ABSTRACT
The complex population of microbes in the human gastrointestinal (GI) tract interacts with itself and with the host, exerting a deep influence on health and disease development. The development of modern sequencing technology has enabled us to gain insight into GI microbes. Helicobacter pylori colonization significantly affects the gastric microenvironment, which in turn affects gastric microbiota and may be correlated with colonic microbiota changes. Crosstalk between H. pylori and GI commensal flora may play a role in H. pylori–related carcinogenicity and extragastric manifestations. We review current knowledge on how H. pylori shapes GI microbiota with a specific focus on its impact on the stomach and colon. We also review current evidence on colonic microbiota changes attributed to eradication therapy based on the clinical studies performed to date.
Introduction
Trillions of microorganisms reside in the human gastrointestinal (GI) tract and form a symbiotic relationship with the host, playing an important role in health and disease. The GI microbiome and the host generate a complex network of interactions that transcends the boundaries of the GI tract, forging intimate connections with all aspects of human physiology, including metabolic, immune, and neuroendocrine systems. The crosstalk is mediated by microbial-derived biochemical signals that are absorbed into the blood and circulated throughout the human body; by signals relayed by the enteric nervous system that transmit microbiota-derived cues to the central nervous system; and by immune cells that perceive local microbial signals in the GI tract and are trafficked throughout the body.Citation1–3
As a GI tract microbe, Helicobacter pylori is one of the most-studied bacteria. It is highly adapted to the human gastric mucosa and thrives in the stomach niche, having co-evolved with humans over tens of thousands of years.Citation4 Chronic infection can lead to either hypo- or hyperchlorhydria, depending on the anatomic distribution and severity of the resulting inflammation.Citation5 Although the majority of H. pylori–infected persons remain asymptomatic, chronic infection has been linked to peptic ulcer disease, gastric cancer, gastric mucosa-associated lymphoid tissue lymphoma, and a multitude of extragastric diseases. Current studies suggest that eradication of H. pylori can effectively reduce gastric cancer incidence and treatment should be considered for all H. pylori–infected persons to reduce the risk of peptic ulcers and gastric cancers.Citation6–8 However, there are still debates regarding the beneficial effects of H. pylori colonization, including regression in childhood asthma and other atopic disorders.Citation9,Citation10 It has been concluded that H. pylori is a common flora, or at least a harmless bacterium. Additionally, the mass eradication of H. pylori with antibiotic treatment as a preventive measure for gastric cancer and peptic ulcers raises several concerns, including the emergence of antibiotic resistance and perturbations in gut microbiota following H. pylori eradication.Citation11,Citation12 Being part of the GI ecosystem, H. pylori infection and its impact on gastric acid secretion may alter the GI microbiome and host health status. Here, we review current understandings of the impact of H. pylori infection on the GI microbiome and how it influences human health.
Helicobacter pylori and the esophageal microbiome
The esophageal microbiome in the normal esophagus
Although the esophagus serves as the beginning of the digestive tract, the esophageal microbiome has long been overlooked and little is known about it relative to our understanding of the composition and function of the gut microbiome. Early culture-based studies using esophageal washing demonstrated a high proportion of Streptococcus viridans and a pattern resembling that of the oral microbiome.Citation13,Citation14 The first culture-independent investigation of the distal esophageal microbiome identified a far more complex microbial community, comprising six major phyla (Firmicutes, Bacteroides, Actinobacteria, Proteobacteria, Fusobacteria, and TM7), with Streptococcus as the most prevalent genus.Citation15 Streptococcus, Hemophilus, Neisseria, Prevotella, and Veillonella are considered to be the core microbes in the normal esophagus.Citation16 However, bacterial composition may differ depending on various factors, such as age, use of proton pump inhibitors, and disease.Citation16–18
The esophageal microbiome in reflux esophagitis, Barrett’s esophagus, and esophageal adenocarcinoma
Chronic gastric acid exposure or duodenal bile in the distal esophagus is considered to be the primary factor in the pathogenesis of reflux esophagitis. It was widely accepted that reflux may cause chronic esophageal injury and promote carcinogenesis in Barrett’s esophagus. A culture-independent study by Yang et al. classified the esophageal microbiota into two distinct types.Citation19 The healthy esophagus harbored Gram-positive taxa from the Firmicutes phylum, of which Streptococcus was the dominant genus (Type I microbiome), while an inflamed esophagus (reflux esophagitis or Barrett’s esophagus) was dominated by Gram-negative taxa from the Bacteroidetes, Proteobacteria, and Fusobacteria phyla (Type II microbiome). These findings are consistent with other studies,Citation18,Citation20,Citation21 reliably demonstrating a change in esophageal microbiota in cases of reflux disease that most likely reflects physiological changes due to excess gastric acid. Studies investigating the microbiota in cases of esophageal adenocarcinoma (EAC) are rare. The studies by Elliott et al. and Snider et al. identified reduced microbial diversity in EAC samples compared with controls.Citation22,Citation23 Some EAC samples were dominated by a single bacterial species belonging to the order Lactobacillales in the study by Elliott et al., while Snider et al. found more Enterobacteriaceae and Akkermansia muciniphila in patients with high-grade dysplasia or EAC. Both studies had relatively small sample sizes and further research is required before an EAC microbiome signature can be defined.
Helicobacter pylori, the esophageal microbiome, and esophageal diseases
The incidences of gastroesophageal reflux disease, Barrett’s esophagus, and EAC have been rising over the past several decades in developed countries and are inversely associated with H. pylori infection prevalence.Citation24–27 Previous research describes the existence of a core esophageal microbiota and has shown that its composition in healthy controls differs at the phylum and genus levels from patients with reflux esophagitis or Barrett’s esophagus. The altered bacterial microenvironment may contribute substantially to esophageal mucosa injury and further carcinogenesis. One of the hypotheses explaining the protection by H. pylori against Barrett’s esophagus and EAC may relate to the fact that at the population-level it reduces acid secretion. H. pylori also influences colonization by other important organisms. Amir et al. and Deshpande et al. determined that the administration of proton pump inhibitors influences microbial composition in the esophagus, and this effect is thought to be related to acid levels.Citation18,Citation28 The H. pylori–positive stomach produces less acid and the microbial community in the distal esophagus is probably altered when reflux occurs. It would be interesting to determine whether H. pylori interacts with the esophageal microbiota to confer protection against Barrett’s esophagus or EAC. However, this is a current gap in esophageal microbiome research, and no studies have assessed whether hosts’ H. pylori status contributes to different esophageal microbial communities. It is imperative to study the impact of H. pylori on host physiology and the ensuing effect on the esophageal microbiome, although this may become increasingly difficult due to a declining prevalence of H. pylori.
Helicobacter pylori and the gastric microbiome
The normal gastric microbiome
Although Gillespie isolated 24 different organisms from the stomach through a stomach tube in 1893, the stomach was still considered sterile due to its acidic environment. Microbes cultured from gastric fluid were generally considered to be transient or passing luminal microbes until the discovery of H. pylori in 1982.Citation29 For the next few decades, H. pylori was considered to be the only organism capable of surviving in the hostile gastric environment because culturing was the mainstay of microbial research.Citation30,Citation31 However, the majority of bacteria are difficult to culture or are uncultivable.
