ABSTRACT
In the present study, we characterized the involvement of Lon protease in bacterial virulence and intracellular survival in Salmonella under abiotic stress conditions resembling the conditions of a natural infection. Wild type (JOL401) and the lon mutant (JOL909) Salmonella Typhimurium were exposed to low temperature, pH, osmotic, and oxidative stress conditions and changes in gene expression profiles related to virulence and metal ion uptake were investigated. Expression of candidate genes invF and hilC of Salmonella Pathogenicity Island (SPI)-1 and sifA and sseJ of SPI-2 revealed that Lon protease controls SPI-1 genes and not SPI-2 genes under all stress conditions tested. The lon mutant exhibited increased accumulation of hydroxyl (OH·) ions that lead to cell damage due to oxidative stress. This oxidative damage can also be linked to an unregulated influx of iron due to the upregulation of ion channel genes such as fepA in the lon mutant. The deletion of lon from the Salmonella genome causes oxidative damage and increased expression of virulence genes. It also prompts the secretion of host pro-inflammatory cytokines leading to early clearance of the bacteria from host cells. We conclude that poor bacterial recovery from mice infected with the lon mutant is a result of disrupted bacterial intracellular equilibrium and rapid activation of cytokine expression leading to bacterial lysis.
Introduction
Salmonella enterica causes a spectrum of diseases in animals and humans, ranging between mild infection to severe infection that can be fatalCitation1. Different strains of Salmonella enterica Typhimurium cause enteric infections in humans and other warm-blooded animals, whereas the serovar Typhi is restricted to humans causing typhoid fever. All Salmonella species possess Salmonella Pathogenicity Island I (SPI-1) despite variations in virulence factors among different serotypes. Coordinated expression of genes belonging to SPI-1 and SPI-2 are important for initial entry and the subsequent establishment of infection in a target host. The SPI-1 encodes proteins including a Salmonella type III secretion system (TTSS) that is responsible for injecting effector molecules that alter cytoskeletal arrangement in the host and help bacterial internalization. Once the bacterium is in the host cell, genes in SPI-2 are expressed that encodes another type of TTSS that secretes effectors diverting endocytic trafficking to prevent lysosome recruitment and fusion of Salmonella-containing vacuoles. This process helps host-adapted Salmonella reach extraintestinal organs and establish a systemic infection. Salmonella invasion is a complex and intricate process that essentially controls virulence. Concerning invasion and virulence of Salmonella, studies have revealed that Lon protease plays a significant role during early infection and colonization of Salmonella.Citation2 As a member of the AAA+ superfamily of proteases that also includes ClpA, ClpB, and FtsH, Lon has been extensively studied in Escherichia coli.Citation3–Citation5 The Lon protease plays a vital role in protein quality control by eliminating faulty proteins during stress responses and activating pathways to prevent any further damage to the bacterial cell.Citation6 Moreover, recent reports indicate the involvement of Lon protease in antimicrobial resistance, further broadening its role in bacterial survival.Citation7
The involvement of Lon in Salmonella SPI-1 gene expression has been previously reported, which proposed a link between the SPI-1 regulator HilA and Lon protease. HilA is an OmpR/ToxR family proteinCitation8 that is activated by the transcriptional regulator InvF, which in turn regulates the transcriptional activation of effector genes. In Salmonella, hilA expression is modulated by the HilC and HilD transcriptional regulators.Citation9 Therefore, the interaction of Lon with the SPI-1 axis of virulence modulation is worth further elaboration under various environmental and physiological stress conditions to investigate the survival and virulence behavior of Salmonella.Citation2 During the natural infection cycle, Salmonella encounters various stress conditions including a transition to increased body temperature, acidic pH in the stomach, high salt and osmolarity in bile salts, and oxidative stress in the lumen as a result of the host inflammatory response.Citation10 During systemic infection, the host attempts to starve Salmonella by limiting nutrient and metal ion uptake; in response, Salmonella manipulates ion and nutrient uptake gene expression to counter the host restrictions.Citation11,Citation12 Further, Salmonella attempts to circumvent the recruitment of antigen-presenting cells to the site of infection by interacting with the host immune pathwaysCitation13 and exploits the metabolic pathways of the host for its survival.Citation14
Apart from manipulating these host mechanisms, the bacteria modulate their biosynthesis machinery that determines their fitness and virulence.Citation14,Citation15 As a facultative anaerobe, Salmonella undergoes metabolic reprogramming depending on the presence or absence of oxygen. One of the principal determinants of this process is redox homeostasis.Citation16 On the other hand, reactive oxygen species (ROS) produced during microbial metabolism and the stress response can alter the redox state of a cell.Citation17 Gene deletions or mutations hindering intracellular iron regulation is known to cause the accumulation of excess free cytosolic iron, leading to iron-catalyzed production of ROS.Citation18,Citation19 This endogenous oxidative stress is regarded as an unwanted and damaging by-product of cellular metabolism.
Here, we describe the quantification of gene expression involved in pathogenesis including genes that encode SPI effector proteins, genes regulating iron uptake, and levels of hydroxyl (OH·) radical accumulation in lon- mutant Salmonella; JOL 909 after exposure to various conditions including acidic, cold, osmotic, and oxidative stress treatments. The changes in gene expression were compared with that of parental wild type strain Salmonella Typhimurium; JOL 401 and lon complemented Salmonella strain JOL 909::lon. We analyzed gene expression after 5 h of exposure and found that JOL 909 exhibited the upregulation of SPI-1 virulence-associated and iron uptake genes, followed by increased accumulation of OH· in the lon mutant suggesting the participation of Lon in modulating bacterial internal stability. Further, a distinct pro-inflammatory cytokine response in the host shows that Lon alters bacterial interaction with the host TLR; a hypothesis that needs further validation.Citation20
Although a previous study reports the inability of a lon mutant to survive phagocytic lysis, the lon-specific molecular mechanism was not elucidatedCitation1. Here, our observations show that lon regulates intracellular homeostasis when Salmonella maneuvers through the host cell to establish an infection. In the absence of this regulator, an unchecked influx of iron cations occurs that reacts with H2O2 produced within the bacteria, as well as inflow from the host cells, leading to the formation of OH· radicals. Hydroxyl ion-mediated oxidative stress leads to DNA damage and subsequent bacterial lysis. The present work adds a new level of information about Lon protease and its role in tightly regulated bacterial stress-response pathways. Here, we show that Lon in Salmonella is essential for mediating regulated virulence behavior that may ensure the successful establishment of systemic infection.
