ABSTRACT
Human milk glycans present a unique diversity of structures that suggest different mechanisms by which they may affect the infant microbiome development. A humanized mouse model generated by infant fecal transplantation was utilized here to evaluate the impact of fucosyl-α1,3-GlcNAc (3FN), fucosyl-α1,6-GlcNAc, lacto-N-biose (LNB) and galacto-N-biose on the fecal microbiota and host–microbiota interactions. 16S rRNA amplicon sequencing showed that certain bacterial genera significantly increased (Ruminococcus and Oscillospira) or decreased (Eubacterium and Clostridium) in all disaccharide-supplemented groups. Interestingly, cluster analysis differentiates the consumption of fucosyl-oligosaccharides from galactosyl-oligosaccharides, highlighting the disappearance of Akkermansia genus in both fucosyl-oligosaccharides. An increment of the relative abundance of Coprococcus genus was only observed with 3FN. As well, LNB significantly increased the relative abundance of Bifidobacterium, whereas the absolute levels of this genus, as measured by quantitative real-time PCR, did not significantly increase. OTUs corresponding to the species Bifidobacterium longum, Bifidobacterium adolescentis and Ruminococcus gnavus were not present in the control after the 3-week intervention, but were shared among the donor and specific disaccharide groups, indicating that their survival is dependent on disaccharide supplementation. The 3FN-feeding group showed increased levels of butyrate and acetate in the colon, and decreased levels of serum HDL-cholesterol. 3FN also down-regulated the pro-inflammatory cytokine TNF-α and up-regulated the anti-inflammatory cytokines IL-10 and IL-13, and the Toll-like receptor 2 in the large intestine tissue. The present study revealed that the four disaccharides show efficacy in producing beneficial compositional shifts of the gut microbiota and in addition, the 3FN demonstrated physiological and immunomodulatory roles.
Introduction
Microbial colonization of the infant gastrointestinal tract plays a fundamental role in maintaining a healthy status.Citation1,Citation2 Disruption or inappropriate development of the neonatal gut microbial community is linked to diseases such as necrotizing enterocolitis, diarrhea and allergies.Citation3–5 The postnatal microbiota structure of the infant gut is closely related to the mother’s breast milk microbiota and the unique human milk glycan composition.Citation6–8 This includes free oligosaccharides (HMOs), mucins, glycoproteins and glycolipids.Citation9,Citation10 HMOs constitute the third largest solid component of human milk and over 200 different structures have already been identified.Citation9 Fecal microbiota of breastfed infants is generally dominated by the phylum Actinobacteria, with Bifidobacterium as the main genus, and Firmicutes, with diverse representation from numerous genera.Citation11 Several studies have demonstrated that the fermentation of HMOs and the glycan moiety of mucins stimulate the growth of specific strains belonging to the genus Bifidobacterium and to a lesser extent to Lactobacillus.Citation10 HMOs are also associated with various beneficial effects, including immunomodulation, protection against infectious diseases, stimulation of intestine barrier functions and brain development.Citation12–14 Compared to human milk, infant formula contains low amounts of oligosaccharides with limited structural diversity and fucosylation.Citation15 Indeed, the gut microbiome composition and health outcomes of formula-fed infants differ markedly from that of exclusively breastfed infants.Citation16 Therefore, it will be important to identify glycan structures that are crucial for developing a beneficial gut microbiota composition, and that can be easily produced for using individually or combined in a glycan mix.
A few studies using in vivo animal models highlight the ability of individual HMOs to modify the composition of the gastrointestinal microbiota. The levels of Barnesiella in the gut increased in a baby mouse model exposured to 2ʹ-fucosyllactose (2ʹFL) and 3-fucosyllactose (3FL), and these changes in the microbiota affected the susceptibility of mice to dextran sulfate sodium-induced colitis.Citation17 Supplementation of 2ʹFL also changed the composition of cecal microbiota, improved metabolic profiles and gut-brain signaling, and downregulated the expression of pro-inflamatory cytokines in a high-fat fed mouse model.Citation18 Mice fed with a diet containing 3ʹ-sialyllactose (3’SL) and 6ʹ-sialyllactose (6’SL) have lesser colonic microbiota alterations and better anxiety-like behavior than control mice fed with a standard diet.Citation19 Humanized microbiome animal models are emerging as powerful tools for analyzing human microbiota in a controlled mode. The microbial populations of fecal and jejunal content of germ-free mice inoculated with a model of human baby microbiota, which comprises seven bacterial strains, were modulated with galactosyl-oligosaccharides co-administrated with the probiotics Lactobacillus paracasei or Lactobacillus rhamnosus.Citation20 An increase in the number of specific Bifidobacterium species and a reduction of Clostridium perfringens was observed. In addition, host metabolic pathways as lipid profiles and gluconeogenesis among others were modulated.Citation20 An important reduction of clostridia numbers was observed recently in a germ-free mouse model with fecal microbiota from infants born by cesarean section and in the presence of a combination of Bifidobacterium infantis and human milk or HMOs.Citation21
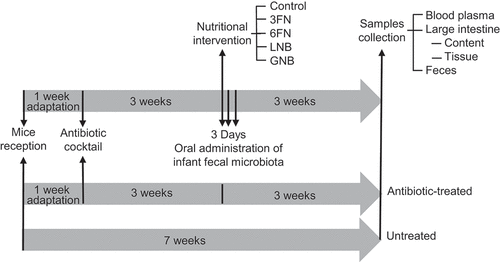
The disaccharides fucosyl-α1,3-GlcNAc (3FN) and fucosyl-α1,6-GlcNAc (6FN) that form part of HMOs and core-fucosylated N-glycans, respectively, have been synthetized in our laboratory.Citation22,Citation23 In vitro fermentation analysis, using pure culturesCitation23 and batch cultures with infant fecal microbiota,Citation24 has demonstrated that 3FN stimulates the growth of the species Lactobacillus casei and Bifidobacterium breve, respectively. We have also produced lacto-N-biose (LNB; Gal- β1,3-GlcNAc), the main building block of type-1 HMOs, and galacto-N-biose (GNB; Gal- β1,3-GalNAc), the core type-1 sugar from mucins.Citation25 LNB is also present as free sugar in human milk.Citation26 Both disaccharides have been shown to efficiently promote in vitro the growth of specific Bifidobacterium species.Citation24,Citation27 In this work, a humanized mouse model, which was generated by infant fecal microbiota transplantation, was utilized to evaluate the effects of 3FN, 6FN, LNB and GNB on the development of the gut microbiota composition (). Short-chain fatty acids production and interactions host–microbiota, through the assessment of serum lipid profile and cytokines expression, were also measured.
Results
Modification of the fecal murine microbiota by infant fecal microbiota transplantation
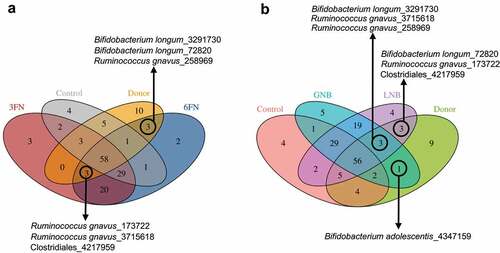
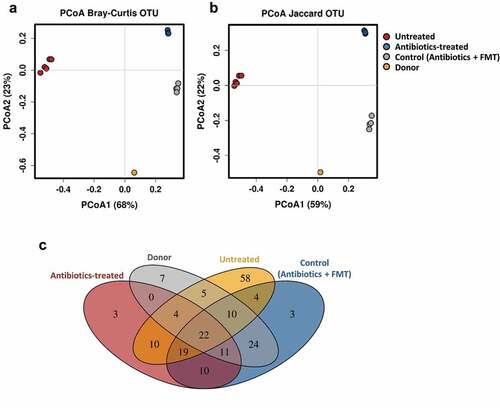
We first assessed if the antibiotic treatment followed by infant fecal microbiota transplantation shifts the murine microbiota composition toward the donor microbiota profile. A PCoA plot of the 16S rRNA fecal microbial composition data using Bray–Curtis and Jaccard distance metrics showed that the mice groups, untreated, treated with antibiotics and treated with antibiotics followed by oral administration of infant fecal microbiota (control), and the pooled donor feces clustered separately (). Both mice groups, treated with antibiotics and control, were separated from the untreated mice group along the PCoA1 axis and from each other along the PCoA2 axis. The significant separation between the antibiotics-treated and control mice groups was confirmed by statistical analysis (ANOSIM R = 1, p = 0.025; Adonis R2 = 0.919, p = 0.029). The control mice group has a lower distance to the donor microbiota than the mice group only treated with antibiotics (, b). A Venn diagram showed that 24 OTUs were absent from untreated and antibiotics-treated mice groups and shared by the pooled infant donor and the control humanized mice group ( and Supplemental Table 1). They belonged to the genera Bacteroides, Parabacteroides, Phascolarctobacterium, Klebsiella and Eggerthella, and the families Ruminococcaceae and Enterobacteriaceae. In addition, ten OTUs from the genera Bacteroides, Parabacteroides and Phascolarctobacterium were present in all the groups except in the antibiotic-treated group, indicating that their presence in the control group, which is also treated with antibiotics, is due to the transplanted infant microbiota and not due to the indigenous bacteria. All these results revealed that antibiotic treatment followed by oral inoculation led to successful engraftment of the infant microbiota. However, a limitation of this humanized animal model is that seven OTUs (belonging to the genera Bifidobacterium, Collinsella, Lactobacillus and Streptococcus, and to the order Clostridiales) specific to the donor were not transferred to the recipient mice. As well 19 OTUs (belonging to the genera Lactococcus and SMB53, the families Clostridiaceae, Peptostreptococcaceae and Rikenellaceae, and to the order Clostridiales) were shared by the untreated, antibiotic-treated group and the control humanized group, indicating that a proportion of the indigenous bacteria remain in the transplanted mice ( and Supplemental Table 1). It cannot be ruled out that some of the residual microbiota influence the effects of the disaccharides, described below, on the infant gut microbiota.