Culture-independent methods, particularly next-generation sequencing (NGS) technology, have broadened the horizons in human microbial research.Citation32 Studies employing NGS reveal that human gastric microbes are more diverse than initially anticipated.Citation33–35 Published studies show significant heterogeneity of gastric microbiota, which may be attributed to inter-individual variability, ethnicity, different sample types, different gastric pathologies, and the use of different technical approaches. In a review article, Rajilic‐Stojanovic et al. compared the studies that investigated the gastric microbiota using NGS. Based on an arbitrary cut‐off value requiring genera to be present in more than 20% of the included studies, the typical gastric microbiota consists of 57 bacterial genera distributed among eight phyla, including Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria, Spirochetes, Tenericutes, and TM7.Citation36 The six most common genera reported were Prevotella, Streptococcus, Neisseria, Hemophilus, Fusobacterium, and Veillonella. Helicobacter was detected in 23 of 36 studies. The bacterial community of the normal stomach has not been extensively characterized; only four studies have reported on the microbiota present in healthy adults, and these provide us with a snapshot of healthy gastric microbiota.Citation37–40 All studies reported the presence of Prevotella, Streptococcus, Megasphaerae, Capnocytophaga, Oribacterium, and Propionibacterium. It is noteworthy that around half of the 266 reported genera were only found in one study, indicating that these groups are most likely of low biological relevance or due to artifacts from the sequencing technique or bioinformatic processing.Citation41
Effect of Helicobacter pylori on the gastric microbiome
H. pylori employs several enzymatic machineries that permit its survival in the harsh acidic conditions of the stomach.Citation42 When H. pylori is present, it is the most abundant organism of the gastric microbiota, representing 40%–90% of the gastric microbiota.Citation34,Citation43–47 The alpha diversity of bacteria in the stomach is negatively associated with the presence of H. pylori.Citation34,Citation43,Citation47–50 Studying the impact of H. pylori status on beta diversity, we observed that if H. pylori is present in the gastric mucosa it gains a clear predominance, which alters the gastric microbial composition in H. pylori–infected individuals.Citation47–51 Most reports show that H. pylori–positive and H. pylori–negative individuals’ microbiota are mainly dominated by the same phyla but with different percentages of relative abundance.Citation34,Citation47,Citation52 H. pylori–positive individuals have a higher abundance of Proteobacteria, probably resulting from the contribution of H. pylori, while there is a lower abundance of Actinobacteria, Bacteroidetes, and Firmicutes.Citation34,Citation43,Citation44,Citation48,Citation49,Citation52 Only one human study discusses the taxonomic differences between H. pylori–positive and H. pylori–negative groups after H. pylori sequence reads were removed.Citation34 When H. pylori sequences were left out of the analysis, the phylotype evenness and diversity of H. pylori–positive individuals were higher than that of H. pylori–negative individuals. Further examination of the phylum distribution of all non–H. pylori phylotypes of individuals based on H. pylori status revealed no gross differences in taxonomic patterns. Martin et al. assessed the impact of H. pylori on the preexisting gastric microbial community in a rhesus macaque model. There was no significant difference in the average relative abundance of non-Helicobacter taxa in pre- and post-inoculation samples after removing Helicobacter reads.Citation53 The rhesus model suggests the rhesus gastric microbial community is largely stable despite the immunological and physiological changes that occur due to H. pylori infection. In human studies, the gastric microbial diversity changes associated with H. pylori seem to be reversible to some degree. Eradication of H. pylori infection may increase the diversity of gastric microbiota.Citation44,Citation54–56
Helicobacter pylori, the gastric microbiome, and gastric cancer
H. pylori is well-recognized as a class I carcinogen for gastric cancer.Citation57,Citation58 Infection initiates chronic gastric inflammation and destroys the hydrochloric acid-secreting glands of the stomach, ultimately leading to the precancerous changes of atrophic gastritis (AG) and intestinal metaplasia (IM).Citation5,Citation59,Citation60 Although H. pylori infection is known to precipitate this cascade, cohort studies show that only 1%–2% of H. pylori–infected individuals develop gastric cancer.Citation61 Moreover, the point of no return that leads to gastric cancer in the carcinogenesis cascade is reportedly associated with IM and dysplasia, independent of H. pylori status.Citation62 H. pylori virulence, host genetics, and environmental factors all contribute to the development of gastric cancer.Citation63
Before H. pylori was discovered in 1982, it had repeatedly been shown that bacteria multiply during gastric diseases, such as peptic ulcer diseases and gastric cancer. Hewetson et al. seem to have been the first to study material taken directly from the stomach during surgery.Citation64 They took cultures from the stomach in 36 cases and a variety of bacteria were isolated. They concluded that 72% of the cases with gastric ulcers were positive for bacteria, compared with 17% of the cases without gastric ulcers. Later studies consistently showed the percentage of sterile stomach samples was lower in patients with gastric ulcers than in patients with duodenal ulcers, which is probably associated with the acidity and mucosal atrophy in the stomach.Citation65–67 Several studies have investigated the bacteriology of patients with gastric cancer and found that patients with gastric carcinoma have higher bacterial counts and are colonized with higher numbers of different species than patients with other gastric diseases.Citation65–68 Oropharyngeal or intestinal commensals (Streptococcus, Bifidobacterium, Lactobacillus, Veilonella, Klebsiella, Escherichia, Pseudomonas, Neisseria, Staphylococcus, and Bacillus) were reported to be associated with gastric cancer.Citation65,Citation68 The results of culture-based studies associated with gastric disease in English literature are summarized in .Citation64–70 It has been hypothesized that the hypochlorhydria associated with AG allows for bacterial overgrowth in the stomach, and this may play a role in gastric carcinogenesis.Citation71 However, research on the microbiota and gastric cancer remained relatively unexplored until the development of NGS.
Table 1. Studies analyzing the role of gastric microbiota in gastric diseases using culture-based methods
Dicksved et al. conducted one of the first DNA-based studies investigating the gastric microbiota in patients with gastric cancer using terminal restriction fragment length polymorphisms in combination with 16S rRNA gene cloning and sequencing.Citation72 They found an enrichment of Streptococcus, Lactobacillus, Veilonella, and Prevotella, and a low abundance of H. pylori in ten patients with gastric cancer. This was followed by 16 studies that assessed the role of the gastric microbiota in gastric cancer (, )).Citation44,Citation46,Citation73–86 Most of these studies observed a reduction in bacterial diversity or richness in the shift from non-atrophic gastritis to gastric cancer, while five studies showed different results. Dicksved et al., Wang et al., and Jo et al. did not find a significant difference in diversity indices between gastric cancer patients and controls.Citation72,Citation75,Citation76 However, two of the studies were small in size and underpowered, which made it difficult to detect potential differences in microbiota diversity between groups.Citation72,Citation75 Eun et al. reported an increase in microbial diversity from gastritis to cancer, but provided this result without a supporting statistical analysis.Citation74 Castaño-Rodríguez et al. utilized an RNA rather than DNA-based analysis and their findings cannot be directly compared with other studies.Citation78 In addition to sample size and differences in methodology, Cocker et al. and Stewart et al. concluded that the discrepancies in the published studies may result from demographic characteristics, including gender, age, H. pylori infection status, and ethnicity.Citation80,Citation87
Table 2. Summary of studies examining the relationships between gastric cancer and gastric microbiota
Figure 1. The interplay between Helicobacter pylori and gastrointestinal (GI) microbiota
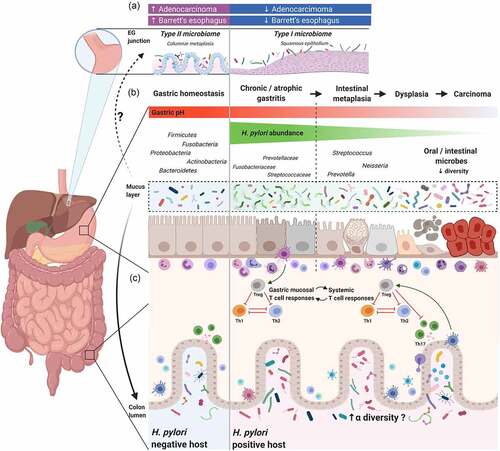
Previously published studies show reduced H. pylori abundance in tumor tissue compared with adjacent non-neoplastic areas,Citation77,Citation79,Citation82,Citation88 suggesting that bacteria other than H. pylori may play a role in the development of gastric cancer. To determine whether changes in gastric microbiota play a role in the development of gastric cancer or are secondary to the changes in the gastric environment, studies of rodent model systems have helped to identify important drivers and modifiers of diseases related to the microbiome. Studies using the insulin-gastrin (INS-GAS) transgenic mouse model demonstrated that mice infected with H. pylori together with the colonization of commensal flora developed more severe gastric lesions and had earlier development of GI intraepithelial neoplasia compared with H. pylori–infected germ-free INS-GAS mice, highlighting the idea that the gastric microbiota may participate in the cascade of events leading to gastric cancer following H. pylori infection.Citation89,Citation90 Although a consensus has not yet been reached regarding the dominant bacteria potentially involved in human gastric cancer development, an increase in several oral and intestinal commensal bacteria has been reported in several studies. Ferrairi et al. reported the enrichment of Achromobacter, Citrobacter, Lactobacillus, Clostridium, Rhodococcus, and Phyllobacterium in gastric cancer microbiota.Citation46 Using a co-occurrence/co-exclusion network analysis, Coker et al. identified the enrichment of Peptostreptococcus stomatis, Streptococcus anginosus, Parvimonas micra, and Slackia exigua in gastric cancer and determined that Dialister pneumosintes was crucial to the gastric cancer occurrence network, and these findings were successfully validated in the Inner Mongolian cohort.Citation80
The majority of the reported studies are based on cross-sectional comparisons of individuals with and without histological changes in the gastric mucosa. This approach only provides a unique snapshot in time, which does not allow us to derive information about gastric carcinogenesis. A recent systemic review failed to find significant differences in microbiota profiles between individuals with superficial gastritis, atrophic gastritis, and IM.Citation36 Defining a gastric cancer microbial signature without considering the underlying mechanism of the ensuing dysbiosis provides a limited perspective with limited therapeutic potential. A recent study carried out in Shandong, China analyzed 102 paired gastric biopsy samples taken before and one year after H. pylori eradication.Citation56 Sung et al. demonstrated Roseburia and Sphingomonas were depleted in patients with persistent inflammation one year after H. pylori eradication. The emergence and persistence of gastric atrophy and IM one year following H. pylori eradication were associated with a cluster of oral bacteria comprising Peptostreptococcus, Streptococcus, Parvimonas, Prevotella, Rothia, and Granulicatella. This study supports the hypothesis that the presence of H. pylori provides various microbiome niches contributing to gastric cancer development. A larger multicenter, multicultural, prospective study focusing on the gastric microbiota during gastric carcinogenesis is warranted to validate the results and to explore underlying mechanisms.