Results
Characterization of a lon mutant in Salmonella
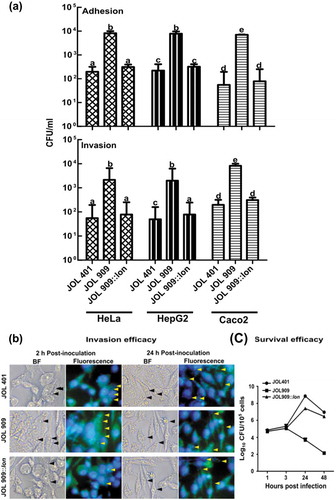
The Lon protease is a global regulator that regulates the expression of early virulence genes located in SPI-1. Mechanistic studies have revealed Lon as a negative regulator of SPI-1 that regulates and ensures the successful establishment of Salmonella in a target host cell. Herein, we investigated the involvement of the lon gene in virulence modulation of Salmonella upon exposure to various abiotic stress conditions. To examine this phenomenon, we used wild type strain ST JOL 401 and a lon deleted strain ST JOL 909. To further confirm that the ability to adhere and invade epithelial cells and metal ion uptake associated genes in JOL 909 are modulated as a result of lon knockout, a functional lon gene was cloned in a low copy-number vector, pWSK 29, and the resultant plasmid was used to transform JOL 909, resulting in strain JOL 909::lon. To assess whether the lon deletion causes growth defects in Salmonella, a growth curve analysis of all three strains was carried out. Results demonstrated that the lon deletion did not significantly alter the growth characteristics of Salmonella (Supplementary Figure 1). The JOL 909 strain exhibited enhanced adhesion and invasion compared to JOL 401. Such enhanced adhesion and invasion were inhibited by lon complementation in JOL 909, showing that Lon plays a crucial role in regulating both adhesion and invasion. The adhesive and invasive phenotype of the lon mutant JOL 909 was demonstrated to be similar in all three cell lines HeLa, HepG2, and Caco-2 (). However, when the JOL 401 and JOL 909 were exposed to anaerobic conditions their adhesion and invasion ability increased drastically. The increased invasion by JOL 909 was further evident in fluorescent microscopy conducted using CellTracker-labeled Salmonella; however, their population was significantly reduced in both non-phagocytic (data not shown) and phagocytic cells (,) earlier than the wild type and complemented strains.
Figure 1. Role of lon in Adhesion and Invasion of HeLa and HepG2 and macrophage survival.
Actin rearrangement by Salmonella
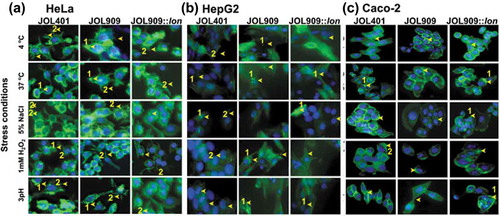
Salmonella induced actin rearrangement in a host cell is essential for Salmonella entry and survival. Such manipulation helps Salmonella in better vacuolization and prevents lysosomal engagement allowing them to thrive in the host. Such phenotypic changes in the host cell induced by JOL 401 and JOL 909 were recorded during the entry phase, characterized by the trigger or zipper mechanisms. In HeLa cells (), both JOL 401 and JOL 909 induced lamellipodia formation characterized by membrane ruffling and formation of appendages engulfing the bacteria. However, in HepG2 cells (), the strains did not induce the formation of membrane extensions most likely due to the engagement of the zipper mechanism. Although no lamellipodia formation was evident in Caco-2 cells, a mild yet distinct membrane ruffling was observed when infected with JOL 401 and JOL 909 (). These results show that the regulation of host actin polymerization is independent of a Lon-mediated regulatory pathway. We hypothesize that this rearrangement is also partly dependent on the host cell type. To further ensure independent behavior on actin rearrangement, microscopic observations were conducted at varying time intervals of 15, 30, 45, 60, 90, and 150 min post-inoculation.
Figure 2. Actin polymerization is a distinctive attribute of individual host cells.
Effect of lon on bacterial hydroxyl ion generation
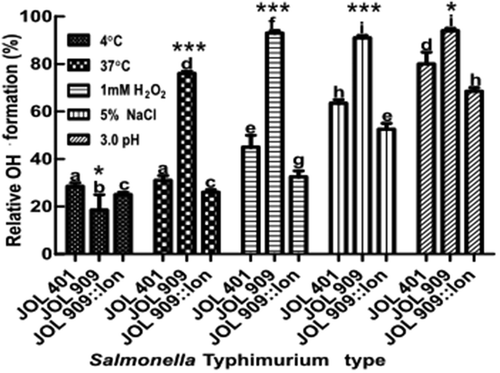
The generation of reactive oxygen species (ROS) is a hallmark of the bacterial stress response. In bacteria, ROS are readily detoxified by enzymatic conversion of ROS (OH·) into hydrogen peroxide and subsequently to water and oxygen. However, in the presence of iron ions, the Fenton reaction can generate OH· that are lethal to bacterial cells.Citation21 To elucidate the kinetics of OH· generation in JOL 401, JOL909, and JOL909::lon, cells were exposed to various naturally-occurring stress conditions for 5 h. JOL 401 was able to effectively mitigate OH· generation under cold and oxidative stress; however, the production of OH· was significantly higher under osmotic and acidic pH conditions. Under all stress conditions, JOL 909 appeared to have produced higher levels of OH·. The maximum level of OH· production in JOL 909 was observed under oxidative stress, suggesting the role of Lon protease in mitigating oxidative stress. The involvement of Lon seems to be comparatively less with osmotic and acidic stress conditions. The results were corroborated with JOL 909::lon, which effectively diminished OH· production. These results imply the critical involvement of Lon protease activity in mitigating oxidative stress in the phagosomes of host cellsCitation22 (). It is worthwhile to note that JOL 909::lon exhibited lower levels of OH· compared to JOL 401. As the overproduction of Lon can prove fatal for bacterial cells, these low levels may be due to slightly higher levels of Lon protein in JOL 909 complemented in trans by a low-copy-number plasmid.
Figure 3. Endogenous Hydroxyl radical (OH·) formation during stress treatments in the lon mutant.
Effect of lon deletion on metal ion uptake and virulence gene expression
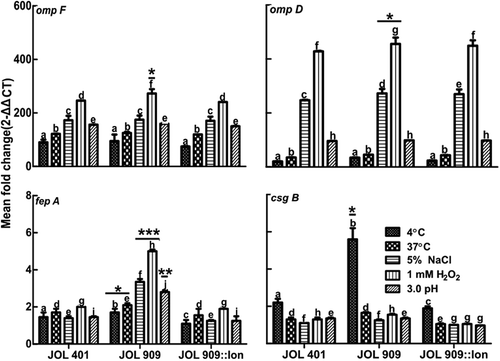
To investigate the role of lon in Salmonella virulence under the aforementioned stress conditions, the expression profile of four genes encoding outer membrane proteins, namely ompD, ompF, fepA, and csgB was studied by qRT-PCR. Among these genes, csgB gene expression was significantly upregulated under cold stress, which lead us to believe that Lon protease, a member of the heat shock protein (Hsp) family, also regulates the expression of genes that are primarily activated below 37ºC. The other three outer membrane genes were downregulated in response to cold stress. Under osmotic and acidic stress conditions, only fepA expression appeared to be regulated by Lon. When exposed to oxidative stress, ompF, and fepA expression were upregulated ().
