ABSTRACT
Humans live in symbiosis with a diverse community of microorganisms, which has evolved to carry out many specific tasks that benefit the host, including protection against invading pathogens. Within the chemical diversity of the gastrointestinal tract, small molecules likely constitute chemical cues for the communication between the microbiota and pathogens. Therefore, we sought to investigate if molecules produced by the human gut microbiota show biological activity against the human pathogen Vibrio cholerae. To probe the effects of the gut metabolome on V. cholerae, we investigated its response to small-molecule extracts from human feces, from a complex bacterial community cultivated in vitro, and from culture supernatants of Enterocloster citroniae, Bacteroides thetaiotaomicron, and Bacteroides vulgatus. Using RNA sequencing, we determined the impact of the human gut metabolome on V. cholerae global gene expression. Among the genes downregulated in the presence of the fecal extract, the most overrepresented functional category was cell motility, which accounted for 39% of repressed genes. Repression of V. cholerae motility by the fecal extract was confirmed phenotypically, and E. citroniae extracts reproduced this phenotype. A complex in vitro microbial community led to increased motility, as did extracts from B. vulgatus, a species present in this community. Accordingly, mucin penetration was also repressed by fecal and E. citroniae extracts, suggesting that the phenotypes observed may have implications for host colonization. Together with previous studies, this work shows that small molecules from the gut metabolome may have a widespread, significant impact on microbe–microbe interactions established in the gut environment.
Introduction
The importance of the gut microbiota for human health is well known, but the molecular mechanisms involved are not fully elucidated. The microbiota contributes to many of the biochemical pathways found in the human gut, aiding in the utilization of proteins and indigestible polysaccharides as carbon and energy sources by breaking them into essential amino acids and short chain fatty acids.Citation1 As the majority of microbiota members are metabolically active, they are constantly producing and secreting small molecules.Citation2 These molecules are part of the gut metabolome, the collection of all small molecules (<3000 Da) present in the gastrointestinal tract, and form a chemical framework that can exert many functions in this environment.Citation3–5 Interactions between host and gut microbiota contribute significantly to the production of small molecules that can be excreted into the feces or reach various organs and tissues through systemic blood circulation.Citation6,Citation7 The effects of such chemical interactions are still mostly unknown, although some compounds can affect host cell differentiation and metabolism.Citation8,Citation9
Over the past few years, studies have demonstrated the interference of small molecules in the survival and behavior of pathogenic species, thus providing a new tool for understanding their relationships with the human host.Citation10 Our group has recently shown that aromatic compounds from the human gut metabolome have a significant impact on the virulence of Salmonella enterica serovar Typhimurium.Citation11,Citation12 Other groups have made similar observations with other enteric pathogens, such as Clostridioides difficile and enteropathogenic Escherichia coli.Citation13–15 However, many other human pathogens are likely to have evolved mechanisms to respond to chemical cues from the resident microbiome, although there have been very few studies investigating the effect of the gut metabolome on other pathogens.
Vibrio cholerae is the causative agent of the intestinal disease, cholera. The main source of infection is contaminated water and food, and thus cholera is an important disease in many developing countries within Africa, Asia and South America that suffer from poor water supplies and sanitation.Citation16 V. cholerae is a noninvasive bacterium that affects the small intestine through epithelial cell adhesion and production of an enterotoxin, cholera toxin (CT).Citation17 ToxR and other regulatory proteins regulate CT expression, and several virulence genes within the ToxR regulon are involved in cholera disease. In addition to the essential role of CT in cholera, the toxin-coregulated pilus (TCP) is pivotal for the colonization of the intestinal epithelium and helps in microcolony formation on the epithelial surface.Citation16,Citation18
Humans, their microbiota, and intestinal pathogens have co-evolved for hundreds of thousands of years. Thus, it is probable that chemical signaling between the intestinal microbiota and pathogens represent not an exception, but rather the rule governing the interactions between these beneficial microbial communities and invaders. The role of the gut microbiota in protection against invading pathogens has been studied for decades, and several studies suggest that small molecules may be involved in this phenomenon.Citation11,Citation12,Citation19,Citation20 In this study, we set out to determine whether the human gut metabolome has biological activity that mitigates V. cholerae virulence by examining pathogen gene expression and behavior in response to small molecules produced by members of the gut microbiota. Our results showed that the gut metabolome exerts specific effects on pathogen gene expression, and that these effects translate into phenotypic changes. How each member of the microbiota and the molecules they produce integrate to control the dynamics of microbiota–pathogen interactions in this complex environment will be an exciting area of research for years to come.