Helicobacter pylori and colonic microbiota
The microbial component of the human digestive tract is at its highest in the colon, with nearly a 107-fold increase in number compared with the stomach.Citation91 The GI tract is a complex and dynamic network with interplay between intestinal epithelial cells, the immune system, food, host metabolism, and commensal microbes. Numerous studies have attempted to define the microbial signatures of various diseases and possible microbial therapeutic interventions. Considering the commensal microbiota and the host form a unique entity in a continuum along the GI tract, any changes in the GI microenvironment may influence the homeostasis of the entire system. The studies described in the previous section reveal that H. pylori colonization has a great impact on the gastric microbiome. Nevertheless, the effect of H. pylori on colonic microbiota remains largely unexplored.
Helicobacter pylori and colonic microbiota in rodent models
Theoretically, H. pylori may influence colonic microbiota through crosstalk with the host immune system or through changes in the local gastric environment. Kienesberger et al. infected neonatal C57Bl/6 mice with H. pylori strain PMSS1 at four or six weeks of age. The study demonstrated that H. pylori not only influences the gastric microbial community structure but also has systemic effects and alters the distal gut microbiota.Citation92 Studies have shown H. pylori infection acts as an immunoregulator of regulatory T cell induction through the downregulation of IL-18 in H. pylori–infected mice, which results in immunotolerance and the facilitation of H. pylori persistence.Citation92,Citation93 H. pylori may regulate microbial composition in the distal intestine in a similar fashion. The most significant route of impact would possibly be through H. pylori–induced hypochlorhydria in the stomach. It is plausible that hypochlorhydria may promote the entrance of acid-sensitive bacteria into the distal GI tract, resulting in the alteration of the colonic microbiome. Heimesaat et al. investigated the GI microbiota changes in Mongolian gerbils after 14 months of infection with H. pylori and reported distinct shifts in microbiota composition of the distal uninflamed GI tract of wildtype H. pylori–infected animals.Citation94 Gastric immunopathology with reduced gastric acid and hypergastrinemia during H. pylori infection has been put forward as a hypothetical explanation for the distal gut microbiota changes. Additionally, reduced leptin and ghrelin secretion in H. pylori–infected individuals may indirectly influence the GI microenvironment by modulating gastric acid secretion and the immune response, which in turn alters the microbial composition of the GI tract.Citation92,Citation95–97
Helicobacter pylori and colonic microbiota in humans
Compared to studies investigating the effect of H. pylori on human gastric microbiota, relatively few studies have addressed the influence of H. pylori on colonic microbiota (summarized in , )).Citation47,Citation54,Citation98–112 Most studies have focused on the consequences of H. pylori eradication therapy.Citation98–101 Earlier studies using culture-based approachesCitation98-100 or fluorescent in situ hybridizationCitation99 suggested different compositions of gut microbiota among H. pylori–infected and uninfected individuals. Bühling et al. and Myllyluoma et al. concluded that the total number of anaerobes was significantly lower in H. pylori–positive individuals compared with H. pylori–negative individuals.Citation98,Citation99 The advent of culture-independent approaches, high-throughput sequencing coupled with advances in computational methods, have enabled genome-wide dissection of H. pylori and gut microbiota interactions. Eleven studies have assessed the gut microbiota in H. pylori–infected individuals (). The majority of these studies were in Asian populations and children were included in three studies. Microbiota composition was assessed from fecal specimens by DNA amplification (in nine studies) or by shotgun sequencing (in one study).Citation54 One study used reverse‐transcribed RNA for 16S rRNA gene sequencing to assess microbial communities in fecal and colon biopsy specimens.Citation47
Table 3. Summary of studies examining the effect of Helicobacter pylori infection on colonic microbiota
Except for one study,Citation111 most reports show higherCitation54,Citation101,Citation106,Citation108,Citation110 or unchangedCitation47,104,105,Citation107,109 alpha diversity indices from the gut microbiota of H. pylori–infected individuals compared to H. pylori–negative controls. The two largest cohorts enrolled 214 H. pylori–infected Japanese participants and 212 H. pylori–infected German participants and both showed higher alpha diversity compared with matched H. pylori–negative controls,Citation106,Citation110 while Wang et al. reported no differences in alpha diversity indices between 128 H. pylori–infected individuals and 158 H. pylori–negative controls.Citation107 High microbial diversity is usually regarded as an indicator of a healthy gut microbiome, while a reduction in diversity is associated with poorer health or diseases. The reason why H. pylori infection is associated with higher diversity is not fully understood. It may reflect the fact that H. pylori is ancestral and has co-evolved with humans over tens of thousands of years.Citation4 It has been suggested that H. pylori infection strengthens the host’s resilience against microbiome perturbations or GI infections, which results in higher fecal microbiota diversity in hosts.Citation110 Another possible explanation for this phenomenon is that chronic H. pylori infection alters the acidic environment in the stomach, permitting more microorganisms to pass through the gastric acid barrier and reach the distal gut.
Seventeen studies reported differences when comparing fecal microbiota compositions of H. pylori–infected and non-infected individuals. Among thirteen studies using NGS technology, six studies observed differences in beta diversity between H. pylori–infected and non-infected populations,Citation54,Citation107,109–Citation111 while five studies showed no differences in fecal microbiota composition.Citation47,104,105,Citation108,Citation112 It is possible that the small sample size of the studies left them statistically underpowered, and potential differences in microbiota composition between groups would be difficult to detect. Chen et al. conducted the first study employing NGS technology to assess fecal microbiota composition in patients infected with H. pylori.Citation101 The study revealed a significant difference of 22 bacterial genera between H. pylori–positive and negative populations. However, the differential taxa of colonic microbiota between infected and uninfected groups have not been well characterized in the published literature (Supplementary Table 1). A higher abundance of Haemophilus, Howardella, Gemella, and Streptococcus, alongside a lower abundance of Pseudoflavonifractor, Fecalibactrium, Ruminococcus, and Eubacterium ventriosum in fecal samples has been reported in H. pylori–infected patients (Supplementary Table 1). The inconsistency in differential taxa in fecal microbiota associated with H. pylori infection may reflect the heterogeneity of age, ethnicity, dietary habits, and gastric pathology in the study populations. Iino et al. demonstrated that Streptococcus was significantly more abundant in feces of H. pylori–infected individuals with severe gastric atrophy, compared with that in H. pylori–infected individuals without atrophic gastritis.Citation106 This suggests H. pylori infection and the extent of gastric mucosal atrophy may affect the composition of the gut microbiota in Japanese populations. In addition, Gao et al. showed that alterations in the fecal microbiota, especially the dominant phyla of Bacteroidetes, Firmicutes, and Proteobacteria, may be associated with H. pylori–related gastric lesion progression in a Chinese population.104 The impacts of gastric pathology severity on fecal microbiota require further investigation because the evidence is still limited.
Colonic microbiota and consequences of H. pylori eradication
Antibiotics break the homeostasis of gut microbiota and result in short-term alterations in the healthy gut microbiota and potentially long-lasting changes in its composition and function.Citation115 One of the ways that H. pylori influences the colonic microbiome would be through H. pylori eradication therapies. Jakobsson et al. revealed that a short‐term antibiotic treatment for H. pylori eradication delivered a profound insult to the GI flora and resulted in a perturbed oral and colonic microbiome observed one week after treatment and persisting up to four years later.Citation116 Several articles have reported short‐term and long‐term changes in gut microbiota after H. pylori eradication and are reviewed and summarized in and .Citation54,Citation55,Citation101,Citation112,Citation116–124 Most of the studies used triple therapy or bismuth quadruple therapy. The short-term changes in gut microbiota after these therapies have been reported in nine studies using culture-independent approaches.Citation54,Citation101,Citation112,Citation119–124 All of these studies showed significant perturbations in the diversity and composition of gut microbiota immediately after H. pylori eradication. Long‐term changes (over six months) were reported in seven studies, although most had low numbers of cases. Of the seven studies that assessed the long‐term changes in gut microbiota at least six months after H. pylori eradication, most reported full recovery of bacterial diversity. However, He et al. reported higher alpha diversity after eradication therapy in children,Citation54 and the largest cohort from Liou et al. demonstrated reduced alpha diversity one year after eradication therapies in patients that received regimens containing metronidazole (quadruple therapy or concomitant therapy).Citation122 Additionally, some studies observed notable changes in abundance at the genus level over six months following H. pylori eradication. A recent meta-analysis compared the taxa changes at three different follow‐up periods after H. pylori eradication.Citation125 In general, Actinobacteria populations decreased compared with baseline levels. Proteobacteria populations increased during short‐term follow‐up and then returned to baseline levels. Enterobacteriaceae and Enterococcus increased in the short‐term and interim follow‐up. However, there were no consistent changes in Firmicutes, Bacteroidetes, Bifidobacterium, or Lactobacillus, probably due to sample size, ethnicity, and eradication regimens.