Figure 4. Expression of bacterial cell surface protein genes during stress treatments.
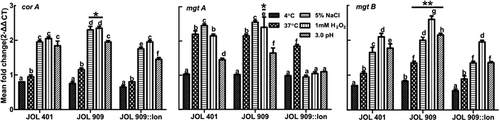
The upregulation of iron uptake regulator genes (fur regulon and sitA) and iron sequestration gene (dps) under various stress conditions was distinct. In particular, the fur regulon was highly upregulated in JOL 909 under osmotic and oxidative stress conditions. The increase in the mRNA level was similar to the increased expression levels of fepA, which is an iron ion channel on the bacterial cell surface (). Also, JOL 909 exhibited a surge in the expression of corA, mgtA, and mgtB compared to JOL 401 (), all three of which are transporters of divalent magnesium ions while corA is also associated with the uptake of ferrous ions.
Figure 5. Expression of regulatory genes that participate in iron uptake and sequestration during stress exposure.
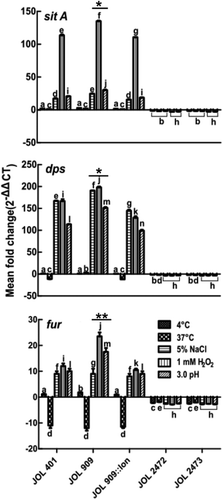
Figure 6. Regulation of Magnesium uptake associated genes.
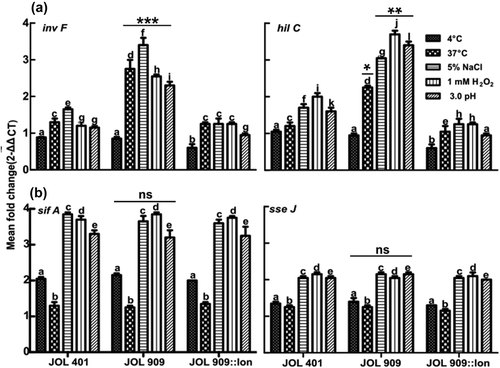
Apart from modulating gene expression levels of outer membrane proteins under various stress conditions, Lon regulates the expression of SPI-1 transcriptional factors. The invF and hilC () genes were significantly upregulated under all stress conditions in JOL 909 compared to JOL 401, attributing it to be Lon-dependent. On the other hand, the SPI-2 genes sifA and sseJ were upregulated uniformly in both JOL 401 and JOL 909 (). The levels of sifA were observed to have increased up to 4-fold during oxidative and osmotic stress treatments. A similar trend was observed for sseJ, whose levels were upregulated 2-fold during oxidative, osmotic, and acidic stress. These observations suggest that the regulation of a cluster of genes in SPI-2 is partly independent of pathways involving Lon protease. SPI-1 is essential for invasion whereas SPI-2 is crucial for intracellular survival and systemic persistence.Citation23,Citation24
Figure 7. Expression of a virulence-associated gene from SPI 1 and 2 gene clusters.
fepA knockout and lon expression
Based on the aforementioned observations, we theorized that fepA is transcriptionally regulated by lon under stress conditions especially during oxidative stress. To further understand the fepA-lon interaction, we constructed JOL 401ΔfepA and JOL 909ΔfepA and refer to them as JOL 2472 and JOL 2473, respectively (Supplementary Figure 2). To study the role of fepA in the regulation of lon expression, JOL 401 and JOL 2472 were exposed to the above-mentioned stress conditions for 5 h and their RNA was isolated to evaluate lon expression (Supplementary Figure 3). The expression of lon was upregulated in JOL 401 by ~10-fold under all the stress treatments, peaking at 15-fold changes on average during oxidative stress. On the other hand, lon expression increased up to 2-fold on average in JOL 2472 after treatment (Supplementary Figure 3). This was further followed by the downregulation of iron uptake and sequestration genes in JOL 2472 and JOL 2473 (). Additionally, under the anaerobic condition, the lon expression decreased by 2-fold in both JOL 401 and JOL 2473.
In vitro bacterial survival and recovery from mice
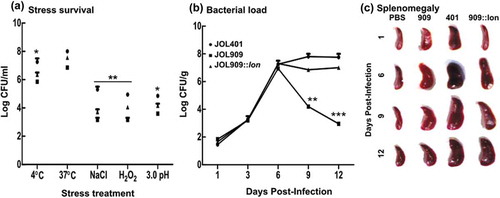
The presence of colony-forming units (CFU) on BGA agar showed the ability of JOL 401 and JOL 909 to survive in vitro after exposure to abiotic stress conditions. Oxidative stress caused characteristic damage to JOL 909 wherein <4 log CFU/mL bacteria were observed compared to a log CFU close to 6 per mL for JOL 401 (,). Although all the stress conditions hampered JOL 909 survival, the oxidative stress-induced damage was distinct. To study if the in vitro observations were consistent with the in vivo bacterial clearance, the spleens of infected mice were aseptically collected and bacterial load was evaluated. The results corroborated with the in vitro survival pattern wherein JOL 909 seemed to be eliminated by host phagocytosis and exerting oxidative stress on the invading bacteria. By day 12 post-infection, splenomegaly was evident, and <3 log CFU/mL JOL 909 were recovered from the spleen ().
Figure 8. Bacterial recovery and survival to study the involvement of lon in bacterial longevity.
Host cytokine responses
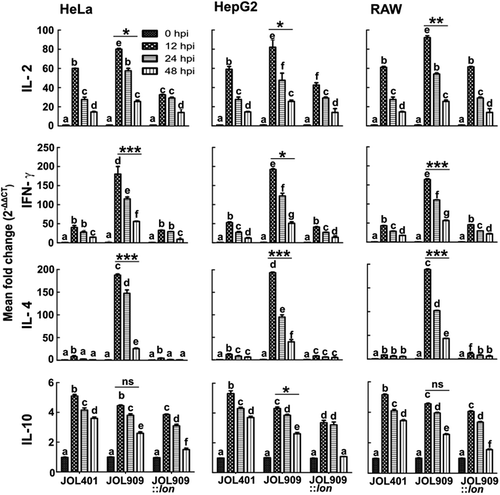
The role of Lon in eliciting the host immune response is crucial as it determines the fate of invading bacteria. The in vitro studies showed that JOL 909 upregulated the expression of pro-inflammatory cytokines up to 48 hours post-infection (hpi). The levels of IL-2 and IFN-γ increased by about 80- and 200-fold on average in both non-phagocytic (HeLa and HepG2) and phagocytic (RAW) cells infected with JOL 909. Levels of the potent anti-inflammatory cytokine IL-10 were within the same range (up to 12-fold change) in cells infected with JOL 401 and JOL 909 (). The levels of IL-4 increased in cells infected with JOL 909, which activates the adaptive immune response proving advantage to the host.