Materials and methods
A scheme displaying the methods employed in this work is presented in Figure S1.
Ethics statement
This study was carried out in accordance with the recommendations and approval of the Research Ethics Committee of the Escola Nacional de Saúde Pública Sergio Arouca, Fundação Oswaldo Cruz (protocol number 69742817.6.0000.5240).
Bacterial strains and growth conditions
The V. cholerae strain used in this work was VC833, a multidrug-resistant V. cholerae O1 strain isolated from human feces during a cholera outbreak in Nigeria in 2010.Citation21 The strain was obtained from the Bacteria Culture Collection of Environment and Health at the Fundação Oswaldo Cruz. VC833 is resistant to streptomycin, trimethoprim, trimethoprim/sulfamethoxazole, cefotin, cefuroxime, ampicillin, nalidixic acid, ciprofloxacin, sulfonamide, sulfamethoxazole, and chloramphenicol.Citation21 V. cholerae was routinely grown in Brain Heart Infusion broth (BHI; Sigma-Aldrich) supplemented with hemin (5 mg/mL; Sigma-Aldrich) at 37°C with shaking (225 rpm).
Bacteroides thetaiotaomicron ATCC29741 and Bacteroides vulgatus ATCC8482 were obtained from the American Type Culture Collection. Enterocloster (previously Clostridium) citroniae FM-V5-E was isolated from a stool sample from a healthy 38-year old female.Citation11 Bacteroides and E. citroniae strains were cultured in an anaerobic chamber (80% N2, 10% H2, 10% CO2) in BHI (Sigma-Aldrich) supplemented with hemin (5 mg/mL; Sigma-Aldrich) at 37°C.
Chemostat
A 124-strain, 69-species community was cultured in vitro within a continuously fed bioreactor and was compiled from a collection of previously-isolated strains originating from a stool sample of a 41-year-old female.Citation22 Strains were routinely grown on Fastidious Anaerobe Agar (Neogen) supplemented with 5% v/v defibrinated sheep’s blood (Hemostat Laboratories) (FAA) within an AS-580 Anaerobe Chamber (Anaerobe Systems) in an atmosphere of N2/CO2/H2 90:5:5. Before inclusion into the community, strains were streaked to purity on FAA, followed by V3-V4 variable region 16S rRNA gene Sanger sequencing for species verification (Table S2). Individual strains were then frozen at −80°C for storage in skim milk freezing media supplemented with dimethyl sulfoxide (Thermo Scientific). Community stocks for chemostat seeding were created by combining equivalent volumes of each strain grown in a lawn before freezing at −80°C for storage in skim milk freezing media without dimethyl sulfoxide.
The chemostat, a 500-mL Multifors bioreactor with a working volume of 400 mL (Infors), had conditions set to mimic the human colonic lumen: 37°C, pH 7, gentle agitation, anaerobic under nitrogen gas bubbling, and supplied continually with a medium designed to emulate the effluent from the small intestine (Table S3).Citation23 The 5 mL community stock was thawed from −80°C to room temperature within the Anaerobe Chamber before adding to 400 mL of degassed medium within the chemostat. The community was allowed 24 h in batch culture within the chemostat before fresh medium was added or waste removed, to allow an increase in biomass. The bioreactor vessel was operated and maintained as previously described.Citation23
The vessel was maintained for 21 days before the entire contents were harvested. Aliquots of 35 mL of this material were centrifuged at 15,000 g and 4°C for 30 min prior to 0.22 µm filter-sterilization. After that, 330 mL of the community filtrate was aliquoted and snap-frozen in ethanol and dry ice followed by overnight lyophilization in a FreeZone 4.5 Liter Benchtop Freeze Dry System (Labconco). The lyophilized community filtrate was reconstituted in water before further investigation.
Human fecal samples
Fecal samples were collected from a 35-year-old female with no history of significant gastrointestinal problems and who did not undergo antibiotic treatment for at least six months before sample collection. Fresh feces were collected using a sterile container, refrigerated and brought to the laboratory within 24 h, where they were immediately used or frozen at −20°C until used.
Small-molecule extraction
To extract small molecules from human feces, thereby producing the fecal extracts used in this work, stool samples were first weighed and transferred to a sterile glass vessel. Then, ethyl acetate (Sigma-Aldrich) was added in a 1:1 (w/v) ratio. The mixture was vortexed for 5 min and then incubated with shaking (100 rpm) for 18 h. After the incubation, the liquid phase was collected in a glass flask and the extract was evaporated in a rotary evaporator (Heidolph) and then frozen at −20°C until used. As a control, ethyl acetate was also dried in a rotary evaporator and the sediment was used in experiments.