Figure 2. Fecal microbial diversity of mice in response to antibiotic-treatment and infant fecal transplantation. Principal coordinates analysis (PCoA) plot of fecal microbiota composition using Bray–Curtis (a) and Jaccard (b) distance metrics. (c) Venn diagram of shared OTUs between the infant donor fecal mix and the fecal microbiota of the untreated, antibiotic-treated and control mice groups. FMT, fecal microbiota transplantation
Human milk-associated disaccharides produce compositional changes in the gut microbiota
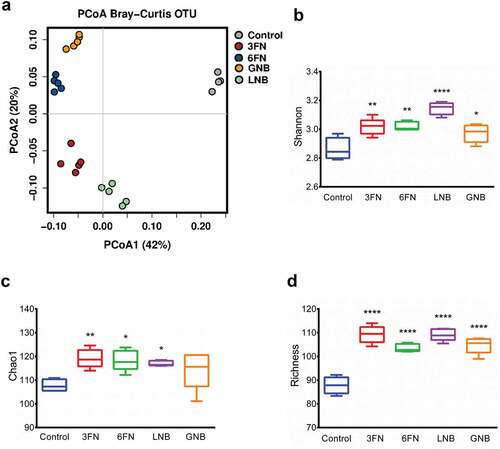
The effect of the consumption of the disaccharides 3FN, 6FN, LNB and GNB was tested in mice pre-treated with antibiotics followed by infant fecal transplantation. To understand the global changes of fecal microbial composition due to the consumption of these disaccharides, PCoA analysis was performed using Bray–Curtis distance metrics. The results showed that the control and each disaccharide feeding groups clustered separately (). The consumption of the disaccharides has a strong effect on the fecal microbiota as PCoA 1 and PCoA 2 axes explain 42% and 20% of the total variance, respectively, and it was confirmed by statistical analysis (ANOSIM R = 0.992, p = 0.001; Adonis R2 = 0.829, p = 0.0003). The microbiota α-diversity of all disaccharide-supplemented mice groups was significantly higher than that in control mice, with the only exception of the Chao1 index for the GNB group (, c, d). Therefore, these results indicate that each of the disaccharides evaluated here is able to modify the fecal bacterial community diversity and richness.
Figure 3. Fecal microbial diversity and richness of infant fecal transplanted mice in response to disaccharide-supplemented diets. Principal coordinates analysis (PCoA) plot of fecal microbiota composition using Bray–Curtis (a), Shannon index (b), Chao1 index (c) and absolute richness (d) at OTU level. Box plots present the median (interquartile range) and min/max. 3FN (fucosyl-α1,3-GlcNAc), 6FN (fucosyl-α1,6-GlcNAc), LNB (lacto-N-biose) and GNB (galacto-N-biose). Statistical significant differences compared to control are indicated: *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001
Analysis of the relative abundance of fecal microbiota at the family level identified 12 families that were differentially abundant between at least one of the four disaccharide-fed mice groups and the control group (Supplemental Figure 1). Notably, LNB supplementation significantly increased the abundance of Bifidobacteriaceae, Lachnospiraceae, Ruminococcaceae, S247 and Turicibacteraceae. The four disaccharides increased Clostridiaceae and unclassified Clostridiales, and significantly decreased Enterobacteriaceae and Erysipelotrichaceae with respect to the control without carbohydrate supplementation. Regarding the relative abundance of fecal microbiota at the genus level, the results showed differences among the five mice groups (). Interestingly, a clustered bar-chart analysis with the top 30 more abundant genera showed two main clusters that differentiate control group from disaccharide-treated groups, and within these two main clusters the analysis clearly differentiates the consumption of fucosyl-oligosaccharides from the consumption of galactosyl-oligosaccharides (). Relative abundance changes of 19 genera were significantly associated with at least one of the four disaccharide-supplementation mice groups with respect to the control group (). The relative abundance of Bibidobacterium and unclassified S247 was significantly increased by LNB and Lactobacillus abundance decreased with GNB. Coprococcus abundance increased by 3FN, and an unclassified genus of the Lachnospiraceae family was significantly increased by both, LNB and GNB. Mice fed with any of the fucosyl-disaccharides tested here showed a significantly decrease in abundance of Akkermansia while in GNB-fed mice its abundance increased. 3FN and LNB-supplemented mice groups showed an increase in Turicibacter abundance, and 6FN and GNB a decrease in unclassified Erysipelotrichae. The abundance of Megasphaera, Enterococcus, SMB53, unclassified Peptostreptococcaceae and unclassified Ruminococcaceae decreased in three out of the four disaccharide-fed mice groups. Certain bacterial genera were affected by all four treatments. Thus, the relative abundance of Ruminococcus, Oscillospira and an unclassified genus of the Clostridiaceae family was significantly increased with the four disaccharides tested (). Contrarily, the genus Eubacterium, an unclassified genus of the Enterobacteriaceae family and Clostridium decreased with the four disaccharides.
Figure 4. Fecal microbial composition of infant fecal transplanted mice in response to disaccharide-supplemented diets. (a) Fecal microbial relative abundances at genus level. Bars represent each diet group and values are mean relative abundance of each bacterial genus. (b) Clustered bar-chart analysis of fecal mice samples at genus level. Bars represent each mouse. Control group (n = 4); diet groups (n = 5). 3FN (fucosyl-α1,3-GlcNAc), 6FN (fucosyl-α1,6-GlcNAc), LNB (lacto-N-biose) and GNB (galacto-N-biose)
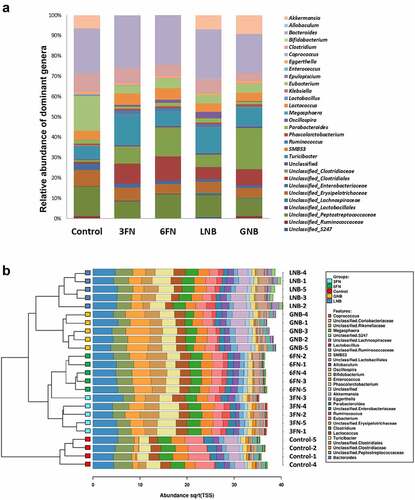
Figure 5. Abundances of the fecal bacterial genera that showed statistically significant differences for at least one of the disaccharide-supplemented mice groups compared to control group. 3FN (fucosyl-α1,3-GlcNAc), 6FN (fucosyl-α1,6-GlcNAc), LNB (lacto-N-biose) and GNB (galacto-N-biose). Box plots present the median (interquartile range) and min/max. n = 4 (control group); n = 5 (diet group). Statistical significant differences compared to control are indicated: #p < 0.1, *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001
In order to evaluate if the disaccharides promote selectively the growth of bacteria from the donor, we compared the fecal microbiota data in a Venn diagram ( and Supplemental Table 2). The results showed that six OTUs corresponding to the species Bifidobacterium longum and Ruminococcus gnavus, and to the order Clostridiales are shared among the donor and specific disaccharide groups, indicating that their survival possibly relies upon disaccharide supplementation. As well, one OTU belonging to Bifidobacterium adolescentis species is only shared by the donor and the GNB group, and it is not present in the control group (). This result suggests that the persistence of this species depends on the GNB supplementation.