Figure 9. The kinetics of cytokine gene expression in the phagocytic and non-phagocytic epithelial cells.
Discussion
The present study investigates the involvement of the Lon protease in Salmonella virulence and intracellular survival with respect to the ability to establish systemic infection. To dissect the details of Lon-mediated Salmonella virulence, we exposed a lon mutant JOL 909, wild type JOL 401, and lon complemented Salmonella JOL 909::lon to a range of abiotic stress conditions including temperature, osmolarity, oxidative and pH-induced stress and quantitatively assessed the expression of virulence and nutrient uptake genes, their interaction with host cells, and fate during an in vivo infection.Citation1 Survival during stress requires proficient manipulation of a myriad of genes; thus, the effect of Lon may be better elucidated under stress-induced virulence studies. Growth analysis experiments revealed that deletion of the lon gene was not detrimental to host cells and did not cause significant growth rate alteration as compared to the wild type strain. Therefore, the mutant could be utilized in subsequent studies comparable to the wild type strain. During the infection process, Salmonella will make contact with the host cell by attaching to the host cell through their receptors, and the level of adhesion and invasion are the very first virulence determinants for successful infection.Citation25,Citation26 The adhesion and invasion studies conducted on non-phagocytic HeLa, HepG2 and Caco-2 cells revealed the lon deletion significantly increased (approximately 2-fold increase) both adhesion and invasion into the host cell. This was confirmed by CFU counting and by fluorescence microscopy. When we examined persistence of the various Salmonella strains within host cells, it was evident that the lon mutant was significantly defective in intracellular survival compared to the wild type strain. Such a dramatic increase in virulence yet defective intracellular fitness in lon mutant hints at a link between Lon protease and SPI-1 genes that are important for the early stage of infection,Citation27,Citation28 but not with SPI-2 genes that are primarily responsible for Salmonella survival in the host cell.Citation29,Citation30 The SPI-1 gene region encodes elements of Type III secretory system (TTSS) machinery that bacteria use to inject effector proteins at early stages of infection that cause actin rearrangement in the host cytoskeleton facilitating Salmonella entry into the host cell.Citation31,Citation32 This phenomenon is demarcated by membrane ruffling and the formation of protrusions called lamellipodia (Trigger mechanism). In certain cells like the HepG2, actin condensation is less profound, and the process is termed the Zipper mechanism. A similar observation was evident in Caco-2 cells wherein distinct membrane ruffling was seen without the formation of lamellipodia. These studies conducted on HeLa, HepG2, and Caco-2 cells for entry mechanism did not show significant alteration due to lon deletion. This suggests, even though Lon causes enhancement of Salmonella virulence, Lon may not influence the entry mechanism into the host cell.
Under normal physiological conditions, cells produce ROS that is harmful to cellular components.Citation33–Citation35 In healthy cells, the levels of ROS are maintained at harmless levels by enzymes such as superoxide dismutase, peroxidase, and catalase that convert reactive oxygen into H2O2 or water as the terminal product. However, in the presence of iron ions, H2O2 can revert to form OH· free radicals that are again harmful to cellular components. On the other hand, mammalian phagocytic cells use H2O2 as a defense tool against invading pathogens.Citation35,Citation36 The results of the present study suggest that the lon mutant extensively produced OH· free radicals relative to the wild type counterpart, demonstrating that the mutant lost the ability to mitigate oxidative stress. It also strengthens the notion that the lon mutant may enhance internal and host-derived oxidative stress due to an enhanced virulence phenotype. One strategy bacteria utilize to avoid the accumulation of H2O2 is active secretion via outer membrane proteins.Citation37 The expression of genes encoding outer membrane proteins was elevated in the lon mutant as we measured four representative outer membrane protein-encoding genes ompF, ompD, fepA, and csgB.Citation38–Citation40 The encoded proteins are important for nutrient and ion uptake. Interestingly, the lon mutant resulted in a sharp increase in csgB at cold shock (4°C) that encodes for a minor curling subunit in Salmonella that confirms Lon protease function as a cold-shock protein similarly to what has been demonstrated in E. coli in previous studies.Citation41,Citation42
Herein, a link between the Lon protease and iron uptake regulation can be proposed due to the hyperexpression of the fepA gene in the lon mutant. To investigate the connection between lon and iron uptake regulation, we evaluated the level of expression of iron homeostasis related genes fur (ferric uptake regulator), sitA (Iron/manganese ABS transporter substrate-binding protein), and dps (DNA protection during starvation protein) genes. Expression kinetics demonstrated significant upregulation of these genes in the lon mutant ST, suggesting that the mutation may result in an influx of iron ions. These observations corroborate the previously described phenotype of increased OH· generation by the lon mutant (Fenton reaction). This suggests lon is involved in iron-dependent oxidative stress response pathways in Salmonella. Interestingly, a fepA deletion in the lon mutant ST (JOL2472) and the ST wild type strain (JOL2473) completely abolished the expression of fur, sitA, and dps in both strains, possibly due to “lon-mediated” and “fepA-mediated” iron uptake is controlled by two independent pathways or these genes may activate upon a certain threshold of iron in the cytoplasm that is never reached in the absence of fepA. In addition to iron, the magnesium ion transportersCitation43,Citation44 mgtA, mgtB, and corA also demonstrated an increasing trend in the lon mutant. Herein, corA expression is particularly important due to its involvement in iron transport along with magnesium. Therefore, it can be concluded that the lon mutant naturally experiences an increased level of oxidative stress that is generated by its cellular machinery.