Figure 2. The impact of Helicobacter pylori eradication on the gut microbiome
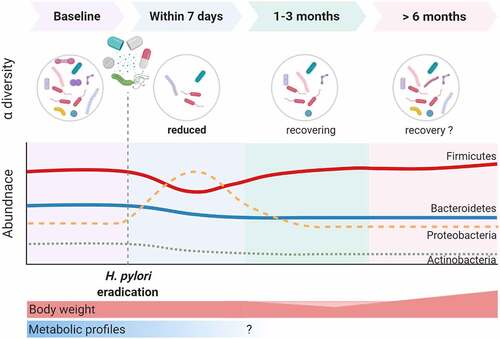
Table 4. Summary of studies examining the impact of Helicobacter pylori eradication therapy on the gut microbiota
In summary, the human digestive tract is a complex ecosystem and H. pylori infection alters not only gastric acidity but also host-microbe interactions, which may result in changes in colonic microbiome composition. Antibiotics are a double-edged sword. The antimicrobial agents (including bismuth) used for H. pylori eradication and gastric cancer prevention have direct effects on the colonic microbiota during short-term and possibly also long-term evaluations.
Helicobacter pylori, gut microbiota, and H. pylori–related extragastric disease
H. pylori has been associated with multiple extragastric diseases, such as cardiovascular diseases, neurological diseases, obesity, metabolic syndromes, and chronic immune-mediated disorders.Citation126 The underlying pathogenic mechanisms are not yet understood. The gut microbiota are involved in nutrient absorption, metabolism, and development and stimulation of the host immune system and digestive tract. It is hypothesized that gut microbiota may play a role in H. pylori–associated diseases. A large-scale cross-sectional study in Japan demonstrated significantly higher low-density lipoprotein levels and significantly lower high-density lipoprotein levels in men who were H. pylori seropositive, compared with H. pylori seronegative men.Citation127 Studies have shown a significant increase in body mass index and body weight after eradication of H. pylori,Citation122,Citation128 which may be partially explained by the restoration of ghrelin secretion, the relief of dyspepsia,Citation129 or a reduced Bacteroidetes-to-Firmicutes ratio.Citation55 In contrast to weight gain, studies showed improvement in insulin resistance, fasting glucose, total cholesterol, and triglyceride levels following eradication therapy.Citation122,Citation130 The improvement in these metabolic parameters may be attributed to gut microbiota alteration. He et al. demonstrated H. pylori infection resulted in alterations of gut microbiota and metabolic phenotypes consistent with those observed in a high-fat diet mouse model.Citation131 This study suggests there is complex crosstalk between H. pylori and the microbiota. Treatment of H. pylori may be beneficial for patients with impaired glucose tolerance in addition to diet control.
As for autoimmune disorders, there is growing evidence that H. pylori may protect hosts from chronic immune-mediated disorders such as asthma,Citation9 atopic disease,Citation132 and inflammatory bowel disease,Citation133,Citation134 which have been previously attributed to the activation of Th1 cells and inhibition of the Th2 allergic response by H. pylori.Citation10
An animal study showed that gut microbes belonging to the families Turicibacteraceae, Erysipelotrichaceae, and Desulfobirionaceae, which have been linked to changes in the host immune response, are influenced by the presence of H. pylori in mice.Citation92 Evidence suggested that the maturation of the human gut microbiota progresses by accruing microbes, followed by subsequent development and enrichment of the microbiome ecosystem throughout early childhood.Citation135 Chen et al. identified a negative association between H. pylori and asthma only in the younger age group of children 3–13 years old.Citation9 Malaty et al. examined the age of H. pylori seroconversion in a prospective cohort and suggested the peak period for newly acquired H. pylori infection was highest among children aged 4–5 years.Citation136 Since the gut microbiome gradually develops its structure and function during childhood,Citation137 further exploration is required to determine whether H. pylori by itself or in combination with the gut microbiota altered by infection protects the host against chronic immune-mediated illnesses. Targeted studies examining the impact of H. pylori during early childhood are urgently needed to help address its specific role in subsequent microbial colonization.
Conclusions
The advances in GI microbiota research allow investigators and clinicians to explore the role of the microbiome in various diseases including, but not limited to, GI diseases. Culture-independent techniques, particularly those based on high‐throughput or NGS technology, have revolutionized our knowledge of the GI microbiota. H. pylori, as one of the most important microbial members of the human GI tract, has been a significant focus for a long time due to its importance within the pathophysiology of peptic ulcer disease and gastric cancer. It is undisputed that significant differences exist in the microbiota of individuals with different gastric pathology, atrophic gastritis, IM, and gastric cancer, highlighting that dysbiosis in the stomach is a dynamic process and correlates with gastric carcinogenesis. The gastric cancer microbiota has drawn researchers’ attention and has been found to be enriched with intestinal or oral taxa. However, most studies on gastric microbiota and gastric cancer development are retrospective and correlational in nature. Longitudinal and prospective studies are needed to identify the presence of specific bacterial species or microbial consortia and the underlying pathways as the microbiota changes during gastric cancer carcinogenesis. It is possible that the presence of certain changes could be used to develop biomarkers to monitor disease progression and to develop disease-modifying therapies to manipulate the gastric microbiota and prevent the risk of developing gastric cancer.
The GI tract is a complex and dynamic ecosystem with interplay between various gut mucosal cells and their defense molecules, the immune system, food particles, and resident microbes. The harsh acidic environment of the stomach serves as a gated entrance to the GI system. H. pylori infection reduces gastric acid and changes the gastric microenvironment, which may in turn influence subsequent GI commensal microbiota colonization. Scientific efforts have been focused on the benefits of treating and eradicating H. pylori, and its relative absence provides us an opportunity to investigate a more complex gut-microbial–host-immune/metabolic axis. The current investigations on the complex crosstalk between H. pylori and the gut microbiota are far from conclusive. Most of the studies have been association studies and the exact underlying mechanisms need to be unraveled further. Longitudinal studies with a focus on the gut microbiota and host phenotype changes during H. pylori infection in humans are missing, as well as studies specifically evaluating the possible long‐term effects of eradication therapies on the GI microbiota. Multiomics approaches employing shotgun sequencing or long-read sequencing technology, in combination with metabolomics, are needed to clarify the long-term implications of gut microbiota and host physiology alterations following H. pylori eradication. The newly acquired knowledge in this field will provide insight into host-microbial crosstalk and will make microbial-directed therapies against diseases possible.
Supplemental Material
Download MS Word (46.7 KB)Acknowledgments
The authors would like to express special thanks to the staff of the Eighth Core Lab of the Department of Medical Research at the National Taiwan University Hospital. Schematic representations were generated with the support of BioRender and Pei-ju Chuang.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Donia MS, Fischbach MA. Small molecules from the human microbiota. Science. 2015;349(6246):1254766. doi:10.1126/science.1254766.
- Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–712. doi:10.1038/nrn3346.
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi:10.1126/science.1223490.
- Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136(6):1863–1873. doi:10.1053/j.gastro.2009.01.073.
- Sipponen P, Kekki M, Seppala K, Siurala M. The relationships between chronic gastritis and gastric acid secretion. Aliment Pharmacol Ther. 1996;10(Suppl 1):103–118. doi:10.1046/j.1365-2036.1996.22164011.x.
- Graham DY. The only good Helicobacter pylori is a dead Helicobacter pylori. Lancet. 1997 author reply 2;350(9070):70–71. doi:10.1016/S0140-6736(05)66278-2.
- Lee YC, Chiang TH, Chou CK, Tu YK, Liao WC, Wu MS, Graham DY. Association between Helicobacter pylori eradication and gastric cancer incidence: a systematic review and meta-analysis. Gastroenterology. 2016;150(5):1113–24 e5. doi:10.1053/j.gastro.2016.01.028.