Effects of human milk-associated disaccharides on short-chain fatty acid (SCFA) concentrations in the colon
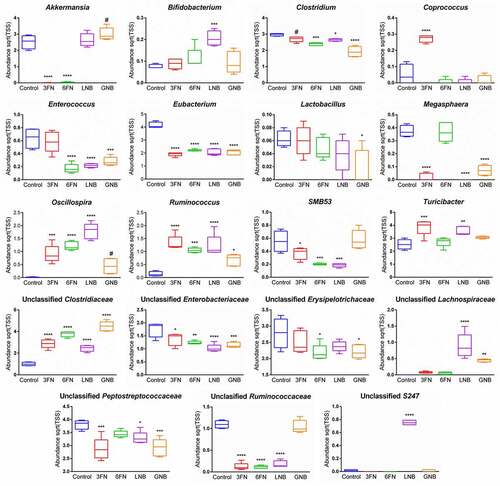
SCFAs are the major end products of the carbohydrate fermentation processes by the intestinal microbiota. Acetate, propionate and butyrate represent more than 90% of the total SCFA produced in the colon.Citation28 To determine whether supplementation of the specific human milk disaccharides tested here affected microbial metabolic activity, those colonic SCFAs were measured (). Formate, a main fermentation end-product,Citation29 was also analyzed. None of the assayed disaccharides affected significantly the production of formate and propionate by the microbiota. However, acetate levels increased significantly in the 3FN supplemented mice group compared to the control group. Interestingly, butyrate was not detected in the fecal samples of the control group; however, consumption of the disaccharides evaluated here resulted in butyrate production by the microbiota, being statistically significant for the 3FN feeding group ().
Figure 7. Effect of 3FN (fucosyl-α1,3-GlcNAc), 6FN (fucosyl-α1,6-GlcNAc), LNB (lacto-N-biose) and GNB (galacto-N-biose) on short-chain fatty acid concentrations in large intestine content of infant fecal transplanted mice. Box plots present the median (interquartile range) and min/max. n = 5 (control group); n = 5 (diet groups). Statistical significant differences compared to control are indicated: *p < 0.05
Impact of human milk-associated disaccharides on lipid metabolism
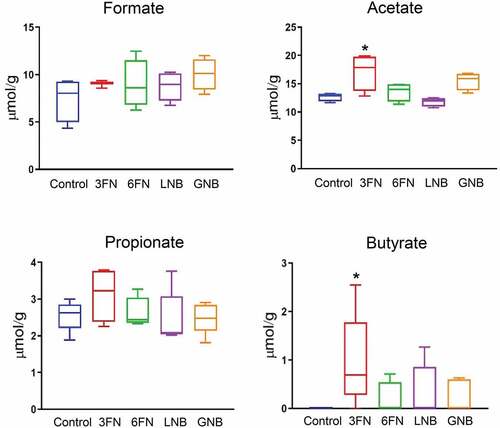
Changes in the gut microbial composition and derived metabolites have been shown to influence lipid metabolism.Citation30,Citation31 Therefore, we analyzed the effect of the oligosaccharides on body weight, and concentrations of serum triglycerides and cholesterol. At the end of the experimental procedure (week 7), the mice groups supplemented with 3FN and GNB had significantly higher weight gain as percentage of initial body weight in comparison with control mice group (Supplemental ). The 6FN-feeding group presented significantly higher levels of triglycerides than the control group, and mice consuming 3FN showed significant decreased levels of HDL-cholesterol (). None of the tested disaccharides resulted in a significant reduction of total cholesterol and LDL-cholesterol levels. The results obtained here may be relevant since breastfed infants have higher levels of triglycerides, total cholesterol and LDL-cholesterol than formula-fed infants.Citation32,Citation33 Unexpectedly, in spite of their high serum lipid concentration, long-term breastfed infants have lower cardiovascular risk in adulthood than their formula-fed counterparts.Citation34
Figure 8. Effect of 3FN (fucosyl-α1,3-GlcNAc), 6FN (fucosyl-α1,6-GlcNAc), LNB (lacto-N-biose) and GNB (galacto-N-biose) on serum lipid profile of infant fecal transplanted mice. Box plots present the median (interquartile range) and min/max. n = 5 (control group); n = 5 (diet groups). Statistical significant differences compared to control are indicated: #p < 0.1, *p < 0.05; **p < 0.01
Effects of human milk-associated disaccharides on immunological biomarkers expression at intestinal tissue level
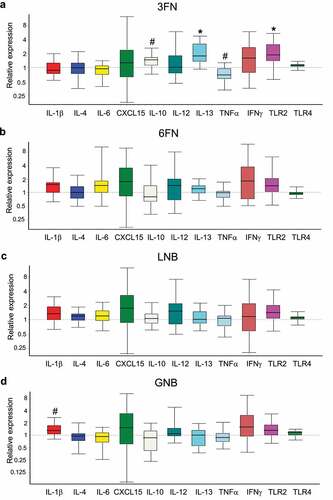
The expression of genes encoding pro-inflammatory (IL-1β, IL-6, Cxcl15 (IL-8), IL-12, TNF-α, IFN-γ) and anti-inflammatory (IL-4, IL-10 y IL-13) cytokines, and Toll-like receptors (TLR2 y TLR4) were evaluated in the large intestine tissue (). Supplementation with 6FN or LNB did not modify significantly the expression of any of those genes involved in the activity of the immune system. However, the supplementation of the disaccharide 3FN resulted in a significant increase in expression of IL-10 and IL-13 and a significant decrease of TNF-α with respect to the control mice group. Moreover, the expression of the gene encoding TLR2, which is involved in the immune response mediated by gut microbiota, is significantly increased in the 3FN mice group (). The other three feeding groups, 6FN, LNB and GNB, showed a trend (p = 0.182, 0.164 and 0.171, respectively) toward increased expression levels of that TLR gene (, c, d). The cytokine IL-1β was up-regulated in the GNB supplemented mice group compared to the control group ().
Figure 9. Effect of 3FN (fucosyl-α1,3-GlcNAc) (a), 6FN (fucosyl-α1,6-GlcNAc) (b), LNB (lacto-N-biose) (c) and GNB (galacto-N-biose) (d) on gene expression of cytokines and Toll-like receptors in the large intestine tissue of infant fecal transplanted mice. The values, expressed as fold-changes, represent relative expression in treated mice groups compared to control group. Box plots present the median (interquartile range) and min/max. n = 5 (control group); n = 5 (diet groups). Statistical significant differences compared to control are indicated: #p < 0.1, *p < 0.05
Discussion
The gut commensal microbiota and associated metabolic products from breastfed infants have long been considered as contributors to infant health.Citation16 However, substantial differences have been found between the gut microbiota composition of formula-fed and breastfed infants.Citation16,Citation35 Those differences are due in part to the low concentrations and different structures of oligosaccharides found in infant formulas compared to human milk.Citation15 Infant formulas are based in bovine milk, which does not contain type-1 oligosaccharides,Citation15 lacking therefore the predominant LNB structural unit of HMOs. Another difference with human milk is that oligosaccharides from bovine milk are highly sialylated and to a lesser extent fucosylated. While about 70% of HMOs are fucosylated, only 1% of the oligosaccharides from bovine milk are fucosylated.Citation36 Therefore, there is a need to search for functional carbohydrates that can fulfill the oligosaccharide scarcity from infant formulas. In this work, we established a humanized mouse model to evaluate the impact of the disaccharides LNB, GNB and two fucosyldisaccharides, 3FN and 6FN, on the gut microbiota composition and host–microbiota interactions. Germ-free mice colonized with infant fecal microbiota have been previously used to evaluate the effect of probiotics and synbiotic mixes.Citation20,Citation21,Citation37 Since germ-free mice have many biological and technological limitations,Citation38,Citation39 a strategy using conventional mice treated with antibioticsCitation40 followed by oral administration of infant fecal microbiota was successfully used here. There are some host factors that may impact the mice bacterial colonization from infant donors, indeed intestinal mucin glycans that are utilized by some bacterial species as growth substrates differ between humans and mice.Citation41 As well, the mucus thickness and many immunological functions are affected by the age of the mice.Citation42 In our study, we have used young mice, but whether differences in the mucus layer and the immune system due to mice age would affect the ability of infant gut bacteria to colonize mice merits further research. In the mice model used here, 3 weeks after the infant fecal transplantation about 41% OTUs of the infant donor pool remain in the transplanted mice (). Additionally, seven OTUs were just present in the donor sample and in at least one of the disaccharide-supplemented groups (). These results evidenced the suitability of the infant microbiota-associated mice as a model to study the role of human milk glycans on infant gut microbiota development.