When examining the virulence gene expression profile of the lon mutant and the wild type strain, it is possible to propose that Lon protease activity is linked to SPI-1 rather than SPI-2,Citation45 as indicated by a sharp increase in the SPI-1 candidate invF and hilC genes but not the SPI-2 sifA and sseJ candidate genes. This allows us to hypothesize that Lon activity is essential during the early phases of infection to maintain the virulence phenotype of Salmonella until the cells have entered and established infection in the host. This was also evident in the lon mutant whereby it lost virulence regulation, which is a key failure for a successful intracellular pathogen. In turn, such an enhanced virulence phenotype may elicit enhanced cytokine response from the host perspective, which was confirmed by assaying cytokine profiles in phagocytic (RAW 264.7) and non-phagocytic (HeLa and HepG2) cells.Citation46,Citation47 Results confirmed that the lon mutant is hyper immunogenic relative to the wild type strain, possibly due to the aforementioned overexpressed virulence genes. However, according to our hypothesis, the lon mutant should be unable to mitigate stress-induced damage and must be rapidly eliminated from the host cell compared to the wild type strain due to weakened virulence homeostasis. To further investigate the idea of early clearance, we exposed Salmonella mutant and wild type strains to all the tested stress conditions for a predetermined time (5 h) and verified their survival by plate counting. Besides, in vivo inoculation studies were also conducted to evaluate bacterial stress survival using a mouse model. As expected, the mutant resulted in rapidly dwindling numbers in both in vitro and in vivo conditions, suggesting that the activity of Lon protease is indispensable for Salmonella systemic infection and phagocytic survival during dissemination stages in the host.Citation48–Citation50
In the present study, the expression of lon was studied under normal laboratory conditions where the Salmonella was allowed to grow aerobically. Oxygen deprivation is yet another virulence determinant of wild type Salmonella. Being a facultative anaerobe, Salmonella can effectively survive under partial or complete anaerobic conditions in the gut. Low oxygen tension in the anaerobic environment upregulates the SPI-1 genes which in turn enhances the invasiveness of the bacteria.Citation51 On the other hand, such a reducing environment causes downregulation of lon expression. Absence of oxygen hampers the synthesis of ATP thereby decreasing the activity of Lon protease (AAA+ ATPase). Once the Salmonella maneuvers through the gut and reaches an oxidizing environment, the Lon protease activity increases. Our observation of a reduction in lon expression under anaerobic condition corroborates with the earlier studies in Escherichia coli. Lon activity is controlled by a redox switch which adjusts the size of the enzyme’s exit pore depending on the presence or absence of oxygenCitation52 and thereby regulates the lon-dependent pathways in Salmonella. Additionally, the exposure to the anaerobic environment helps Salmonella to survive in the extra-intestinal oxygen-rich conditions.Citation53 Further, Salmonella virulence is influenced by multiple genes and pathways other than lon.Citation54,Citation55
Summarizing the results presented here, we conclude that Lon protease is essential for Salmonella oxidative stress regulation followed by acid stress, both of which are important aspects of Salmonella survival in macrophages and the hostile gastric tract. The role of Lon protease goes beyond its cellular homeostasis and quality control role; rather it is a key element in Salmonella pathogenesis and is indispensable for systemic infection. We propose that Lon protease is a mediator of virulence genes hosted in the SPI-1 genomic region. This study adds mechanistic insight to the fate of Salmonella in the absence of Lon protease and highlights its essentiality as a virulence mediator on top of its already known function as a housekeeping protease.
Materials and methods
Bacterial strains, plasmids, and primers
The bacterial strains, plasmids, and primers used in this study are listed in and . Salmonella strains were routinely grown in Luria broth (LB) at 37°C with constant agitation or on LB agar. For all stress-induced experiments, Salmonella Typhimurium (ST) grown to log phase was utilized unless otherwise indicated.
Table 1. List of bacterial strains and plasmids used in this study.
Table 2. List of primers used in this study.
Construction of a fepA Salmonella mutant and a lon complemented strain
The mutant strain was constructed as previously described.Citation57 Briefly, prior to the target gene deletion from the host strain, JOL 401 and JOL 909 were electroporated with the helper plasmid pKD46, which provides the inducible red lambda components required for recombination. The target gene fepA was replaced with the catR gene contained on a linear PCR product amplified from the pkD3 plasmid. Recombinant clones were selected by plating on LB agar containing 25 µg/mL chloramphenicol. Successful fepA gene deletion was confirmed using flanking (fepA-F) and inner (fepA-I) PCR primers (). The lon gene was amplified for complementation experiments using the Lon-C primers (). The general PCR steps used for amplification consisted of a 3-min denaturation step at 95°C, 30 cycles of denaturation for 45 s at 90°C, annealing for 45 s at 55°C, an extension for the 60 s at 73°C, followed by a final elongation of 10 min at 73°C. The PCR product was cloned into the pWSK29 low copy number plasmid using EcoRI and HindIII as restriction enzymes.
Cell culture
The human epithelial cancer cell lines HeLa, HepG2, and Caco-2 were procured from the American Type Culture Collection (Manassas, VA, USA) and were maintained in Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, MA, USA) containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 µg/mL streptomycin, and L-glutamine (0.3 mg/mL). When the cells reached 60–80% confluence, cells were harvested and seeded in 96-well tissue culture plates at a rate of 2.5x105/mL in 10% DMEM. Plates were incubated at 37°C in a 5% CO2 atmosphere.
Growth curve analysis
Growth of the ST wild type strain (JOL 401), ST lon mutant (JOL 909), and ST lon complemented strain (JOL 909::lon) was analyzed by sub-culturing overnight grown cultures at a 1:100 ratio in LB broth. Cultures were incubated at 37°C on a shaking platform (225 rpm). Aliquots of 1 mL were collected every hour for a period of 12 h and OD600 was determined.
Bacterial growth in stress conditions
Log phase bacterial cultures were subjected to various stress conditions with slight modifications.Citation58 Briefly, bacterial cells were grown to mid-log phase (0.4 OD600) and cells were collected by centrifugation at 3,500 x g for 15 min. Bacterial cells were re-suspended in pre-warmed LB medium at pH 3.0 (adjusted with hydrochloric acid) for acid stress, into pre-chilled LB medium at 4°C for cold stress, into pre-warmed LB with 5 mM H2O2 for oxidative stress, and LB with 5% NaCl (wt/v) for osmotic stress. Bacteria were exposed to respective stress conditions for 5 h.
Adhesion and invasion assays
Adhesion and invasion assays were conducted using HeLa, HepG2, and Caco-2 cell lines according to a previously described procedure.Citation59 Confluent cell monolayers were infected with Salmonella strains JOL401, JOL909, and the JOL909::lon complemented strain at 20 multiplicity of infection (MOI) in 24-well tissue culture plates. After 45 min of incubation, cells were washed three times with PBS, and adhered bacterial cells were enumerated by lysis of monolayers with 0.1% Triton X-100 for 10 min. For invasion assays, bacterial interaction with cell monolayers was conducted for 2 h and subsequently treated with gentamycin (100 μg/mL) for 1.5 h to eliminate any extracellular bacteria. Then, the cells were lysed with Triton X-100 for 10 min. Invaded cells were enumerated by plating on LB agar using decimal dilutions. The data from three independent experiments are presented as CFU/mL with standard deviation.