- Chiang TH, Chang WJ, Chen SL, Yen AM, Fann JC, Chiu SY, Chen Y-R, Chuang S-L, Shieh C-F, Liu C-Y, et al. Mass eradication of Helicobacter pylori to reduce gastric cancer incidence and mortality: a long-term cohort study on Matsu Islands. Gut. pp.gutjnl-2020-322200. 2020. doi:10.1136/gutjnl-2020-322200
- Chen Y, Blaser MJ. Helicobacter pylori colonization is inversely associated with childhood asthma. J Infect Dis. 2008;198(4):553–560. doi:10.1086/590158.
- Amedei A, Codolo G, Del Prete G, De Bernard M, D’Elios MM. The effect of Helicobacter pylori on asthma and allergy. J Asthma Allergy. 2010;3:139–147. doi:10.2147/JAA.S8971.
- Liou JM, Lee YC, El-Omar EM, Wu MS. Efficacy and long-term safety of H. Pylori Eradication for Gastric Cancer Prevention Cancers (Basel). 2019;11(5):593.
- Liou JM, Lee YC, Wu MS. Treatment of Helicobacter pylori infection and its long-term impacts on gut microbiota. J Gastroenterol Hepatol. 2020;35(7):1107–1116. doi:10.1111/jgh.14992.
- Gagliardi D, Makihara S, Corsi PR, Viana Ade T, Wiczer MV, Nakakubo S, Mimica LMJ. Microbial flora of the normal esophagus. Dis Esophagus. 1998;11(4):248–250. doi:10.1093/dote/11.4.248.
- Pajecki D, Zilberstein B, Dos Santos MA, Ja U, Ag Q, Cecconello I. Megaesophagus microbiota: a qualitative and quantitative analysis. J Gastrointest Surg. 2002;6(5):723–729. doi:10.1016/S1091-255X(02)00028-8.
- Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci U S A. 2004;101(12):4250–4255. doi:10.1073/pnas.0306398101.
- Park CH, Lee SK. Exploring Esophageal Microbiomes in Esophageal diseases: a systematic review. J Neurogastroenterol Motil. 2020;26(2):171–179. doi:10.5056/jnm19240.
- Nardone G, Compare D, Rocco A. A microbiota-centric view of diseases of the upper gastrointestinal tract. Lancet Gastroenterol Hepatol. 2017;2(4):298–312. doi:10.1016/S2468-1253(16)30108-X.
- Deshpande NP, Riordan SM, Castano-Rodriguez N, Wilkins MR, Kaakoush NO. Signatures within the esophageal microbiome are associated with host genetics, age, and disease. Microbiome. 2018;6(1):227. doi:10.1186/s40168-018-0611-4.
- Yang L, Lu X, Nossa CW, Francois F, Peek RM, Pei Z. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137(2):588–597. doi:10.1053/j.gastro.2009.04.046.
- Liu N, Ando T, Ishiguro K, Maeda O, Watanabe O, Funasaka K, Nakamura M, Miyahara R, Ohmiya N, Goto H, et al. Characterization of bacterial biota in the distal esophagus of Japanese patients with reflux esophagitis and Barrett’s esophagus. BMC Infect Dis. 2013;13(1):130. doi:10.1186/1471-2334-13-130.
- Gall A, Fero J, McCoy C, Claywell BC, Sanchez CA, Blount PL, Li X, Vaughan TL, Matsen FA, Reid BJ, et al. Bacterial composition of the human upper gastrointestinal tract microbiome is dynamic and associated with genomic instability in a Barrett’s Esophagus Cohort. PLoS One. 2015;10(6):e0129055. doi:10.1371/journal.pone.0129055.
- Elliott DRF, Walker AW, O’Donovan M, Parkhill J, Fitzgerald RC. A non-endoscopic device to sample the oesophageal microbiota: a case-control study. Lancet Gastroenterol Hepatol. 2017;2(1):32–42. doi:10.1016/S2468-1253(16)30086-3.
- Snider EJ, Compres G, Freedberg DE, Khiabanian H, Nobel YR, Stump S, Uhlemann A-C, Lightdale CJ, Abrams JA. Alterations to the Esophageal Microbiome associated with progression from Barrett’s Esophagus to Esophageal Adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2019;28(10):1687–1693. doi:10.1158/1055-9965.EPI-19-0008.
- Anderson LA, Murphy SJ, Johnston BT, Watson RG, Ferguson HR, Bamford KB, Ghazy A, McCarron P, McGuigan J, Reynolds JV, et al. Relationship between Helicobacter pylori infection and gastric atrophy and the stages of the oesophageal inflammation, metaplasia, adenocarcinoma sequence: results from the FINBAR case-control study. Gut. 2008;57(6):734–739. doi:10.1136/gut.2007.132662.
- Peek RM Jr., Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2(1):28–37. doi:10.1038/nrc703.
- Corley DA, Kubo A, Levin TR, Block G, Habel L, Zhao W, Leighton P, Rumore G, Quesenberry C, Buffler P, et al. Helicobacter pylori infection and the risk of Barrett’s oesophagus: a community-based study. Gut. 2008;57(6):727–733. doi:10.1136/gut.2007.132068.
- Wang Z, Shaheen NJ, Whiteman DC, Anderson LA, Vaughan TL, Corley DA, El-Serag HB, Rubenstein JH, Thrift AP. Helicobacter pylori infection is associated with reduced risk of Barrett’s Esophagus: an analysis of the Barrett’s and Esophageal Adenocarcinoma consortium. Am J Gastroenterol. 2018;113(8):1148–1155. doi:10.1038/s41395-018-0070-3.
- Amir I, Konikoff FM, Oppenheim M, Gophna U, Half EE. Gastric microbiota is altered in oesophagitis and Barrett’s oesophagus and further modified by proton pump inhibitors. Environ Microbiol. 2014;16(9):2905–2914. doi:10.1111/1462-2920.12285.
- Yang I, Nell S, Suerbaum S. Survival in hostile territory: the microbiota of the stomach. FEMS Microbiol Rev. 2013;37(5):736–761. doi:10.1111/1574-6976.12027.
- Stockbruegger RW. Bacterial overgrowth as a consequence of reduced gastric acidity. Scand J Gastroenterol Suppl. 1985;111(sup111):7–16. doi:10.3109/00365528509093749.
- Thorens J, Froehlich F, Schwizer W, Saraga E, Bille J, Gyr K, Duroux P, Nicolet M, Pignatelli B, Blum AL, et al. Bacterial overgrowth during treatment with omeprazole compared with cimetidine: a prospective randomised double blind study. Gut. 1996;39(1):54–59. doi:10.1136/gut.39.1.54.
- Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi:10.1038/nature11234.
- Monstein HJ, Tiveljung A, Kraft CH, Borch K, Jonasson J. Profiling of bacterial flora in gastric biopsies from patients with Helicobacter pylori-associated gastritis and histologically normal control individuals by temperature gradient gel electrophoresis and 16S rDNA sequence analysis. J Med Microbiol. 2000;49(9):817–822. doi:10.1099/0022-1317-49-9-817.
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103(3):732–737. doi:10.1073/pnas.0506655103.
- Li XX, Wong GL, To KF, Wong VW, Lai LH, Chow DK, Lau JYW, Sung JJY, Ding C. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One. 2009;4(11):e7985. doi:10.1371/journal.pone.0007985.
- Rajilic-Stojanovic M, Figueiredo C, Smet A, Hansen R, Kupcinskas J, Rokkas T, Andersen L, Machado JC, Ianiro G, Gasbarrini A, et al. Systematic review: gastric microbiota in health and disease. Aliment Pharmacol Ther. 2020;51(6):582–602. doi:10.1111/apt.15650.
- Bashir M, Prietl B, Tauschmann M, Mautner SI, Kump PK, Treiber G, Wurm P, Gorkiewicz G, Högenauer C, Pieber TR, et al. Effects of high doses of vitamin D3 on mucosa-associated gut microbiome vary between regions of the human gastrointestinal tract. Eur J Nutr. 2016;55(4):1479–1489. doi:10.1007/s00394-015-0966-2.
- Stearns JC, Lynch MD, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD. Bacterial biogeography of the human digestive tract. Sci Rep. 2011;1(1):170. doi:10.1038/srep00170.
- Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, Beck JM, Curtis JL, Huffnagle GB. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio. 2015;6(2):e00037. doi:10.1128/mBio.00037-15.
- Al-Momani H, Perry A, Stewart CJ, Jones R, Krishnan A, Robertson AG, Bourke S, Doe S, Cummings SP, Anderson A, et al. Microbiological profiles of sputum and gastric juice aspirates in Cystic Fibrosis patients. Sci Rep. 2016;6(1):26985. doi:10.1038/srep26985.