Unlike formula-fed infants, the fecal microbiota of breastfed infants is dominated by bifidobacteria.Citation43 Using in vitro fermentation analysis, our group and others have demonstrated that LNB is metabolized by infant-gut associated bifidobacteria.Citation24,Citation27 Here, this disaccharide is tested for the first time using an in vivo model and it has been demonstrated that it significantly increased the relative abundance of the genus Bifidobacterium in feces. However, the absolute levels of this genus in the LNB mice group were similar to the control group (Supplemental ), suggesting that the high relative abundance of Bifidobacterium in LNB could be due to reductions in the relative abundances of other taxa. Regarding GNB, two OTUs belonging to B. longum and B. adolescentis species, respectively, that were present in the infant donor sample, persisted in the GNB supplemented group but not in the control group. These results suggest that this disaccharide might have a role in the survival of particular species or strains of bifidobacteria. B. longum species persisted also in the LNB and 6FN diet groups. All the B. longum strains tested in vitro fermented LNB and all of them contain the gene lpnA encoding the GNB/LNB phosphorylase specific not only to LNB if not also to GNB.Citation44 Indeed, a B. longum strain isolated from fermented cultures with infant gut microbiota was able to grow in the presence of GNB.Citation24 The positive effect of LNB, GNB or 6FN in the persistence of B. longum species in the gastrointestinal tract is a relevant outcome, since this bacterium has been widely associated with prevention and fighting of several intestinal and immune diseases.Citation45,Citation46 Regarding B. adolescentis species, it has been shown previously that this species is not able to metabolize 3FN and 6FN.Citation23 Unlike these, the utilization of LNB and GNB by B. adolescentis has not been tested. Then, we analyzed here if the type strain B. adolescentis ATCC 15703 utilizes those disaccharides as substrates and the results showed that this strain can be cultured in the presence of both LNB and GNB (data not shown). Survival of B. adolescentis in the GNB diet group might be important for serious infant gastrointestinal disorders such as necrotizing enterocolitis, for which a protection effect has been shown by that species.Citation47
R. gnavus is a human gut symbiont present at early and adult life stages,Citation48,Citation49 and various studies have pointed toward a key role of this species in modulating gut inflammatory responses.Citation50,Citation51 The four disaccharides tested here significantly increased the relative abundance of the Ruminococcus genus in feces and allowed the persistence of three OTUs corresponding to the species R. gnavus (). The genomes of strains belonging to this species harbor several genes encoding for glycosidases potentially implicated in the breakdown of mucin-derived glycans (https://www.ncbi.nlm.nih.gov/genome). Four α-L-fucosidases from two R. gnavus species have been recently characterized and they catalyzed the release of α-1,2, α-1,3 or α-1,4-linked fucose.Citation52 The presence of fucosidases with α-1,3 specificity in this species is in agreement with its survival in the 3FN supplemented group. The activity of α-fucosidases on 3FN and 6FN will produce the constituent monosaccharides fucose and GlcNAc. The action of specific β-galactosidases on LNB and GNB would generate galactose and the corresponding N-acetylhexosamines GlcNAc and GalNAc, respectively. R. gnavus has been shown to metabolize all those monosaccharides with the exception of GalNAc.Citation53
In recent years, the Akkermansia genus has received much attention because of its controversial involvement in human health and disease.Citation5,Citation54 Low levels of Akkermansia muciniphila in the human intestine have been associated with several diseases, including inflammatory bowel disease, atopic dermatitis and type-2 diabetes.Citation54 Conversely, a recent study showed that the relative abundance of this genus in a group of infants with allergic diseases is significantly higher than that in the healthy group.Citation5 A. muciniphila is known as an intestinal mucin-degrading bacterium, and possibly because of this, the members of this genus have been linked to reduced integrity of the intestinal barrier and infiltration of allergens through the intestinal wall.Citation55 Alternatively, a role in maintaining intestinal integrity has also been claimed for that bacterium.Citation56 A striking difference has been shown between the galactosyl-disaccharides and the fucosyl-disaccharides tested here in relation to the relative abundance of Akkermansia genus in feces (). While with LNB and GNB (a mucin-derived disaccharide) the levels of these bacteria remained and significantly increased, respectively, with both fucosyl-disaccharides were significantly reduced. This latest result is in agreement with a previous work that showed that the gut of newborn mice fed with regular core-fucosylated milk N-glycan had less abundance of members of the Akkermansia spp. than those fed with low-core-fucosylated milk N-glycan.Citation57
A desirable effect of HMOs is also to protect children from pathogenic diarrhea caused by the intestinal viruses rotavirus and norovirus. Interestingly, we have demonstrated in previous studies that those bacterial groups (Akkermansia spp. and Ruminococcus spp.) had divergent effects in rotavirus and norovirus susceptibility. Ruminococcus spp. correlated negatively with both rotavirus and norovirus IgA antibody titters, showing a lower susceptibility to these two virus infections in individuals with higher amounts of Ruminococcus spp. (revealing its potential as antiviral bacteria). Contrarily, the IgA antibody titer to rotaviruses positively correlated with the amounts of Akkermansia spp., pointing to this bacterial group as a facilitator of rotavirus infections.Citation58
SCFAs, the end products of carbohydrate fermentation by the intestinal microbiota, are efficiently absorbed by the gut mucosa and have important effects on host physiology though their involvement in gene expression regulation and action as signaling molecules.Citation30 Acetate, propionate and butyrate are the most abundant SCFAs in the colon, and they are substrates for colonocytes and peripheral tissues.Citation30,Citation59 In addition, SCFAs decrease the luminal pH helping to inhibit potential pathogens growth and to increase nutrients absorption.Citation60 Butyrate has also been associated with protection against colorectal cancer and atherosclerosis, and it showed immune-modulatory activities.Citation61,Citation62 Compared with formula-fed infants, exclusively breastfed infants present a higher proportion of acetate with respect to other SCFAs in the gut,Citation63 and this may provide protection against intestinal pathogens and allergic disease.Citation64,Citation65 Therefore, it is particularly relevant to provide specific carbohydrates that shift the microbiota toward the production of those catabolic products. In this study, the supplementation with 3FN significantly increased the levels of butyrate and acetate. Interestingly, a significant increment of the abundance of Coprococcus genus presented in the 3FN-feeding group clearly differentiates this group from the other three disaccharide-feeding groups (). The species belonging to that genus have been described to ferment carbohydrates and produce butyrate and acetate as end products.Citation66,Citation67 However, other possibilities for the increment of those SCFAs in the 3FN group cannot be discarded, including cross-feeding of intermediary and end metabolites between different gut bacteria.Citation66
HMOs are known to affect the mucosal and systemic immunity of newborns, either directly by interacting with the immune cells or indirectly through the microbiota.Citation68 Immunomodulatory activity has been previously described for fucosylated HMOs such as 2ʹFLCitation69 and lacto-N-fucopentaose.Citation70 Here, the supplementation of 3FN significantly decreased the expression of the pro-inflammatory cytokine TNF-α and enhanced the expression of the anti-inflammatory cytokines IL-10 and IL-13 in the mice large intestine tissue (). Therefore, these results suggested an anti-inflammatory potential for 3FN. This oligosaccharide enhanced the production of butyrate, which has been previously involved in immune homeostasis, for example, by reducing the expression of pro-inflammatory cytokines, including TNF-α.Citation71 We observed that the butyrate levels correlated positively with the expression of the cytokine IL-10 (r = 0.6), while it correlated negatively with TNF-α (r = −0.7), showing a trend toward statistical significance (p = 0.175 and 0.233, respectively). Interestingly, 3FN forms part of the Lewis x antigen and the presence of Lewis x-type oligosaccharides on the human milk glycoprotein mucin 1 have been shown to interact with the dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN), a specific C-type lectin on dendritic cells that binds fucose.Citation72 DC-SIGN is expressed in the entire gastrointestinal tract of neonates and its interaction with fucosylated oligosaccharides has been suggested to be an important mechanism of human milk to shape the infant immune system.Citation72,Citation73 Whether butyrate and/or C-type lectins are involved in the immunomodulatory effects of 3FN requires further investigations. The 3FN feeding group also showed a significant increment of the expression of the gene encoding TLR2. The other three feeding groups, 6FN, LNB and GNB, tended to increase the expression of that TLR compared to the control group, although they did not reach statistical significance. TLR2 stimulation has an important role in protecting gut epithelial barrier function.Citation74 This immunological receptor recognizes microorganism-associated molecular patterns, such as peptidoglycan, lipoteichoic acid and exopolysaccharide from Gram-positive bacteria.Citation75 Indeed, its activation has been demonstrated for Bidifobacterium strains isolated from breastfed infant feces.Citation76
Even knowing the many benefits of HMOs in infant health, only 2ʹFL and lacto-N-neotetraose are currently added to infant formula, possible because their synthesis is still difficult and expensive.Citation12 In this study, we demonstrated that four different disaccharides (3FN, 6FN, LNB and GNB) that are present in HMOs and glycoconjugates of human milk and mucosa, may play a role on infant microbiome building. However, this study may have limitations due to the variability usually observed in animal experimentation, and should therefore be corroborated by further research. Within the four disaccharides, it is important to highlight that LNB increased the relative abundance of Bifidobacterium genus, whose high levels is the most outstanding differential characteristic of the gut microbiota in breastfed infants. In addition, the efficacy of 3FN in changing the microbiota concomitant with an increase in SCFA levels and an immunomodulation activity was also demonstrated. These results allowed to gain insights in the mechanisms by which human milk glycans are associated with infant health benefits, and the simple structure of those disaccharides, which facilitates their synthesis, make them good candidates for being utilized in infant functional food development.