In order to obtain small-molecule extracts from E. citroniae, B. thetaiotaomicron and B. vulgatus cultures, bacterial strains were inoculated in 250 mL of BHI and incubated in anaerobiosis at 37°C for 18 h. After incubation, 250 mL of ethyl acetate was added to the cultures and the vials were shaken vigorously and then allowed to stand for 10 min. The vials were then vigorously mixed again and left for another 10 min at rest to allow phases to separate. The organic phase was collected and dried in a rotary evaporator. As a control, BHI medium without bacterial growth was also extracted, as described. Dried extracts were maintained at −20°C until used. Chemostat effluent was processed in the same manner. First, freeze-dried effluent was reconstituted in the original volume of water to bring metabolites up to their original concentration. Then, one volume of ethyl acetate was added and the molecules were extracted as described above for bacterial cultures. As a control, chemostat media without bacterial growth was also extracted, as described.
For experiments using small-molecule extracts, the dried material was suspended directly in BHI. The suspension was filtered (0.45-μm pore size) to remove larger particles that did not solubilize and the pH was adjusted to 7.2, followed by filtration again using 0.2-μm filters to sterilize extracts. Fecal extracts were used in experiments at a 1x relative concentration, to approximate the metabolite concentrations found in feces. Bacterial culture extracts were used at a 2x relative concentration in all experiments.
Bacterial growth curves
An overnight culture of V. cholerae was diluted 1:200 in BHI broth, with or without the addition of extracts from human feces, B. thetaiotaomicron, E. citroniae, B. vulgatus or chemostat cultures, and added to a 96-well microplate. The growth curve was performed using the Infinite® F50 spectrophotometer (TECAN) with readings every 30 min for 12 h at an optical density of 620 nm (OD620).
RNA sequencing and data analysis
V. cholerae was grown in BHI broth with and without the fecal extract. After approximately 4 h of growth (mid-log phase), bacterial RNA was stabilized by the addition of 2 volumes of RNAprotect bacterial reagent (Qiagen) to bacterial cultures, which were thoroughly mixed and incubated at room temperature for approximately 5 min. Cells were pelleted by centrifugation, and RNA was isolated using the RNeasy Mini Kit (Qiagen), according to the manufacturer’s recommendations. RNA was quantified using a NanoVue spectrophotometer (GE HealthCare). RNA was treated with Turbo DNase (2 U/μL) according to the manufacturer’s manual (Life Technologies) to eliminate residual DNA, followed by purification using the RNeasy Mini kit (Qiagen) according to the manufacturer’s recommendations. rRNA depletion was performed with the Ribo-Zero rRNA Removal Kit (Illumina), according to the manufacturer´s recommendations (Document #15,066,012 v02). Library construction was performed with the TruSeq Stranded mRNA Sample Preparation Kit, according to the manufacturer´s recommendations (catalog #RS-122-9004DOC). The final libraries obtained were assessed for quality using a DNA 1000 chip on an Agilent Technologies 2100 Bioanalyzer (Agilent) and quantified using Qubit 2.0 kit High Sensitivity (ThermoFisher). Sequencing was performed on an Illumina HiSeq 2500 (Illumina) at the Plataforma de Sequenciamento de Ácidos Nucleicos de Nova Geração – RPT01J of the Fundação Oswaldo Cruz (Rio de Janeiro, Brazil).
Data processing and analyses were performed at the Plataforma de Bioinformática – RPT04A of the Fundação Oswaldo Cruz, as previously described.Citation24 In summary, raw sequencing read files (bcl files) were converted to fastq files with bcl2fastq software version 2.17 (Illumina). Technical and low-quality sequences were removed using Trimmomatic (v 2.2.0) with the following parameters: ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:40.Citation25 Read quality was assessed using FastQC (Babraham Bioinformatics), and filtered reads were mapped with Bowtie2 (–no-mixed – no-discordant) to the two chromosomes of V. cholerae O1 biovar El Tor str. N16961 (GenBank accession no. NC_002505 and NC_002506) as references.Citation26 SAMtools was used to extract and sort mapped reads (-G 77, -G 141), and the Cufflinks suite was used to compare global gene expression between conditions.Citation27–29 Genes that showed a differential expression of 2-fold with p< .05 between conditions (absence or presence of fecal extract) were considered significantly regulated. Genes were functionally annotated and had their functional abundance profiles created with EggNOG (v. 5.0.0) (http://eggnog5.embl.de).Citation30 Protein-protein association networks of significantly altered genes (≥10-fold change) were analyzed using STRING v11.0 (https://string-db.org/).Citation31
Motility assays
Motility assays were carried out in BHI medium containing 0.3% agar with or without fecal, E. citroniae, B. thetaiotaomicron, B. vulgatus or chemostat extracts. Culture medium was poured into Petri dishes and allowed to solidify overnight. Plates were then inoculated with a single colony using a sterile toothpick and incubated at 37°C. The diameter of the motility halo was measured after 18 h.