- Edgar RC. Accuracy of microbial community diversity estimated by closed- and open-reference OTUs. PeerJ. 2017;5:e3889. doi:10.7717/peerj.3889.
- Scott DR, Marcus EA, Weeks DL, Sachs G. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology. 2002;123(1):187–195. doi:10.1053/gast.2002.34218.
- Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L, Ahmed N. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3(7):e2836. doi:10.1371/journal.pone.0002836.
- Li TH, Qin Y, Sham PC, Lau KS, Chu KM, Leung WK. Alterations in gastric microbiota after H. Pylori Eradication and in Different Histological Stages of Gastric Carcinogenesis Sci Rep. 2017;7:44935.
- Klymiuk I, Bilgilier C, Stadlmann A, Thannesberger J, Kastner M-T, Hogenauer C, Püspök A, Biowski-Frotz S, Schrutka-Kölbl C, Thallinger GG, et al. The human gastric microbiome is predicated upon infection with Helicobacter pylori. Front Microbiol. 2017;8:2508. doi:10.3389/fmicb.2017.02508.
- Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, Figueiredo C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67(2):226–236. doi:10.1136/gutjnl-2017-314205.
- Vasapolli R, Schutte K, Schulz C, Vital M, Schomburg D, Pieper DH, Vilchez-Vargas R, Malfertheiner P. Analysis of transcriptionally active bacteria throughout the gastrointestinal tract of healthy individuals. Gastroenterology. 2019;157(4):1081–92 e3. doi:10.1053/j.gastro.2019.05.068.
- Llorca L, Perez-Perez G, Urruzuno P, Martinez MJ, Iizumi T, Gao Z, Sohn J, Chung J, Cox L, Simón-Soro A, et al. Characterization of the gastric microbiota in a pediatric population according to Helicobacter pylori status. Pediatr Infect Dis J. 2017;36(2):173–178. doi:10.1097/INF.0000000000001383.
- Miftahussurur M, Waskito LA, El-Serag HB, Ajami NJ, Nusi IA, Syam AF. Gastric microbiota and Helicobacter pylori in Indonesian population. In: Helicobacter. 2020. p.e12695.
- Miao R, Wan C, Wang Z. The relationship of gastric microbiota and Helicobacter pylori infection in pediatrics population. Helicobacter. 2020;25(1):e12676. doi:10.1111/hel.12676.
- Zhao Y, Gao X, Guo J, Yu D, Xiao Y, Wang H, Li Y. Helicobacter pylori infection alters gastric and tongue coating microbial communities. Helicobacter. 2019;24(2):e12567. doi:10.1111/hel.12567.
- Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, Brodie EL, Dominguez-Bello MG. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. Isme J. 2011;5(4):574–579. doi:10.1038/ismej.2010.149.
- Martin ME, Bhatnagar S, George MD, Paster BJ, Canfield DR, Eisen JA, Solnick JV. The impact of Helicobacter pylori infection on the gastric microbiota of the rhesus macaque. PLoS One. 2013;8(10):e76375. doi:10.1371/journal.pone.0076375.
- He C, Peng C, Wang H, Ouyang Y, Zhu Z, Shu X, Zhu Y, Lu N. The eradication of Helicobacter pylori restores rather than disturbs the gastrointestinal microbiota in asymptomatic young adults. Helicobacter. 2019;24:e12590. doi:10.1111/hel.12590.
- Guo Y, Zhang Y, Gerhard M, Gao JJ, Mejias-Luque R, Zhang L. Effect of Helicobacter pylori on gastrointestinal microbiota: a population-based study in Linqu, a high-risk area of gastric cancer. Gut. 2019;69(9):1598–1607.
- Sung JJY, Coker OO, Chu E, Szeto CH, Luk STY, Lau HCH, Yu J. Gastric microbes associated with gastric inflammation, atrophy and intestinal metaplasia 1 year after Helicobacter pylori eradication. Gut. 2020;69(9):1572–1581. doi:10.1136/gutjnl-2019-319826.
- IARC working group on the evaluation of carcinogenic risks to humans. some industrial chemicals. IARC Monogr Eval Carcinog Risks Hum. Lyon, 15-22 February 1994. 1994;60:1–560.
- Correa P. Helicobacter pylori and gastric carcinogenesis. Am J Surg Pathol. 1995;19:S37–43.
- Valle J, Kekki M, Sipponen P, Ihamaki T, Siurala M. Long-term course and consequences of Helicobacter pylori gastritis results of a 32-year follow-up study. Scand J Gastroenterol. 1996;31(6):546–550. doi:10.3109/00365529609009126.
- Sipponen P, Kekki M, Siurala M. Atrophic chronic gastritis and intestinal metaplasia in gastric carcinoma. Comparison with a Representative Population Sample Cancer. 1983;52:1062–1068.
- Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345(11):784–789. doi:10.1056/NEJMoa001999.
- Chen HN, Wang Z, Li X, Zhou ZG. Helicobacter pylori eradication cannot reduce the risk of gastric cancer in patients with intestinal metaplasia and dysplasia: evidence from a meta-analysis. Gastric Cancer. 2016;19(1):166–175. doi:10.1007/s10120-015-0462-7.
- Amieva M, Peek RM Jr. Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology. 2016;150(1):64–78. doi:10.1053/j.gastro.2015.09.004.
- Hewetson JT. Report LXXXVIII: the bacteriology of certain parts of the human alimentary canal and of the inflammatory processes arising therefrom. Br Med J. 1904;2(2291):1457–1460. doi:10.1136/bmj.2.2291.1457.
- Barber M, Franklin RH. Bacteriology of peptic ulcer and gastric carcinoma. Br Med J. 1946;1(4459):951–953. doi:10.1136/bmj.1.4459.951.
- Cregan J, Dunlop EE, Hayward NJ. The bacterial content of human small intestine in disease of the stomach. Br Med J. 1953;2(4848):1248–1251. doi:10.1136/bmj.2.4848.1248.
- Gatehouse D, Dimock F, Burdon DW, Alexander-Williams J, Keighley MR. Prediction of wound sepsis following gastric operations. Br J Surg. 1978;65(8):551–554. doi:10.1002/bjs.1800650808.
- Sjostedt S, Heimdahl A, Kager L, Nord CE. Microbial colonization of the oropharynx, esophagus and stomach in patients with gastric diseases. Eur J Clin Microbiol. 1985;4(1):49–51. doi:10.1007/BF02148660.
- Rosenow EC, Sanford AH. The bacteriology of ulcer of the stomach and duodenum in man. Journal of Infectious Diseases. 1915;17(1):219–U12. doi:10.1093/infdis/17.1.219.
- Kato S, Fujimura S, Kimura K, Nishio T, Hamada S, Minoura T, Oda M. Non-Helicobacter bacterial flora rarely develops in the gastric mucosal layer of children. Dig Dis Sci. 2006;51(4):641–646. doi:10.1007/s10620-006-3185-0.
- Engstrand L, Lindberg M. Helicobacter pylori and the gastric microbiota. Best Pract Res Clin Gastroenterol. 2013;27(1):39–45. doi:10.1016/j.bpg.2013.03.016.
- Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol. 2009;58(4):509–516. doi:10.1099/jmm.0.007302-0.
- Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep. 2015;4(1):4202. doi:10.1038/srep04202.
- Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, Song KS, Kim YS, Kim JF. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. 2014;19(6):407–416. doi:10.1111/hel.12145.
- Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, Dong Q. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol. 2016;28(3):261–266. doi:10.1097/MEG.0000000000000542.
- Jo HJ, Kim J, Kim N, Park JH, Nam RH, Seok YJ, Kim Y-R, Kim JS, Kim JM, Kim JM, et al. Analysis of gastric microbiota by pyrosequencing: minor role of bacteria other than Helicobacter pylori in the gastric carcinogenesis. Helicobacter. 2016;21(5):364–374. doi:10.1111/hel.12293.
- Yu G, Torres J, Hu N, Medrano-Guzman R, Herrera-Goepfert R, Humphrys MS, Wang L, Wang C, Ding T, Ravel J, et al. Molecular characterization of the human stomach microbiota in gastric cancer patients. Front Cell Infect Microbiol. 2017;7:302. doi:10.3389/fcimb.2017.00302.
- Castano-Rodriguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. 2017;7(1):15957. doi:10.1038/s41598-017-16289-2.
- Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, Deng Y-F, Hsu W-T, Wu C-S, Li C, et al. Increased abundance of Clostridium and Fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci Rep. 2018;8(1):158. doi:10.1038/s41598-017-18596-0.
- Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJY, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67(6):1024–1032. doi:10.1136/gutjnl-2017-314281.