Material and methods
Animals, infant fecal samples and disaccharides
Thirty-five C57BL/6 J female mice, 6 weeks old, were acquired from Charles River Laboratories (Saint Germain Nuelles, France). They were randomly separated into seven groups of five mice each in individually ventilated cages in an environmentally controlled room, following the standard protocols of the animal facilities and the rules of animal wellness.
The study includes four infants, whose parents were volunteers. Our inclusion criteria were that the infants were healthy, receiving no antibiotic or probiotic treatment, between 1 and 3 months old, and exclusively breastfed. Stool samples were collected in anaerobic jars with Oxoid AnaeroGen anaerobic atmosphere generation system sachets (Thermo Scientific), immediately stored at 4 °C and cryopreserved within the next 12 h as previously described with some modifications.Citation77 Briefly, cryopreservation media contained 80% BHI (Brain Heart Infusion, Pronadisa) 2X concentrated supplemented with 0.1% of cysteine and 20% skim milk (200 g/l) (Scharlab). Feces were diluted in cryopreservation media 1:2 (vol/vol) and stored in aliquots at −80°C.
The disaccharides 3FN, 6FN, LNB and GNB were synthesized in our laboratory by enzymatic transglycosylation reactions and purified by high-performance liquid chromatography (HPLC) as previously described.Citation23,Citation25
Experimental design, fecal transplant and treatments
The experimental protocol is outlined in . The seven groups of mice (untreated, antibiotic-treated, control, 3FN, 6FN, LNB and GNB) were acclimated in the animal facility for 1 week and then an antibiotic cocktail (0.5 g/l vancomycin, 1 g/l neomycin sulfate, 1 g/l metronidazole, 1 g/l ampicillin) was administered in drinking water ad libitum for 3 weeks to all the groups except the untreated group. That antibiotic combination has been previously used.Citation40 The antibiotic cocktail was renewed every 3 d and removed 24 h before infant fecal microbiota transplant. One mouse from the antibiotic-treated group died for unknown reasons during the study. Mice were fed a standard diet until 1 week before fecal transplantation that was substituted by purified-defined germ-free diet (AIN-93 G, Envigo).
For the fecal transplant (control, 3FN, 6FN, LNB and GNB groups) a pool mix was prepared everyday with four fecal samples, one of each infant, and was kept on ice during the process. Each mouse received a volume of 100 μl of the pool mix in 3 consecutive days through oral gavage. Regarding oligosaccharide supplementation, 100 μl of 3FN, 6FN, LNB or GNB at 10 mM were supplied through oral gavage every day to each mouse (5 mice in each feeding group) for 3 weeks. Control mice received water. Feces were collected from each mouse before the sacrifice. After this, intestines content and tissues were also collected. Feces and intestines content were kept frozen at −80°C until analysis. Intestine tissues were preserved in RNA Later (Sigma) and kept the first 24 h at 4°C and then at −80°C until use.
All animal experimentation procedures were validated by the Ethical Committee for Use of Laboratory Animals of the University of Valencia, and the Department of Agriculture, Livestock and Fisheries of the Generalitat Valenciana, with registration number 2018/VSC/PEA/0181. The use of human samples was approved by the Ethical Committee for Human Research of the University of Valencia, with registration number H1544010468380. Written informed consent was obtained from a parent of each of the subjects.
DNA extraction from fecal samples
Total DNA was extracted from fecal samples of each mouse at the end of the experiment () using the MasterPure Complete DNA & RNA Purification Kit (Epicenter) according to the manufacturer’s instructions with some modifications that included a 60 min incubation with 2 μl of lysozyme 20 mg/ml and 1 μl of mutanolysin 10 U/ml followed by mechanical disruption using 3-µm diameter glass beads in a FastPrep 24-5 G Homogenizer (MP Biomedicals, CA, USA). Total DNA concentration was measured using a Qubit® 3.0 Fluorometer (Life Technologies, Carlsbad, CA, United States) and DNA integrity-quality was analyzed by gel electrophoresis. A DNA sample from the control group was discarded for failing to pass the quality control.
16S rRNA amplicon sequencing and data analysis
The amplification of the V3-V4 variable region of the 16S rRNA gene of total DNA from fecal samples was conducted following the Metagenomic Sequencing Library Preparation Illumina protocol (Cod. 15044223 Rev. A). Gene-specific primers (PCR1_f: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′; PCR1_r: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTA
TCTAATCC-3′) containing Illumina adapter overhang nucleotide sequences were selected as previously described.Citation78 A multiplexing step was performed using Nextera XT Index Kit and a Bioanalyzer DNA 1000 chip (Agilent Technologies) was used to verify the amplicons size (~550 bp). Libraries were sequenced using a 2 × 300 pb paired-end run (MiSeq Reagent kit v3) on a MiSeq Sequencer according to manufacturer’s instructions (Illumina) by the Central Service of Research Support of the University of Valencia (Spain).
Sequencing data have been demultiplexed using Illumina bcl2fastq© program. Forward and reverse raw reads were checked for quality, adapter trimmed and filtered using AfterQCCitation79 and FastQC v0.11.8 (http://www.bioinformatics.babraham.ac.uk) tools. QIIME software V1.9.1 was used to analyze the MiSeq sequencing data,Citation80 including forward and reverse reads joining, chimera removal, data filtering and taxonomic annotation. Chimeric sequences were removed from the reads using the USEARCH 6.1 algorithm.Citation81 Reads were clustered into operational taxonomic units (OTUs) based on a 97% identity threshold value. Alignment of the sequences was carried out using PyNASTCitation82 with reference to the Greengenes core reference database (version 13_8).Citation83 Taxonomic assignment was made using the UCLUST classifier.Citation81
Microbiota data were analyzed in the Calypso online platform (v8.84) (http://cgenome.net/wiki/index.php/Calypso/) and data was normalized by the Total-Sum Scaling (TSS) method with square root transformation. Total Richness and alpha diversity indexes (Shannon and Chao1) were determined. Beta diversity was represented by PCoA plot based on Bray–Curtis and Jaccard distance. Analysis of similarities (ANOSIM) and permutational multivariate analysis of variance (Adonis) based on Bray–Curtis distance were also achieved. Linear discriminant analysis effect size (LEfSe) was used to identify differences in microbial genera and families between control and oligosaccharide treated mice groups.
Raw sequences are deposited into the Sequence Read Archive (SRA) of NCIB (http://www.ncbi.nlm.nih.gov/sra) and can be assessed with the accession number PRJNA668130.
