ajcn/55.1.78. PMID: 1728822.
- Higuchi S, Fukushi G, Baba T, Sasaki D, Yoshida Y. New method of testing for carbohydrate absorption in man. Dig Dis Sci. 1986;31(4):369–375. doi:10.1007/bf01311671. PMID: 3956333.
- Danjo K, Nakaji S, Fukuda S, Shimoyama T, Sakamoto J, Sugawara K. Research communication: the resistant starch level of heat moisture–treated high amylose cornstarch is much lower when measured in the human terminal ileum than when estimated in vitro. J Nutr. 2003;133(7):2218,–2221. doi:10.1093/jn/133.7.2218. PMID: 12840182.
- Shimaya S, Shimoyama T, Fukuda S, Matsuzaka M, Takahashi I, Umeda T, Chinda D, Saito D, Sakamoto J, Nagura T, et al. The recovery rate at the human terminal ileum of an orally administered non-digestive oligosaccharide (raffinose). Int J Food Sci Nutr. 2009;60(4):344–351. doi:10.1080/09637480801990454. PMID: 19115124.
- Saito D, Nakaji S, Fukuda S, Shimoyama T, Sakamoto J, Sugawara K. Comparison of the amount of pectin in the human terminal ileum with the amount of orally administered pectin. Nutrition. 2005;21(9):914–919. doi:10.1016/j.nut.2005.01.005. PMID: 16043326.
- Oyama T, Fukuda S, Shimoyama T, Takahashi I, Umeda T, Danjo K, Saito D, Chinda D, Sakamoto J, Nakaji S. The oro-ileal transit of cellulose. J Food Sci. 2008;73(9):H229–H234. doi:10.1111/j.1750-3841.2008.00942.x. PMID: 19021806.
- Kitajima H, Sumida Y, Tanaka R, Yuki N, Takayama H, Fujimura M. Early administration of Bifidobacterium breve to preterm infants: randomised controlled trial. Arch Dis Child. 1997;76(2):F101–F107. doi:10.1136/fn.76.2.f101. PMID: 9135288.
- Fujimoto J, Matsuki T, Sasamoto M, Tomii Y, Watanabe K. Identification and quantification of Lactobacillus casei strain Shirota in human feces with strain-specific primers derived from randomly amplified polymorphic DNA. Int J Food Microbiol. 2008;126(1–2):210–215. doi:10.1016/j.ijfoodmicro.2008.05.022. PMID: 18573558.
- Tanida M, Takada M, Kato-Kataoka A, Kawai M, Miyazaki K, Shibamoto T. Intragastric injection of Lactobacillus casei strain Shirota suppressed spleen sympathetic activation by central corticotrophin-releasing factor or peripheral 2-deoxy-D-glucose in anesthetized rats. Neurosci Lett. 2016;619:114–120. doi:10.1016/j.neulet.2016.03.016. PMID: 26971699.
- Sakai T, Oishi K, Asahara T, Takada T, Yuki N, Matsumoto K, Nomoto K, Kushiro A. M-RTLV agar, a novel selective medium to distinguish Lactobacillus casei and Lactobacillus paracasei from Lactobacillus rhamnosus. Int J Food Microbiol. 2010;139(3):154–160. doi:10.1016/j.ijfoodmicro.2010.03.019. PMID: 20385416.
- Lorenzo-Zúñiga V, Bartolí R, Planas R, Hofmann AF, Viñado B, Hagey LR, Hernández JM, Mañé J, Alvarez MA, Ausina V, et al. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology. 2003;37:551–557. doi:10.1053/jhep.2003.50116. PMID: 12601352.
- Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29(4):625–651. doi:10.1016/j.femsre.2004.09.003. PMID: 16102595.
- Gorbach SL, Tabaqchali S. Bacteria, bile and the small bowel. Gut. 1969;10(12):963–972. doi:10.1136/gut.10.12.963. PMID: 4983639.
- Matsumoto K, Takada T, Shimizu K, Moriyama K, Kawakami K, Hirano K, Kajimoto O, Nomoto K. Effects of a probiotic fermented milk beverage containing Lactobacillus casei strain Shirota on defecation frequency, intestinal microbiota, and the intestinal environment of healthy individuals with soft stools. J Biosci Bioeng. 2010;110:547–552. doi:10.1016/j.jbiosc.2010.05.016. PMID: 20580604.
- Wang C, Chang T, Yang H, Cui M. Antibacterial mechanism of lactic acid on physiological and morphological properties of Salmonella enteritidis, Escherichia coli and Listeria monocytogenes. Food Control. 2015;47:231–236. doi:10.1016/j.foodcont.2014.06.034.
- Matsuki T, Pédron T, Regnault B, Mulet C, Hara T, Sansonetti PJ. Epithelial cell proliferation arrest induced by lactate and acetate from Lactobacillus casei and Bifidobacterium breve. PLoS One. 2013;8:e63053. doi:10.1371/journal.pone.0063053. PMID: 23646174.