Mucin penetration assays
Mucin penetration assays were performed in 1% mucin columns (Mucin from porcine stomach, type II, Sigma) in 1 mL syringes. V. cholerae was grown with or without the addition of extracts from human feces, E. citroniae or B. thetaiotaomicron cultures for approximately 4 h, until the mid-log growth phase was reached. Cells were then pelleted, washed, and reconstituted in phosphate-buffered saline (pH 7.4), and the suspension standardized to an OD600 of 0.3. A 0.1 mL aliquot of the cell suspension was added to the top of 1% mucin columns. Columns were kept at 37°C in vertical position under static conditions. After 30 min of incubation, 500 µL fractions were collected from the bottom of the columns, serially diluted, and plated onto BHI agar to measure CFU after incubation at 37°C for 16 h.Citation32
Biofilm formation assays
Quantification of total growth and biofilm formation was performed as described previously, with some modifications. Briefly, V. cholerae strains were grown overnight in LB broth (1% NaCl) at 37°C with shaking (250 rpm). Following overnight growth, the culture was diluted 1:200 in LB broth with and without the addition of extracts from human feces, E. citroniae or B. thetaiotaomicron cultures and grown as static cultures in a 96-well polystyrene plate (Greiner) at 37°C. After incubation for approximately 48 h, planktonic cell suspensions were collected, leaving only surface-attached cells in each well. The planktonic cell density was determined by measuring the OD600. To quantify biofilm formation, 200 μL of PBS were added to the surface-attached cells remaining in the microplate, and the cells were dispersed by vigorous pipetting and scraping. The amount of biofilm formed was determined by measuring the OD600 of the resulting cell suspension using an Infinite M200 PRO microplate reader (TECAN). The reported total growth was calculated from the sum of the OD600 values measured for planktonic and biofilm cell suspensions.
Results
The human gut metabolome moderately inhibits V. cholerae growth
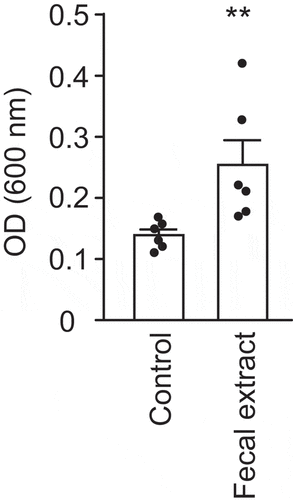
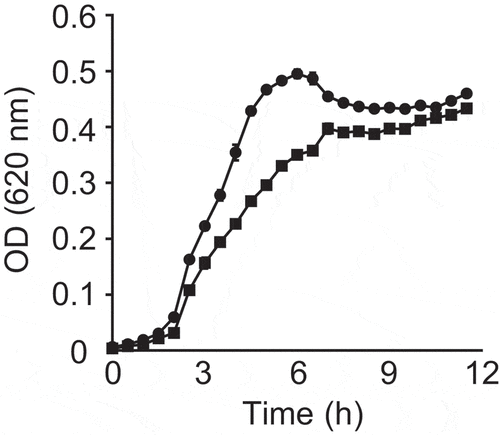
In order to test if small molecules extracted from fresh feces of a healthy donor have any effect on V. cholerae, we compared bacterial growth with or without the fecal extract. As can be observed in , V. cholerae growth was slightly delayed in the presence of the fecal extract. In regular culture medium, V. cholerae reached stationary phase after approximately 5.5 hours of growth. Growth in the fecal extract led to a delay of approximately 1.5 hours, with V. cholerae reaching stationary phase after approximately 7 hours. Although the optical density (OD) reached by the cultures in stationary phase was comparable, a significant difference was observed during exponential growth, with the largest difference being 36.6% at 5 hours of growth (doubling time of 52.72 ± 1.70 minutes for control cultures vs. 61.64 ± 7.03 minutes for cultures containing the extract). This reduction was not caused by either pH differences or solvent impurities, since the pH of the culture medium containing the fecal extract was adjusted to match that of the control medium, and the control cultures contained an ethyl acetate evaporate without fecal material. This result suggests that molecules of low polarity present in the fecal metabolome show moderate biological activity against V. cholerae growth. However, both cultures in the presence and absence of the fecal extract reached similar OD values in the stationary phase.