Quantification of Bifidobacterium genus by specific real-time PCR
Quantitative real-time PCR (qPCR) assays were performed as previously describedCitation24 and using the Bifidobacterium-specific 16S rRNA gene primers Bifido5ʹ (GAT TCT GGC TCA GGA TGA) and Bifido3ʹ (CTG ATA GGA CGC GAC CCC). The qPCR amplification and detection were conducted in a LightCycler 480 Real-Time PCR System (Roche). Each reaction mixture of 10 μl contained NZYSpeedy qPCR Green Master Mix (NZytech), 0.25 μl of each primer (10 μM) and 1 μl of template DNA. All samples were analyzed in triplicate. Standard curves of specific DNA amplicon-fragments obtained with the primers pair were used to calculate bacterial concentration in each sample.
Short-chain fatty acids’ (SCFAs) analysis
SCFAs were extracted from large intestine content and analyzed by gas chromatography mass spectrometry (GC/MS) as described in the Agilent application note.Citation84 Briefly, 30 mg of intestinal content were suspended in 1 ml 10% isobutanol and mechanically homogenized with glass beads The mixtures were centrifuged at 17,000 g for 5 min. Sample supernatants and standards were treated and subjected to the derivatization procedure as described in the Agilent application note, and 3-methylpentanoic acid was used as internal standard. Analysis of SCFAs was performed on an Agilent 7890B GC/5977 MSD (Agilent, Santa Clara, CA, USA) using a Agilent HP-5 ms column (30 m × 0.25 mm × 0.25 µm). The injector temperature was set at 260°C in split mode (10:1) and a volume of 1 µl was automatically set. The column temperature was initially 40°C for 5 min and then increased to 120°C at 10°C/min and then ramped to 310°C at 40°C/min and held for 2 min. The MS transfer line was maintained at 280°C and the ion source at 230°C.
Serum lipid analysis
About 1 ml of blood from the heart of mice was collected at the time of sacrifice and then centrifuged at 1,500 × g for 5 min. The serum was used to determine total cholesterol, triglycerides and low-density lipoproteins (LDL) by Echevarne Laboratories (Spain) using standard methods.
RNA isolation
Five mg of large intestine tissue were homogenized using a Polytron PT10-35 GT (Thermo Fisher Scientific). After tissue disruption, the RNA was purified using the NZY total RNA isolation kit (Nzytech) following the manufacturer instructions. RNA quality was checked by gel electrophoresis and quantified spectroscopically using a NanoDrop ND-1000 system (NanoDrop Technologies).
Analysis of cytokines and Toll-like receptors expression
The expression of genes encoding cytokines (IL-1β, IL-6, Cxcl15 (IL-8), IL-12, TNF-α, IFN-γ, IL-4, IL-10 y IL-13) and Toll-like receptors (TLR2 y TLR4) were evaluated using reverse transcription-quantitative PCR (RT-qPCR). First-strand complementary DNA (cDNA) was obtained from 1 μg of total RNA using Maxima first-strand cDNA synthesis kit (Thermo Scientific™). RT-qPCR was performed for each cDNA in triplicate using the LightCycler 480 System (Roche Technologies). Each qPCR reaction mixture (10 μl) contained 5 μl of NZY Speedy qPCR Green Master Mix 2X (Nzytech), 0.4 μl of each primer (10 mM) and 2 μl of diluted 1:20 cDNA template. Primer sequences are listed in Supplemental Table 3. The expression level of GAPDH and RPLPO housekeeping genes was used as reference.
The qPCR conditions were 95°C for 2 min, followed by 40 cycles of 10 s denaturation at 95°C and 15 s of annealing/extension at 60°C or 65°C (Supplemental Table 3). Relative expression values were calculated using the Relative Expression Software Tool (REST 2009, Qiagen). Linearity and amplification efficiency were determined for each primer pair using LinRegPCR software.Citation85
Culture of Bifidobacterium adolescentis with LNB and GNB
The growth of B. adolescentis ATCC 15703 in the presence of each disaccharide at 10 mM was tested in MRS basal medium as previously described.Citation23
Statistical analysis
The data obtained were analyzed by one-way ANOVA with Dunnett´s multiple comparisons test using GraphPad Prism, version 6.07 (GraphPad Software Inc., San Diego, CA, USA). Correlation of the expression levels of cytokines with butyrate concentrations were analyzed using the Spearman’s correlation coefficient. Statistical significant differences were accepted at different levels # p < 0.1, *p < 0.05; **P < 0.01; ***p < 0.001, ****p < 0.0001.
Disclosure of interest
The authors report no conflict of interest.
Supplemental Material
Download Zip (371.9 KB)Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Jost T, Lacroix C, Braegger C, Chassard C. Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health. Nutr Rev. 2015;73(7):426–20. doi:10.1093/nutrit/nuu016.
- Rautava S. Early microbial contact, the breast milk microbiome and child health. J Dev Orig Health Dis. 2016;7(1):5–14. doi:10.1017/S2040174415001233.
- Sim K, Shaw AG, Randell P, Cox MJ, McClure ZE, Li MS, Haddad M, Langford PR, Cookson WO, Moffatt MF, et al. Dysbiosis anticipating necrotizing enterocolitis in very premature infants. Clin Infec Dis. 2015;60(3):389–397. doi:10.1093/cid/ciu822.
- Goulet O. Potential role of the intestinal microbiota in programming health and disease. Nutr Rev. 2015;73(Suppl 1):32–40. doi:10.1093/nutrit/nuv039.
- Shen X, Wang M, Zhang X, He M, Li M, Cheng G, Wan C, He F. Dynamic construction of gut microbiota may influence allergic diseases of infants in Southwest China. BMC Microbiol. 2019;19(1):123. doi:10.1186/s12866-019-1489-4.
- Newburg DS, Morelli L. Human milk and infant intestinal mucosal glycans guide succession of the neonatal intestinal microbiota. Pediatric Res. 2015;77(1–2):115–120. doi:10.1038/pr.2014.178.
- Wang M, Li M, Wu S, Lebrilla CB, Chapkin RS, Ivanov I,Donovan SM. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J Pediatr Gastroenterol Nutr. 2015;60(6):825–833. doi:10.1097/MPG.0000000000000752.
- Kozak K, Charbonneau D, Sanozky-Dawes R, Klaenhammer T. Characterization of bacterial isolates from the microbiota of mothers’ breast milk and their infants. Gut Microbes. 2015;6(6):341–351. doi:10.1080/19490976.2015.1103425.
- Kobata A Structures and application of oligosaccharides in human milk. Proc Jpn Acad Ser B Phys Biol Sci.. 2010; 86:731–747. doi: 10.2183/pjab.86.731
- Zuniga M, Monedero V, Yebra MJ. Utilization of host-derived glycans by intestinal Lactobacillus and Bifidobacterium species. Front Microbiol. 2018;9:1917. doi:10.3389/fmicb.2018.01917.
- Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, Sears MR, Becker AB, Scott JA, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. Can Med Assoc J. 2013;185(5):385–394. doi:10.1503/cmaj.121189.
- Vandenplas Y, Berger B, Carnielli VP, Ksiazyk J, Lagstrom H, Sanchez Luna M, Migacheva N, Mosselmans JM, Picaud JC, Possner M, et al. Human milk oligosaccharides: 2ʹ-fucosyllactose (2ʹ-FL) and lacto-N-neotetraose (LNnT) in infant formula. Nutrients. 2018;10(9):1161. doi: 10.3390/nu10091161.
- Zuurveld M, Van Witzenburg NP, Garssen J, Folkerts G, Stahl B, Van’t Land B, Willemsen LEM.Immunomodulation by human milk oligosaccharides: the potential role in prevention of allergic diseases. Front Immunol. 2020;11:801. doi:10.3389/fimmu.2020.00801.
- Cheng L, Akkerman R, Kong C, Walvoort MTC, De Vos P. More than sugar in the milk: human milk oligosaccharides as essential bioactive molecules in breast milk and current insight in beneficial effects. Crit Rev Food Sci Nutr. 2021;61(7):1184-1200. doi: 10.1080/10408398.2020.1754756.
- Urashima T, Taufik E, Fukuda K, Asakuma S. Recent advances in studies on milk oligosaccharides of cows and other domestic farm animals. Biosci Biotech Biochem. 2013;77(3):455–466. doi:10.1271/bbb.120810.