Intracellular survival in phagocytic cells
To assess the intracellular survival of the lon mutant in phagocytic cells, the Salmonella strains were labeled using a CFSE Cell Division Tracking Kit (BioLegend, CA, USA). Then, the labeled Salmonella strains were injected into the mouse macrophage cell line RAW and incubated at 105 cells/mL for 24 h in 24-well plates. Cells were observed under fluorescent microscopy (Leica, Wetzlar, Germany) at 400 x magnification. Further, actual CFU counts were taken at 1, 3, 24 and 48 h post-inoculation to confirm the reduction in cell numbers in phagocytic cells.Citation45
Phalloidin staining of actin
HeLa, HepG2 and Caco-2 cells growing on a coverslip were infected with stress exposed JOL 401, JOL 909, or JOL 909::lon cells at 20 MOI. Uninfected cells at t = 0 were considered as the control. Cells were incubated for 20 min and washed three times with PBS. Slides containing infected cells were fixed with 4% paraformaldehyde for 30 min at room temperature and washed three times with 1X PBS. Cells were permeabilized with 0.01% Triton X-100 in PBS for 10 min at room temperature. After washing, cells were blocked with 5% bovine serum albumin. To stain actin filaments, 100 μL of 1X phalloidin (Abnova, Taiwan) was prepared in 1% BSA and incubated with cells for 1 h in the dark. Nuclear staining was carried out using DAPI (Sigma, CA, USA) and observed under fluorescence microscopy (Leica). The extent of actin polymerization by each ST strain was compared in three independent experiments.
Hydroxyl radical (OH·) measurement
Production of OH· was measured following the manufacturer’s protocol (Abcam, Cambridge, UK). After exposure to stress treatments, Salmonella strains were plated in 96-well plates at a concentration of 1 × 10Citation4 cells/90 µL per well. Plates were centrifuged at 800 rpm for 2 min. The supernatant was removed, and wells were supplemented with 100 µL/well OH580 Stain Working Solution and incubated at 37ºC for 1 h. Then, cells were washed three times with DPBS followed by the addition of 100 µl assay buffer to each well. Fluorescence was recorded at Ex/Em = 540/590 nm. As a positive control, HeLa and HepG2 cells treated with 10 µM CuCl2 and 100 µM H2O2 (Fenton reaction) at 37ºC for 1 h. Cells without any treatment were considered as a negative control.
In vitro bacterial survival
To evaluate the ability of bacterial cells to withstand stress treatment, cells were collected by vacuum filtration using a 47-mm filter membrane with 0.45-µm pores (Thermo Fisher Scientific) after 5 h. Filtered cells were resuspended in pre-warmed LB broth by vortexing. The viable bacterial counts were performed through serial dilutions and plating on BGA agar plates.
In vitro bacterial virulence gene expression
The expression of virulence genes in Salmonella strains after stress treatment was investigated at the mRNA level at 5 hpi under each stress condition. The total RNA was isolated, and cDNA was synthesized (Toyobo, Osaka, Japan). Virulence gene expression was quantitatively analyzed using qRT-PCR (). The PCR conditions were 95°C for 5 min, 40 cycles at 95°C for 30 s, 56°C for 30 s, 72°C for 1 min, and final annealing and extension of 55°C for 5 s and 95°C for 30 s. The expression results were normalized against the housekeeping gene rrsG of Salmonella and data are presented in the 2−ΔΔCT method.Citation60
In vitro cytokine gene expression
The cytokine gene expression in HeLa, HepG2, and RAW cells in response to bacterial infection (oxidative stress treated) were carried out for IL-2, IFN- γ, IL-4, and IL-10 (). Herein, infected HeLa and HepG2 cells were cultured for 12, 24, and 48 h. The cells were harvested at each time point and the total RNA was isolated and cDNA was synthesized as described above. The expression of cytokine genes was normalized against GAPDH and non-infected control. The changes in the relative expression of cytokine genes were determined using the 2−ΔΔCT method.Citation60
Survival and growth in epithelial cells
HeLa and HepG2 cells were grown in DMEM complete medium. A total of 4 × 105 cells were seeded per well in 24-well plates. When the cells reached 80% confluence, monolayers were infected with JOL401, JOL909, and JOL909::lon at 10 MOI for 2 h. After incubation, cells were washed three times with PBS and the extracellular bacteria were removed by gentamycin treatment (100 μg/mL) for 2 h. Then, cells were washed three times with PBS and replenished with complete medium. The bacteria-induced cytotoxicity was assessed using IncuCyte (Thermo Fisher Scientific) live-cell imaging system using Cytotox Green reagent at 2 ng/mL.
Assessment of virulence in mice
All animal experimental procedures were approved (CBNU2015-00085) by the Jeonbuk National University Animal Ethics Committee in accordance with the guidelines of the Korean Council on Animal Care and Korean Animal Protection Law, 2007; Article 13 (experiments with animals). Specific pathogen-free, female BALB/c inbred mice (4–5 weeks average age) were purchased from Koatech (Pyeongtaek, Gyeonggi-do, Korea). The animals were provided with water and antibiotic-free food ad libitum. Animals were monitored twice daily for behavioral and physiological signs.
To assess in vivo virulence of Salmonella strains, mice (n = 10) were orally inoculated at 1 × 109 CFU/mice/100 μl of PBS with JOL401, JOL909, or JOL909::lon. Mice were monitored for Salmonella induced clinical signs including ruffled fur, weight loss, reduced feed intake, and mortality. Mice were sacrificed on days 1, 6, 9, and 12 post-infection, and enlargement of the spleen was recorded. Further, the level of bacterial colonization in the spleen was assessed by plating tissue homogenate (1 g) on BGA agar after serial dilutions.
Statistical analysis
Statistical analysis was conducted using GraphPad Prism 7 (GraphPad Software, CA, USA) and IBM SPSS software. Student’s t-test and analysis of variance (ANOVA) followed by Turkey’s multiple comparison test was used to compare means among the treatment groups and to compute the p-value. The difference was considered significant if p was ≤0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download Zip (382.1 KB)Data availability statement
The fluorescent microscopy imaging conducted for Salmonella mediated actin cytoskeleton rearrangement compiled and has been deposited in the Mendeley Data repository and can be accessed using the following link. Lee, John Hwa (2020), “Salmonella mediated actin condensation upon entry into HeLa and HepG2 cells”, Mendeley Data, V1, doi: 10.17632/czng5n4fdk.1.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Takaya A, Suzuki M, Matsui H, Tomoyasu T, Sashinami H, Nakane A, Yamamoto T. Lon, a stress-induced ATP-dependent protease, is critically important for systemic Salmonella enterica serovar Typhimurium infection of mice. Infect Immun. 2003;17:690–696. doi:10.1128/iai.71.2.690-696.2003.
- Takaya A, Tomoyasu T, Tokumitsu A, Morioka M, Yamamoto T. The ATP-dependent lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J Bacteriol. 2002;184:224–232. doi:10.1128/jb.184.1.224-232.2002.