- Hu YL, Pang W, Huang Y, Zhang Y, Zhang CJ. The gastric microbiome is perturbed in advanced gastric adenocarcinoma identified through shotgun metagenomics. Front Cell Infect Microbiol. 2018;8:433. doi:10.3389/fcimb.2018.00433.
- Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, Liu F, Yan C, Li L, Ling Z, et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine. 2019;40:336–348. doi:10.1016/j.ebiom.2018.12.034.
- Gunathilake MN, Lee J, Choi IJ, Kim YI, Ahn Y, Park C, Kim J. Association between the relative abundance of gastric microbiota and the risk of gastric cancer: a case-control study. Sci Rep. 2019;9(1):13589. doi:10.1038/s41598-019-50054-x.
- Park CH, Lee AR, Lee YR, Eun CS, Lee SK, Han DS. Evaluation of gastric microbiome and metagenomic function in patients with intestinal metaplasia using 16S rRNA gene sequencing. Helicobacter. 2019;24(1):e12547. doi:10.1111/hel.12547.
- Wu ZF, Zou K, Wu GN, Jin ZJ, Xiang CJ, Xu S, Wang Y-H, Wu X-Y, Chen C, Xu Z, et al. A comparison of tumor-associated and non-tumor-associated gastric microbiota in gastric cancer patients. Dig Dis Sci. 2020. doi:10.1007/s10620-020-06415-y.
- Gantuya B, El Serag HB, Matsumoto T, Ajami NJ, Uchida T, Oyuntsetseg K, Bolor D, Yamaoka Y. Gastric mucosal microbiota in a Mongolian population with gastric cancer and precursor conditions. Aliment Pharmacol Ther. 2020;51(8):770–780. doi:10.1111/apt.15675.
- Stewart OA, Wu F, Chen Y. The role of gastric microbiota in gastric cancer. Gut Microbes. 2020;11(5):1220–1230. doi:10.1080/19490976.2020.1762520.
- Rodriguez RM, Hernandez BY, Menor M, Deng Y, Khadka VS. The landscape of bacterial presence in tumor and adjacent normal tissue across 9 major cancer types using TCGA exome sequencing. Comput Struct Biotechnol J. 2020;18:631–641. doi:10.1016/j.csbj.2020.03.003.
- Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, Mobley M, Potter A, Varro A, Eibach D, Suerbaum S, et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140(1):210–220. doi:10.1053/j.gastro.2010.09.048.
- Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, Ge Z, Wang TC, Fox JG. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. 2014;63(1):54–63. doi:10.1136/gutjnl-2013-305178.
- Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14(8):e1002533. doi:10.1371/journal.pbio.1002533.
- Kienesberger S, Cox LM, Livanos A, Zhang XS, Chung J, Perez-Perez GI, Gorkiewicz G, Zechner E, Blaser M. Gastric Helicobacter pylori infection affects local and distant microbial populations and host responses. Cell Rep. 2016;14(6):1395–1407. doi:10.1016/j.celrep.2016.01.017.
- Oertli M, Sundquist M, Hitzler I, Engler DB, Arnold IC, Reuter S, Maxeiner J, Hansson M, Taube C, Quiding-Järbrink M, et al. DC-derived IL-18 drives Treg differentiation, murine Helicobacter pylori-specific immune tolerance, and asthma protection. J Clin Invest. 2012;122(3):1082–1096. doi:10.1172/JCI61029.
- Heimesaat MM, Fischer A, Plickert R, Wiedemann T, Loddenkemper C, Gobel UB, Bereswill S, Rieder G. Helicobacter pylori induced gastric immunopathology is associated with distinct microbiota changes in the large intestines of long-term infected Mongolian gerbils. PLoS One. 2014;9(6):e100362. doi:10.1371/journal.pone.0100362.
- Baatar D, Patel K, Taub DD. The effects of ghrelin on inflammation and the immune system. Mol Cell Endocrinol. 2011;340(1):44–58. doi:10.1016/j.mce.2011.04.019.
- La Cava A, Matarese G. The weight of leptin in immunity. Nat Rev Immunol. 2004;4(5):371–379. doi:10.1038/nri1350.
- Muhsen K, Goren S, Cohen D. Helicobacter pylori infection in early childhood and growth at school age. Helicobacter. 2015;20(6):410–417. doi:10.1111/hel.12227.
- Buhling A, Radun D, Muller WA, Malfertheiner P. Influence of anti-Helicobacter triple-therapy with metronidazole, omeprazole and clarithromycin on intestinal microflora. Aliment Pharmacol Ther. 2001;15(9):1445–1452. doi:10.1046/j.1365-2036.2001.01033.x.
- Myllyluoma E, Ahlroos T, Veijola L, Rautelin H, Tynkkynen S, Korpela R. Effects of anti-Helicobacter pylori treatment and probiotic supplementation on intestinal microbiota. Int J Antimicrob Agents. 2007;29(1):66–72. doi:10.1016/j.ijantimicag.2006.08.034.
- Yang YJ, Sheu BS. Probiotics-containing yogurts suppress Helicobacter pylori load and modify immune response and intestinal microbiota in the Helicobacter pylori-infected children. Helicobacter. 2012;17(4):297–304. doi:10.1111/j.1523-5378.2012.00941.x.
- Chen L, Xu W, Lee A, He J, Huang B, Zheng W, Su T, Lai S, Long Y, Chu H, et al. The impact of Helicobacter pylori infection, eradication therapy and probiotic supplementation on gut microenvironment homeostasis: an open-label, randomized clinical trial. EBioMedicine. 2018;35:87–96. doi:10.1016/j.ebiom.2018.08.028.
- Iino C, Shimoyama T, Chinda D, Arai T, Chiba D, Nakaji S, Fukuda S. Infection of Helicobacter pylori and Atrophic Gastritis Influence Lactobacillus in gut microbiota in a Japanese population. Front Immunol. 2018;9:712. doi:10.3389/fimmu.2018.00712.
- Benavides-Ward A, Vasquez-Achaya F, Silva-Caso W, Aguilar-Luis MA, Mazulis F, Urteaga N, Del Valle-mendoza J. Helicobacter pylori and its relationship with variations of gut microbiota in asymptomatic children between 6 and 12 years. BMC Res Notes. 2018;11(1):468. doi:10.1186/s13104-018-3565-5.
- Gao JJ, Zhang Y, Gerhard M, Mejias-Luque R, Zhang L, Vieth M, Ma J-L, Bajbouj M, Suchanek S, Liu W-D, et al. Association between gut microbiota and Helicobacter pylori-related gastric lesions in a high-risk population of gastric cancer. Front Cell Infect Microbiol. 2018;8:202. doi:10.3389/fcimb.2018.00202.
- Osaki T, Zaman C, Yonezawa H, Lin Y, Okuda M, Nozaki E, Hojo F, Kurata S, Hanawa T, Kikuchi S, et al. Influence of intestinal indigenous microbiota on intrafamilial infection by Helicobacter pylori in Japan. Front Immunol. 2018;9:287. doi:10.3389/fimmu.2018.00287.
- Iino C, Shimoyama T, Chinda D, Sakuraba H, Fukuda S, Nakaji S. Influence of Helicobacter pylori infection and atrophic gastritis on the gut microbiota in a Japanese population. Digestion. 2019;101(4):422–432.
- Wang D, Li Y, Zhong H, Ding Q, Lin Y, Tang S, Zong Y, Wang Q, Zhang X, Yang H, et al. Alterations in the human gut microbiome associated with Helicobacter pyloriinfection. FEBS Open Bio. 2019;9(9):1552–1560. doi:10.1002/2211-5463.12694.
- Dash NR, Khoder G, Nada AM, Al Bataineh MT, Yamaoka Y. Exploring the impact of Helicobacter pylori on gut microbiome composition. PLoS One. 2019;14(6):e0218274. doi:10.1371/journal.pone.0218274.
- Yang L, Zhang J, Xu J, Wei X, Yang J, Liu Y. Helicobacter pylori infection aggravates dysbiosis of gut microbiome in children with gastritis. Front Cell Infect Microbiol. 2019;9:375. doi:10.3389/fcimb.2019.00375.
- Frost F, Kacprowski T, Ruhlemann M, Bang C, Franke A, Zimmermann K, Nauck M, Völker U, Völzke H, Biffar R, et al. Helicobacter pylori infection associates with fecal microbiota composition and diversity. Sci Rep. 2019;9(1):20100. doi:10.1038/s41598-019-56631-4.
- Cornejo-Pareja I, Martin-Nunez GM, Roca-Rodriguez MM, Cardona F, Coin-Araguez L, Sanchez-Alcoholado L, Gutiérrez-Repiso C, Muñoz-Garach A, Fernández-García J, Moreno-Indias I, et al. H. pylori eradication treatment alters gut microbiota and GLP-1 secretion in humans. J Clin Med. 2019;8(4):451. doi:10.3390/jcm8040451.