- Ho NT, Li F, Lee-Sarwar KA, Tun HM, Brown BP, Pannaraj PS, Bender JM, Azad MB, Thompson AL, Weiss ST, et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nature Commun. 2018;9(1):4169. doi:10.1038/s41467-018-06473-x.
- Weiss GA, Chassard C, Hennet T. Selective proliferation of intestinal Barnesiella under fucosyllactose supplementation in mice. British J Nutr. 2014;111(9):1602–1610. doi:10.1017/S0007114513004200.
- Lee S, Goodson M, Vang W, Kalanetra K, Barile D, Raybould H. 2ʹ-fucosyllactose supplementation improves gut-brain signaling and diet-induced obese phenotype and changes the gut microbiota in high fat-fed mice. Nutrients. 2020;12:1003. doi: 10.3390/nu12041003.
- Tarr AJ, Galley JD, Fisher SE, Chichlowski M, Berg BM, Bailey MT. The prebiotics 3’Sialyllactose and 6’Sialyllactose diminish stressor-induced anxiety-like behavior and colonic microbiota alterations: evidence for effects on the gut-brain axis. Brain Behav Immun. 2015;50:166–177. doi:10.1016/j.bbi.2015.06.025.
- Martin FP, Wang Y, Sprenger N, Yap IK, Rezzi S, Ramadan Z, Peré-Trepat E, Rochat F, Cherbut C, van Bladeren P, et al. Top-down systems biology integration of conditional prebiotic modulated transgenomic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4(1):205. doi:10.1038/msb.2008.40.
- Musilova S, Modrackova N, Hermanova P, Hudcovic T, Svejstil R, Rada V, Tejnecky V, Bunesova V. Assessment of the synbiotic properties of human milk oligosaccharides and Bifidobacterium longum subsp. infantis in vitro and in humanised mice. Benef Microbes. 2017;8(2):281–289. doi:10.3920/BM2016.0138.
- Rodriguez-Diaz J, Carbajo RJ, Pineda-Lucena A, Monedero V, Yebra MJ. Synthesis of fucosyl-N-acetylglucosamine disaccharides by transfucosylation using alpha-L-fucosidases from Lactobacillus casei. Appl Environ Microbiol. 2013;79(12):3847–3850. doi:10.1128/AEM.00229-13.
- Becerra JE, Coll-Marques JM, Rodriguez-Diaz J, Monedero V, Yebra MJ. Preparative scale purification of fucosyl-N-acetylglucosamine disaccharides and their evaluation as potential prebiotics and antiadhesins. Appl Microbiol Biotechnol. 2015;99(17):7165–7176. doi:10.1007/s00253-015-6666-2.
- Rubio-Del-Campo A, Alcantara C, Collado MC, Rodriguez-Diaz J, Yebra MJ. Human milk and mucosa-associated disaccharides impact on cultured infant fecal microbiota. Sci Rep. 2020;10(1):11845. doi:10.1038/s41598-020-68718-4.
- Bidart GN, Rodriguez-Diaz J, Palomino-Schatzlein M, Monedero V, Yebra MJ. Human milk and mucosal lacto- and galacto-N-biose synthesis by transgalactosylation and their prebiotic potential in Lactobacillus species. Appl Microbiol Biotechnol. 2017;101(1):205–215. doi:10.1007/s00253-016-7882-0.
- Balogh R, Jankovics P, Beni S. Qualitative and quantitative analysis of N-acetyllactosamine and lacto-N-biose, the two major building blocks of human milk oligosaccharides in human milk samples by high-performance liquid chromatography-tandem mass spectrometry using a porous graphitic carbon column. J Chromatog A. 2015;1422:140–146.
- Satoh T, Odamaki T, Namura M, Shimizu T, Iwatsuki K, Nishimoto M,Kitaoka M, Xiao JZ. In vitro comparative evaluation of the impact of lacto-N-biose I, a major building block of human milk oligosaccharides, on the fecal microbiota of infants. Anaerobe. 2013;19:50–57. doi:10.1016/j.anaerobe.2012.12.007.
- Rios-Covian D, Ruas-Madiedo P, Margolles A, Gueimonde M, De Los Reyes-gavilan CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol. 2016;7:185. doi:10.3389/fmicb.2016.00185.
- Moens F, Verce M, De Vuyst L. Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans. Int J Food Microbiol. 2017;241:225–236. doi:10.1016/j.ijfoodmicro.2016.10.019.
- Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7(3):189–200. doi:10.1080/19490976.2015.1134082.
- Joyce SA, Kamil A, Fleige L, Gahan CGM. The cholesterol-lowering effect of oats and oat beta glucan: modes of action and potential role of bile acids and the microbiome. Front Nutr. 2019;6(171). doi:10.3389/fnut.2019.00171.
- Wong WW, Hachey DL, Insull W, Opekun AR, Klein PD. Effect of dietary cholesterol on cholesterol synthesis in breast-fed and formula-fed infants. J Lipid Res. 1993;34(8):1403–1411. doi:10.1016/S0022-2275(20)36969-8.
- Timby N, Lonnerdal B, Hernell O, Domellof M. Cardiovascular risk markers until 12 mo of age in infants fed a formula supplemented with bovine milk fat globule membranes. Ped Res. 2014;76(4):394–400. doi:10.1038/pr.2014.110.
- Owen CG, Whincup PH, Kaye SJ, Martin RM, Davey Smith G, Cook DG, Bergstrom E, Black S, Wadsworth ME, Fall CH, et al. Does initial breastfeeding lead to lower blood cholesterol in adult life? A quantitative review of the evidence. Am J Clin Nutr. 2008;88(2):305–314. doi:10.1093/ajcn/88.2.305.
- Li N, Yan F, Wang N, Song Y, Yue Y, Guan J, Li B, Huo G. Distinct gut microbiota and metabolite profiles induced by different feeding methods in healthy Chinese infants. Front Microbiol. 2020;11:714. doi:10.3389/fmicb.2020.00714.
- Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22(9):1147–1162. doi:10.1093/glycob/cws074.
- Martin FP, Wang Y, Sprenger N, Yap IK, Lundstedt T, Lek P, Rezzi S, Ramadan Z, van Bladeren P, Fay LB, et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4(1):157. doi:10.1038/msb4100190.
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nature Rev Immunol. 2004;4(6):478–485. doi:10.1038/nri1373.
- Johansson ME, Jakobsson HE, Holmen-Larsson J, Schutte A, Ermund A, Rodriguez-Pineiro AM, Arike L, Wising C, Svensson F, Bäckhed F, et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 2015;18(5):582–592. doi:10.1016/j.chom.2015.10.007.
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi:10.1016/j.cell.2004.07.002.
- Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nature Rev Microbiol. 2012;10(5):323–335. doi:10.1038/nrmicro2746.
- Elderman M, Sovran B, Hugenholtz F, Graversen K, Huijskes M, Houtsma E, Belzer C, Boekschoten M, de Vos P, Dekker J, et al. The effect of age on the intestinal mucus thickness, microbiota composition and immunity in relation to sex in mice. PLoS One. 2017;12(9):e0184274. doi:10.1371/journal.pone.0184274.
- Liu Z, Roy NC, Guo Y, Jia H, Ryan L, Samuelsson L, Thomas A, Plowman J, Clerens S, Day L, et al. Human breast milk and infant formulas differentially modify the intestinal microbiota in human infants and host physiology in rats. Journal Nutr. 2016;146(2):191–199. doi:10.3945/jn.115.223552.
- Xiao JZ, Takahashi S, Nishimoto M, Odamaki T, Yaeshima T, Iwatsuki K,Kitaoka M. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl Environ Microbiol. 2010;76(1):54–59. doi:10.1128/AEM.01683-09.
- De Andres J, Manzano S, Garcia C, Rodriguez JM, Espinosa-Martos I, Jimenez E. Modulatory effect of three probiotic strains on infants’ gut microbial composition and immunological parameters on a placebo-controlled, double-blind, randomised study. Benef Microbes. 2018;9(4):573–584. doi:10.3920/BM2017.0132.
- Duar RM, Henrick BM, Casaburi G, Frese SA. Integrating the ecosystem services framework to define dysbiosis of the breastfed infant gut: the role of B. infantis and human milk oligosaccharides. Front Nutr. 2020;7:33. doi:10.3389/fnut.2020.00033.