- Maurizi MR. Proteases and protein degradation in Escherichia coli. Experientia. 1992;48:178–201. doi:10.1007/BF01923511.
- Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi:10.1146/annurev.genet.30.1.465.
- Smith CK, Baker TA, Sauer RT. Lon and Clp family proteases and chaperones share homologous substrate-recognition domains. Proc Natl Acad Sci U S A. 1999;96:6678–6682. doi:10.1073/pnas.96.12.6678.
- Micevski D, Dougan DA. Proteolytic regulation of stress response pathways in Escherichia coli. Subcellular Biochem. 2013:105–128. doi:10.1007/978-94-007-5940-4.
- Nicoloff H, Andersson DI. Lon protease inactivation, or translocation of the lon gene, potentiate bacterial evolution to antibiotic resistance. Mol Microbiol. 2013;90:1233–1248. doi:10.1111/mmi.12429.
- Bajaj V, Hwang C, Lee CA. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi:10.1111/j.1365-2958.1995.mmi_18040715.x.
- Lucas RL, Lee CA. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar typhimurium. J Bacteriol. 2001;183:2733–2745. doi:10.1128/JB.183.9.2733-2745.2001.
- Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol. 2004;180:820–832. doi:10.1038/nrmicro1004.
- Qi L, Hu M, Fu J, Liu Y, Wu M, Yu K, Liu X. Quantitative proteomic analysis of host epithelial cells infected by Salmonella enterica serovar Typhimurium. Proteomics. 2017;17:1700092. doi:10.1002/pmic.201700092.
- Liu Y, Zhang Q, Hu M, Yu K, Fu J, Zhou F, Liu X. Proteomic analyses of intracellular Salmonella enterica serovar typhimurium reveal extensive bacterial adaptations to infected host epithelial cells. Infect Immun. 2015;83:2897–2906. doi:10.1128/IAI.02882-14.
- Wemyss MA, Pearson JS. Host cell death responses to non-typhoidal Salmonella infection. Front Immunol. 2019;26:1758. doi:10.3389/fimmu.2019.01758.
- Bumann D, Schothorst J. Intracellular Salmonella metabolism. Cell Microbiol. 2017;19:e12766. doi:10.1111/cmi.12766.
- Eisenreich W, Rudel T, Heesemann J, Goebel W. How viral and intracellular bacterial pathogens reprogram the metabolism of host cells to allow their intracellular replication. Front Cell Infect Microbiol. 2019;9:42. doi:10.3389/fcimb.2019.00042.
- Shan Y, Lai Y, Yan A. Metabolic reprogramming under microaerobic and anaerobic conditions in bacteria. Reprogramming Microb Metab Pathways. 2012;64:159–179. Springer. doi:10.1007/978-94-007-5055-5_8.
- McBee ME, Chionh YH, Sharaf ML, Ho P, Cai MWL, Dedon PC. Production of superoxide in bacteria is stress-and cell state-dependent: a gating-optimized flow cytometry method that minimizes ROS measurement artifacts with fluorescent dyes. Front Microbiol. 2017;8:459. doi:10.3389/fmicb.2017.00459.
- Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373:1–6. doi:10.1006/abbi.1999.1518.
- Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi:10.1128/jb.177.9.2305-2314.1995.
- Parent MA, Goenka R, Murphy E, LeVier K, Carreiro N, Golding B, Ferguson G, Ii Rm R, Walker GC, Baldwin CL. Brucella abortus bacA mutant induces greater pro-inflammatory cytokines than the wild-type parent strain. Microbes Infect. 2007;9:55–62. doi:10.1016/j.micinf.2006.10.008.
- Farr SB, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi:10.1128/MMBR.55.4.561-585.1991.
- Dupré-Crochet S, Erard M, Nüβe O. ROS production in phagocytes: why, when, and where? J Leukoc Biol. 2013;94:657–670. doi:10.1189/jlb.1012544.
- He L, Luo D, Yang F, Li C, Zhang X, Deng H, Zhang J-R. Multiple domains of bacterial and human Lon proteases define substrate selectivity. Emerg Microbes Infect. 2018;7:1–18. doi:10.1038/s41426-018-0148-4.
- Bustamante VH, Martinez LC, Santana FJ, Knodler LA, Steele-Mortimer O, Puente JL. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad Sci. 2008;105:14591–14596. doi:10.1073/pnas.0801205105.
- Lee HY, Biswas D, Ahn J. In-vitro adhesion and invasion properties of Salmonella typhimurium competing with bacteriophage in epithelial cells and chicken macrophages. Rev Bras Cienc Avic. 2015;17:427–432. doi:10.1590/1516-635X1704427-432.
- Elhadad D, Desai P, Grassl GA, McClelland M, Rahav G, Gal-Mor O. Differences in host cell invasion and Salmonella pathogenicity island 1 expression between Salmonella enterica serovar Paratyphi A and nontyphoidal S. Typhimurium. Infect Immun. 2016;84:1150–1165. doi:10.1128/IAI.01461-15.
- Que F, Wu S, Huang R. Salmonella pathogenicity Island 1(SPI-1) at work. Curr Microbiol. 2013;66:582–587. doi:10.1007/s00284-013-0307-8.
- Lou L, Zhang P, Piao R, Wang Y. Salmonella pathogenicity island 1 (SPI-1) and its complex regulatory network. Front Cell Infect Microbiol. 2019;9:270. doi:10.3389/fcimb.2019.00270.
- Jennings E, Thurston TLM, Holden DW. Salmonella SPI-2 type III secretion system effectors: molecular mechanisms and physiological consequences. Cell Host Microbe. 2017;22:217–231. doi:10.1016/j.chom.2017.07.009.
- Figueira R, Holden DW. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology. 2012;5(8):501–511. doi:10.1046/j.1462-5822.2003.00294.x.
- Velge P, Wiedemann A, Rosselin M, Abed N, Boumart Z, Chaussé AM, Grépinet O, Namdari F, Roche SM, Rossignol A, et al. Multiplicity of Salmonella entry mechanisms, a new paradigm for Salmonella pathogenesis. Microbiologyopen. 2012;1:243–258. doi:10.1002/mbo3.28.
- Boumart Z, Velge P, Wiedemann A. Multiple invasion mechanisms and different intracellular behaviors: a new vision of Salmonella-host cell interaction. FEMS Microbiol Lett. 2014;361:1–7. doi:10.1111/1574-6968.12614.
- Korshunov S, Imlay JA. Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol Microbiol. 2010;75:1389–1401. doi:10.1111/j.1365-2958.2010.07059.x.
- Imlay JA. Diagnosing oxidative stress in bacteria: not as easy as you might think. Curr Opin Microbiol. 2015;24:124–131. doi:10.1016/j.mib.2015.01.004.