- Zhou Y, Ye Z, Lu J, Miao S, Lu X, Sun H. Long-term changes in the gut microbiota after 14-day bismuth quadruple therapy in penicillin-allergic children. Helicobacter. 2020;25(5):e12721. doi: 10.1111/hel.12721. Epub 2020 Jul 12.
- Seley GP, Colp R. The bacteriology of peptic ulcers and gastric malignancies - possible bearing on complications following gastric surgery. Surgery. 1941;10:369–380.
- Sjostedt S, Kager L, Heimdahl A, Nord CE. Microbial colonization of tumors in relation to the upper gastrointestinal tract in patients with gastric carcinoma. Ann Surg. 1988;207(3):341–346. doi:10.1097/00000658-198803000-00020.
- Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016;65(11):1906–1915. doi:10.1136/gutjnl-2016-312297.
- Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L, Ratner AJ. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5(3):e9836. doi:10.1371/journal.pone.0009836.
- Yap TW, Gan HM, Lee YP, Leow AH, Azmi AN, Francois F, Perez-Perez GI, Loke M-F, Goh K-L, Vadivelu J, et al. Helicobacter pylori eradication causes perturbation of the human gut microbiome in young adults. PLoS One. 2016;11(3):e0151893. doi:10.1371/journal.pone.0151893.
- Oh B, Kim BS, Kim JW, Kim JS, Koh SJ, Kim BG, Lee KL, Chun J. The effect of probiotics on gut microbiota during the Helicobacter pylori eradication: randomized controlled trial. Helicobacter. 2016;21(3):165–174. doi:10.1111/hel.12270.
- Yanagi H, Tsuda A, Matsushima M, Takahashi S, Ozawa G, Koga Y, Takagi A. Changes in the gut microbiota composition and the plasma ghrelin level in patients with Helicobacter pylori-infected patients with eradication therapy. BMJ Open Gastroenterol. 2017;4(1):e000182. doi:10.1136/bmjgast-2017-000182.
- Hsu PI, Pan CY, Kao JY, Tsay FW, Peng NJ, Kao SS, Wang H-M, Tsai T-J, Wu D-C, Chen C-L, et al. Helicobacter pylori eradication with bismuth quadruple therapy leads to dysbiosis of gut microbiota with an increased relative abundance of Proteobacteria and decreased relative abundances of Bacteroidetes and Actinobacteria. Helicobacter. 2018;23(4):e12498. doi:10.1111/hel.12498.
- Gotoda T, Takano C, Kusano C, Suzuki S, Ikehara H, Hayakawa S, Andoh A. Gut microbiome can be restored without adverse events after Helicobacter pylori eradication therapy in teenagers. Helicobacter. 2018;23(6):e12541. doi:10.1111/hel.12541.
- Liou JM, Chen CC, Chang CM, Fang YJ, Bair MJ, Chen PY, Chang C-Y, Hsu Y-C, Chen M-J, Chen -C-C, et al. Long-term changes of gut microbiota, antibiotic resistance, and metabolic parameters after Helicobacter pylori eradication: a multicentre, open-label, randomised trial. Lancet Infect Dis. 2019;19(10):1109–1120. doi:10.1016/S1473-3099(19)30272-5.
- Martin-Nunez GM, Cornejo-Pareja I, Coin-Araguez L, Mdm R-R, Munoz-Garach A, Clemente-Postigo M, Cardona F, Moreno-Indias I, Tinahones FJ. H. pylori eradication with antibiotic treatment causes changes in glucose homeostasis related to modifications in the gut microbiota. PLoS One. 2019;14(3):e0213548. doi:10.1371/journal.pone.0213548.
- Hsu PI, Pan CY, Kao JY, Tsay FW, Peng NJ, Kao SS, Chen Y-H, Tsai T-J, Wu D-C, Tsai K-W, et al. Short-term and long-term impacts of Helicobacter pylori eradication with reverse hybrid therapy on the gut microbiota. J Gastroenterol Hepatol. 2019;34(11):1968–1976. doi:10.1111/jgh.14736.
- Ye Q, Shao X, Shen R, Chen D, Shen J. Changes in the human gut microbiota composition caused by Helicobacter pylori eradication therapy: a systematic review and meta-analysis. Helicobacter. 2020;25(4):e12713. doi:10.1111/hel.12713.
- Franceschi F, Zuccala G, Roccarina D, Gasbarrini A. Clinical effects of Helicobacter pylori outside the stomach. Nat Rev Gastroenterol Hepatol. 2014;11(4):234–242. doi:10.1038/nrgastro.2013.243.
- Satoh H, Saijo Y, Yoshioka E, Tsutsui H. Helicobacter Pylori infection is a significant risk for modified lipid profile in Japanese male subjects. J Atheroscler Thromb. 2010;17(10):1041–1048. doi:10.5551/jat.5157.
- Upala S, Sanguankeo A, Saleem SA, Jaruvongvanich V. Effects of Helicobacter pylori eradication on insulin resistance and metabolic parameters: a systematic review and meta-analysis. Eur J Gastroenterol Hepatol. 2017;29(2):153–159. doi:10.1097/MEG.0000000000000774.
- Osawa H, Nakazato M, Date Y, Kita H, Ohnishi H, Ueno H, Shiiya T, Satoh K, Ishino Y, Sugano K, et al. Impaired production of gastric ghrelin in chronic gastritis associated with Helicobacter pylori. J Clin Endocrinol Metab. 2005;90(1):10–16. doi:10.1210/jc.2004-1330.
- tAbdel-Razik A, Mousa N, Shabana W, Refaey M, Elhelaly R, Elzehery R, Abdelsalam M, Elgamal A, Nassar MR, Abu El‐Soud A, et al. Helicobacter pylori and non-alcoholic fatty liver disease: a new enigma? Helicobacter. 2018;23(6):e12537. doi:10.1111/hel.12537.
- He C, Yang Z, Cheng D, Xie C, Zhu Y, Ge Z, Luo Z, Lu N. Helicobacter pylori infection aggravates diet-induced insulin resistance in association with gut microbiota of Mice. EBioMedicine. 2016;12:247–254. doi:10.1016/j.ebiom.2016.09.010.
- Chen Y, Blaser MJ, Blaser MJ. Inverse associations of Helicobacter pylori with asthma and allergy. Arch Intern Med. 2007;167(8):821–827. doi:10.1001/archinte.167.8.821.
- El-omar E, Penman I, Cruikshank G, Dover S, Banerjee S, Williams C, McColl KE. Low prevalence of Helicobacter pylori in inflammatory bowel disease: association with sulphasalazine. Gut. 1994;35(10):1385–1388. doi:10.1136/gut.35.10.1385.
- Castano-Rodriguez N, Kaakoush NO, Lee WS, Mitchell HM. Dual role of Helicobacter and Campylobacter species in IBD: a systematic review and meta-analysis. Gut. 2017;66(2):235–249. doi:10.1136/gutjnl-2015-310545.
- Dogra S, Sakwinska O, Soh SE, Ngom-Bru C, Bruck WM, Berger B, Brüssow H, Lee YS, Yap F, Chong Y-S, et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio. 2015;6(1). doi:10.1128/mBio.02419-14.
- Malaty HM, El-Kasabany A, Graham DY, Miller CC, Reddy SG, Srinivasan SR, Yamaoka Y, Berenson GS. Age at acquisition of Helicobacter pylori infection: a follow-up study from infancy to adulthood. Lancet. 2002;359(9310):931–935. doi:10.1016/S0140-6736(02)08025-X.
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi:10.1038/nature11053.
- Kao JY, Zhang M, Miller MJ, Mills JC, Wang B, Liu M, Eaton KA, Zou W, Berndt BE, Cole TS, et al. Helicobacter pylori immune escape is mediated by dendritic cell-induced Treg skewing and Th17 suppression in mice. Gastroenterology. 2010;138(3):1046–1054. doi:10.1053/j.gastro.2009.11.043.
- Yu Y, Zhu S, Li P, Min L, Zhang S. Helicobacter pylori infection and inflammatory bowel disease: a crosstalk between upper and lower digestive tract. Cell Death Dis. 2018;9(10):961. doi:10.1038/s41419-018-0982-2.
- Arnold IC, Dehzad N, Reuter S, Martin H, Becher B, Taube C, Müller A. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J Clin Invest. 2011;121(8):3088–3093. doi:10.1172/JCI45041.
- Izcue A, Coombes JL, Powrie F. Regulatory T cells suppress systemic and mucosal immune activation to control intestinal inflammation. Immunol Rev. 2006;212(1):256–271. doi:10.1111/j.0105-2896.2006.00423.x.