- Wu W, Wang Y, Zou J, Long F, Yan H, Zeng L, Chen Y. Bifidobacterium adolescentis protects against necrotizing enterocolitis and upregulates TOLLIP and SIGIRR in premature neonatal rats. BMC Pediatr. 2017;17(1):1. doi:10.1186/s12887-016-0759-7.
- Kraal L, Abubucker S, Kota K, Fischbach MA, Mitreva M. The prevalence of species and strains in the human microbiome: a resource for experimental efforts. PLoS One. 2014;9(5):e97279. doi:10.1371/journal.pone.0097279.
- Sagheddu V, Patrone V, Miragoli F, Puglisi E, Morelli L. Infant early gut colonization by Lachnospiraceae: high frequency of Ruminococcus gnavus. Front Pediatr. 2016;4:57. doi:10.3389/fped.2016.00057.
- Nishikawa J, Kudo T, Sakata S, Benno Y, Sugiyama T. Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scand J Gastroenterol. 2009;44(2):180–186. doi:10.1080/00365520802433231.
- Grabinger T, Glaus Garzon JF, Hausmann M, Geirnaert A, Lacroix C, Hennet T. Alleviation of intestinal inflammation by oral supplementation with 2-fucosyllactose in mice. Front Microbiol. 2019;10:1385. doi:10.3389/fmicb.2019.01385.
- Wu H, Rebello O, Crost EH, Owen CD, Walpole S, Bennati-Granier C, Ndeh D, Monaco S, Hicks T, Colvile A, et al. Fucosidases from the human gut symbiont Ruminococcus gnavus.Cell Mol Life Sci. 2021 78(2):675-693. doi:10.1007/s00018-020-03514-x.
- Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One. 2013;8(10):e76341. doi:10.1371/journal.pone.0076341.
- Geerlings SY, Kostopoulos I, De Vos WM, Belzer C. Akkermansia muciniphila in the human gastrointestinal tract: when, where, and how? Microorganisms. 2018;6(3):6. doi:10.3390/microorganisms6030075.
- Zheng H, Liang H, Wang Y, Miao M, Shi T, Yang F,Liu E, Yuan W, Ji ZS, Li DK. Altered gut microbiota composition associated with eczema in infants. PLoS One. 2016;11(11):e0166026. doi:10.1371/journal.pone.0166026.
- Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H,de Vos WM, Satokari R. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol. 2015;81(11):3655–3662. doi:10.1128/AEM.04050-14.
- Li M, Bai Y, Zhou J, Huang W, Yan J, Tao J, et al. Core fucosylation of maternal milk N-glycan evokes B cell activation by selectively promoting the l-fucose metabolism of gut Bifidobacterium spp. and Lactobacillus spp. mBio. 2019;10(2):e00128-19. doi: 10.1128/mBio.00128-19.
- Rodriguez-Diaz J, Garcia-Mantrana I, Vila-Vicent S, Gozalbo-Rovira R, Buesa J, Monedero V,Collado MC. Relevance of secretor status genotype and microbiota composition in susceptibility to rotavirus and norovirus infections in humans. Sci Rep. 2017;7(1):45559. doi:10.1038/srep45559.
- Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217(2):133–139. doi:10.1111/j.1574-6968.2002.tb11467.x.
- Macfarlane GT, Macfarlane S. Bacteria, colonic fermentation, and gastrointestinal health. J AOAC Int. 2012;95(1):50–60. doi:10.5740/jaoacint.SGE_Macfarlane.
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi:10.1038/nature12721.
- Bultman SJ. Bacterial butyrate prevents atherosclerosis. Nature Microbiol. 2018;3(12):1332–1333. doi:10.1038/s41564-018-0299-z.
- Bridgman SL, Azad MB, Field CJ, Haqq AM, Becker AB, Mandhane PJ, Subbarao P, Turvey SE, Sears MR, Scott JA, et al. Fecal short-chain fatty acid variations by breastfeeding status in infants at 4 months: differences in relative versus absolute concentrations. Front Nutr. 2017;4:11. doi:10.3389/fnut.2017.00011.
- Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. doi:10.1126/scitranslmed.aab2271.
- Kumari M, Kozyrskyj AL. Gut microbial metabolism defines host metabolism: an emerging perspective in obesity and allergic inflammation. Obesity Rev. 2017;18(1):18–31. doi:10.1111/obr.12484.
- Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environmental Microbiol. 2017;19(1):29–41. doi:10.1111/1462-2920.13589.
- Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. Acetate utilization and butyryl coenzyme A (CoA): acetate-CoAtransferase in butyrate-producing bacteria from the human large intestine. App Environ Microbiol. 2002;68(10):5186–5190. doi:10.1128/AEM.68.10.5186-5190.2002.
- Donovan SM, Comstock SS. Human Milk Oligosaccharides influence Neonatal Mucosal and systemic immunity. Ann Nutr Metab. 2016;69(Suppl 2):42–51. doi:10.1159/000452818.
- Azagra-Boronat I, Massot-Cladera M, Mayneris-Perxachs J, Knipping K, Van’t Land B, Tims S, Stahl B, Garssen J, Franch À, Castell M, et al. Immunomodulatory and prebiotic effects of 2ʹ-Fucosyllactose in suckling rats. Front Immunol. 2019;10:1773. doi:10.3389/fimmu.2019.01773.
- Srivastava L, Tundup S, Choi BS, Norberg T, Harn D. Immunomodulatory glycan lacto-N-fucopentaose III requires clathrin-mediated endocytosis to induce alternative activation of antigen-presenting cells. Infect Immun. 2014;82(5):1891–1903. doi:10.1128/IAI.01293-13.
- Segain JP, Raingeard De La Bletiere D, Bourreille A, Leray V, Gervois N, Rosales C,Ferrier L, Bonnet C, Blottière HM, Galmiche JP. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut. 2000;47(3):397–403. doi:10.1136/gut.47.3.397.
- Koning N, Kessen SF, Van Der Voorn JP, Appelmelk BJ, Jeurink PV, Knippels LM, Garssen J, Van Kooyk Y. Human Milk Blocks DC-SIGN-Pathogen Interaction via MUC1. Front Immunol. 2015;6:112. doi:10.3389/fimmu.2015.00112.
- Noll AJ, Gourdine JP, Yu Y, Lasanajak Y, Smith DF, Cummings RD. Galectins are human milk glycan receptors. Glycobiology. 2016;26(6):655–669. doi:10.1093/glycob/cww002.
- Ren C, Zhang Q, De Haan BJ, Faas MM, Zhang H, De Vos P. Protective effects of lactic acid bacteria on gut epithelial barrier dysfunction are Toll like receptor 2 and protein kinase C dependent. Food Funct. 2020;11(2):1230–1234. doi:10.1039/C9FO02933H.
- McGuire VA, Arthur JS. Subverting toll-like receptor signaling by bacterial pathogens. Front Immunol. 2015;6:607. doi:10.3389/fimmu.2015.00607.
- Yu R, Zuo F, Ma H, Chen S. Exopolysaccharide-producing Bifidobacterium adolescentis strains with similar adhesion property induce differential regulation of inflammatory immune response in Treg/Th17 Axis of DSS-Colitis Mice. Nutrients. 2019;11(4):11. doi:10.3390/nu11040782.
- Wrzosek L, Ciocan D, Borentain P, Spatz M, Puchois V, Hugot C,Ferrere G, Mayeur C, Perlemuter G, Cassard AM. Transplantation of human microbiota into conventional mice durably reshapes the gut microbiota. Sci Rep. 2018;8(1):6854. doi:10.1038/s41598-018-25300-3.
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41(1):e1. doi:10.1093/nar/gks808.
- Chen S, Huang T, Zhou Y, Han Y, Xu M, Gu J. AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinform. 2017;18(S3):80. doi:10.1186/s12859-017-1469-3.
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi:10.1038/nmeth.f.303.
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi:10.1093/bioinformatics/btq461.
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26(2):266–267. doi:10.1093/bioinformatics/btp636.
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi:10.1128/AEM.03006-05.
- Furuhashi T, Ishihara G. GC/MS detection of short chain fatty acids from mammalian feces using automated sample preparation in aqueous solution. Appl Note Metabolomics. 2019. 1–6. https://www.agilent.com/cs/library/applications/5991-9103EN_Metabolomics_AppNote.pdf
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, Van Den Hoff MJ, Moorman AF. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37(6):e45. doi:10.1093/nar/gkp045.