- Bedard K, Lardy B, Krause K-H. NOX family NADPH oxidases: not just in mammals. Biochimie. 2007;89:1107–1112. doi:10.1016/j.biochi.2007.01.012.
- Clifford DP, Repine JE. Hydrogen peroxide mediated killing of bacteria. Mol Cell Biochem. 1982;49:143–149. doi:10.1007/bf00231175.
- Kumar SR, Imlay JA. How Escherichia coli tolerates profuse hydrogen peroxide formation by a catabolic pathway. J Bacteriol. 2013;195:4569–4579. doi:10.1128/JB.00737-13.
- Pérez-Toledo M, Valero-Pacheco N, Pastelin-Palacios R, Gil-Cruz C, Perez-Shibayama C, Moreno-Eutimio MA, Becker I, Pérez-Tapia SM, Arriaga-Pizano L, Cunningham AF, et al. Salmonella typhi porins OmpC and OmpF are potent adjuvants for T-dependent and T-independent antigens. Front Immunol. 2017;8:230. doi:10.3389/fimmu.2017.00230.
- Nagy TA, Moreland SM, Andrews-Polymenis H, Detweiler CS. The ferric enterobactin transporter fep is required for persistent Salmonella enterica serovar typhimurium infection. Infect Immun. 2013;81(11):4063–4070. doi:10.1128/IAI.00412-13.
- Newman SL, Will WR, Libby SJ, Fang FC. The curli regulator CsgD mediates stationary phase counter-silencing of csgBA in Salmonella Typhimurium. Mol Microbiol. 2018;108(1):101–114. doi:10.1111/mmi.13919.
- Jubete Y, Maurizi MR, Gottesmanf S. Role of the heat shock protein DnaJ in the Lon-dependent degradation of naturally unstable proteins. J Biol Chem. 1996;271:30798–30803. doi:10.1074/jbc.271.48.30798.
- Phillips TA, VanBogelen RA, Neidhardt FC. Lon gene product of Escherichia coli is a heat-shock protein. J Bacteriol. 1984;159:283–287. doi:10.1128/JB.159.1.283-287.1984.
- Sleymi S, Lahbib K, Rahmouni N, Rzaigui M, Besbes-Hentati S, Abid S. Synthesis, characterization, electrochemical investigation and antioxidant activities of a new hybrid cyclohexaphosphate: cu1.5Li(C2H10N2)P6O18·7H2O. J Mol Struct. 2017;1144:406–414. doi:10.1016/j.molstruc.2017.05.071.
- Thomas KJ, Rice CV. Revised model of calcium and magnesium binding to the bacterial cell wall. Biometals. 2014;27:1361–1370. doi:10.1007/s10534-014-9797-5.
- Tsilibaris V, Maenhaut-Michel G, Van Melderen L. Biological roles of the Lon ATP-dependent protease. Res Microbiol. 2006;157:701–713. doi:10.1016/j.resmic.2006.05.004.
- Mizuno Y, Takada H, Nomura A, Jin CH, Hattori H, Ihara K, Aoki T, Eguchi K, Hara T. Th1 and Th1-inducing cytokines in Salmonella infection. Clin Exp Immunol. 2003;131:111–117. doi:10.1046/j.1365-2249.2003.02060.x.
- Franchi L. Role of inflammasomes in Salmonella infection. Front Microbiol. 2011;2:8. doi:10.3389/fmicb.2011.00008.
- Fenlon LA, Slauch JM. Phagocyte roulette in Salmonella killing. Cell Host Microbe. 2014;15(1):7–8. doi:10.1016/j.chom.2014.01.001.
- Drecktrah D, Knodler LA, Ireland R, Steele-Mortimer O. The mechanism of Salmonella entry determines the vacuolar environment and intracellular gene expression. Traffic. 2006;7:39–51. doi:10.1111/j.1600-0854.2005.00360.x.
- Steinberg BE, Grinstein S. Pathogen destruction versus intracellular survival: the role of lipids as phagosomal fate determinants. J Clin Invest. 2008;118:2002–2011. doi:10.1172/JCI35433.
- Jiang L, Feng L, Yang B, Zhang W, Wang P, Jiang X, Wang L. Signal transduction pathway mediated by the novel regulator LoiA for low oxygen tension induced Salmonella Typhimurium invasion. PLoS Pathog. 2017;13:e1006429. doi:10.1371/journal.ppat.1007997.
- Nishii W, Kukimoto-Niino M, Terada T, Shirouzu M, Muramatsu T, Kojima M, Kihara H, Yokoyama S. A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis. Nat Chem Biol. 2015;11:46. doi:10.1038/nchembio.1688.
- Jennewein J, Matuszak J, Walter S, Felmy B, Gendera K, Schatz V, Nowottny M, Liebsch G, Hensel M, Hardt WD. Low oxygen tensions found in Salmonella infected gut tissue boost Salmonella replication in macrophages by impairing antimicrobial activity and augmenting Salmonella virulence. Cell Microbiol. 2015;17:1833–1847. doi:10.1111/cmi.12476.
- Ilyas B, Tsai CN, Coombes BK. Evolution of Salmonella-host cell interactions through a dynamic bacterial genome. Front Cell Infect Microbiol. 2017;7:428. doi:10.3389/fcimb.2017.00428.
- Pilar AVC, Reid-Yu SA, Cooper CA, Mulder DT, Coombes BK. GogB is an anti-inflammatory effector that limits tissue damage during Salmonella infection through interaction with human FBXO22 and Skp1. PLoS Pathog. 2012:8. doi:10.1371/journal.ppat.1002773.
- Birhanu BT, Park N-H, Lee S-J, Hossain MA, Park S-C. Inhibition of Salmonella Typhimurium adhesion, invasion, and intracellular survival via treatment with methyl gallate alone and in combination with marbofloxacin. Vet Res. 2018;49:101. doi:10.1186/s13567-018-0597-8.
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci. 2000;97:6640–6645. doi:10.1073/pnas.120163297.
- Takaya A, Kubota Y, Isogai E, Yamamoto T. Degradation of the HilC and HilD regulator proteins by ATP-dependent Lon protease leads to downregulation of Salmonella pathogenicity island 1 gene expression. Mol Microbiol. 2005;55:839–852. doi:10.1111/j.1365-2958.2004.04425.x.
- Chandrapala D, Kim K, Choi Y, Senevirathne A, Kang DH, Ryu S, Kim KP. Putative inv is essential for basolateral invasion of Caco-2 cells and acts synergistically with OmpA to affect in vitro and in vivo virulence of Cronobacter sakazakii ATCC 29544. Infect Immun. 2014;82:1755–1765. doi:10.1128/IAI.01397-13.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi:10.1006/meth.2001.1262.