- Sato T, Matsumoto K, Okumura T, Yokoi W, Naito E, Yoshida Y, Nomoto K, Ito M, Sawada H. Isolation of lactate-utilizing butyrate-producing bacteria from human feces and in vivo administration of Anaerostipes caccae strain L2 and garacto-oligosaccharides in a rat model. FEMS Microbiol Ecol. 2008;66:528–536. doi:10.1111/j.1574-6941.2008.00528.x. PMID: 18554304.
- Mortensen PB, Clausen MR. Short-chain fatty acids in the human colon: relation to gastrointestinal health and disease. Scand J Gastroenterol. 1996;216:132–148. doi:10.3109/00365529609094568. PMID: 8726286.
- Hague A, Singh B, Paraskeva C. Butyrate acts as a survival factor for colonic epithelial cells: further fuel for the in vivo versus in vitro debate. Gastroenterology. 1997;112(3):1036–1040. doi:10.1053/gast.1997.v112.agast971036. PMID: 9041270.
- Sakata T, Takada M, Nishida K, Kataoka-Kato A, Gondo Y, Ishikawa H, Suda K, Kawai M, Hoshi R, Watanabe O. Stimulatory effect of short-chain fatty acids on epithelial cell proliferation in the rat intestine: a possible explanation for trophic effects of fermentable fiber, gut microbes, and luminal trophic factors. Br J Nutr. 1987;58(1):95–103. doi:10.1079/bjn19870073. PMID: 3620440.
- Shimotoyodome A, Meguro S, Hase T, Tokimitsu I, Sakata T. Short chain fatty acids but not lactate or succinate stimulate mucus release in the rat colon. Comp Biochem Physiol Part A. 2000;125:525–531. doi:10.1016/s1095-6433(00)00183-5. PMID: 10840229.
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721–5732. doi:10.1128/JCM.43.11.5721-5732.2005. PMID: 16272510.
- Moore WE, Moore LV. The bacteria of periodontal disease. Periodontol. 1994;5(1):66–77. 2000. doi:10.1111/j.1600-0757.1994.tb00019.x. PMID: 9673163.
- Yamanaka W, Takeshita T, Shibata Y, Matsuo K, Eshima N, Yokoyama T, Yamashita Y. Compositional stability of a salivary bacterial population against supragingival microbiota shift following periodontal therapy. PLoS One. 2012;7(8):e42806. doi:10.1371/journal.pone.0042806. PMID: 22916162.
- Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. 2017;358:359–365. doi:10.1126/science.aan4526. PMID: 29051379.
- Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. 2019;25(5):716–729. doi:10.1038/s41591-019-0439-x. PMID: 31061539.
- Matsuki T, Watanabe K, Fujimoto J, Takada T, Tanaka R. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl Environ Microbiol. 2004;70(12):7220–7228. doi:10.1128/AEM.70.12.7220-7228.2004. PMID: 15574920.
- Nagara Y, Takada T, Nagata Y, Kado S, Kushiro A. Microscale spatial analysis provides evidence for adhesive monopolization of dietary nutrients by specific intestinal bacteria. PLoS One. 2017;12:e0175497. doi:10.1371/journal.pone.0175497. PMID: 28394924.
- QIIME 2 website. Accessed 2018 December 10. https://qiime2.org/
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from illumina amplicon data. Nat Methods. 2016;13(7):581–583. doi:10.1038/nmeth.3869. PMID: 27214047.
- Fujimoto J, Tanigawa K, Kudo Y, Makino H, Watanabe K. Identification and quantification of viable Bifidobacterium breve strain Yakult in human faeces by using strain-specific primers and propidium monoazide. J Appl Microbiol. 2011;110:209–217. doi:10.1111/j.1365-2672.2010.04873.x. PMID: 21029276.
- Malawer SJ, Powell DW. An improved turbidimetric analysis of polyethylene glycol utilizing an emulsifier. Gastroenterology. 1967;53(2):250–256. doi:10.1016/S0016-5085(19)34232-5.
- Phillips SF, Giller J. The contribution of the colon to electrolyte and water conservation in man. J Lab Clin Med. 1973;81(5):733–746. PMID: 4698660.
- Holloway WD, Tasman-Jones C, Maher K. Pectin digestion in human. Am J Clin Nutr. 1983;37(2):253–255. doi:10.1093/ajcn/37.2.253. PMID: 6297291.
- Takahashi T, Morotomi M. Absence of cholic acid 7 alpha-dehydroxylase activity in the strains of Lactobacillus and Bifidobacterium. J Dairy Sci. 1994;77(11):3275–3286. doi:10.3168/jds.S0022-0302(94)77268-4. PMID: 7814703.
- Irisawa T, Okada S. Lactobacillus sucicola sp. nov., a motile lactic acid bacterium isolated from oak tree (Quercus sp.) sap. Int J Syst Evol Microbiol. 2009;59(11):2662–2665. doi:10.1099/ijs.0.006478-0. PMID: 19625442.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi:10.1016/S0022-2836(05)80360-2. PMID: 2231712.