Figure 1. Impact of a fecal extract on V. cholerae growth in vitro.
Small molecules from human fecal extracts modulate V. cholerae gene expression
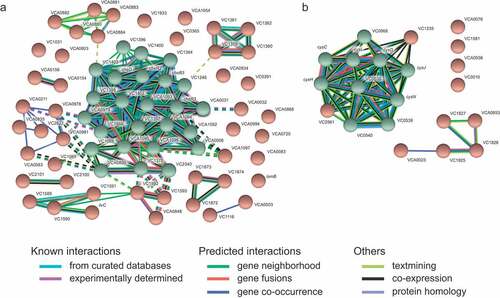
To further investigate the effect of the human gut metabolome on V. cholerae, we compared the transcriptome of cells during mid-log growth in the absence or presence of a human fecal extract. RNAseq results revealed 912 differentially expressed genes, representing 24.4% of all genes in the genome. The results of the transcriptome analysis can be seen in Table S1, which shows expression levels of all genes. Specifically, 410 genes were upregulated, whereas 502 genes were downregulated during growth in the presence of the fecal extract. We grouped these genes into functional categories based on proposed functions using EggNOG. As shown in , among genes upregulated by the fecal extract, the two most altered functional categories were nucleotide transport and metabolism (29%), and translation, ribosomal structure, and biogenesis (28%). Among downregulated genes, cell motility (39%), and signal transduction mechanisms (37%) were the two most regulated functional categories (). Also, to identify gene categories whose genes were most affected by the fecal extract, an interaction network of proteins coded by genes whose expression was altered at least tenfold was constructed. We observed that among the 69 genes downregulated at least tenfold, 25 are involved in V. cholerae chemotaxis (). A network of proteins coded by genes upregulated at least tenfold by the fecal extract was also constructed and revealed that several of them are related to sulfur metabolism ().
Figure 2. Functional analysis of V. cholerae genes differentially expressed during growth in the presence of a human fecal extract
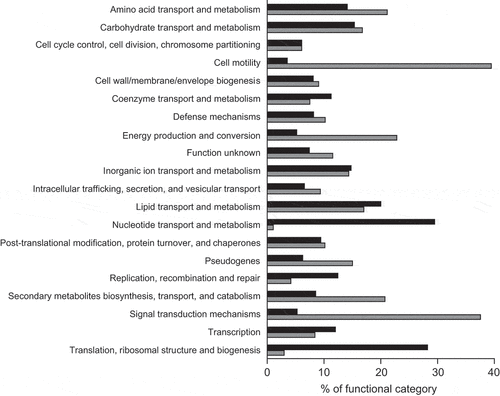
Figure 3. STRING analysis of predicted interactions between proteins encoded by genes differentially regulated by the fecal extract
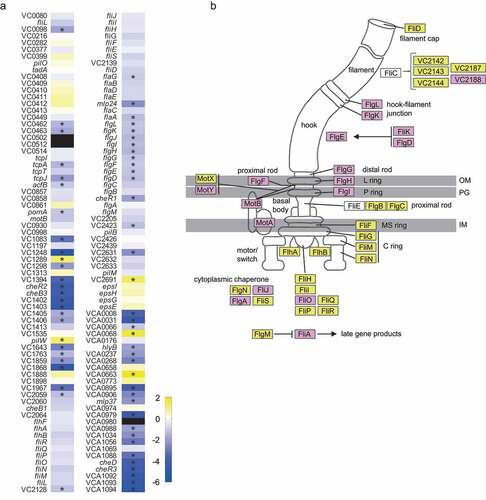
Given that there was a clear overrepresentation of motility and chemotaxis predicted functions within the group of genes regulated by the fecal extract, we next assessed the magnitude of the observed effect on motility and chemotaxis genes. shows a heatmap of 142 genes associated with motility, as predicted by EggNOG, of which 63 genes were significantly (≥2-fold, p< .05) affected. Among them are genes related to chemotaxis, mainly cluster I (cheR2, cheB3, VC1402, VC1403) and cluster III (cheD, cheR3, VCA1092, VCA1093, VCA1094) genes. In addition to genes related to chemotaxis, genes related to the flagellum were also modulated, including flaA, required for filament synthesis. depicts a flagellar model from KEGG where the effect (or lack thereof) of the fecal extract on the expression of V. cholerae genes is indicated, once again showing a marked repressive effect of the human fecal metabolome on flagellar expression.
Figure 4. Effect of the fecal extract on the expression of V. cholerae motility-associated genes
Bioactive molecules from the human gut inhibit V. cholerae motility and mucin penetration
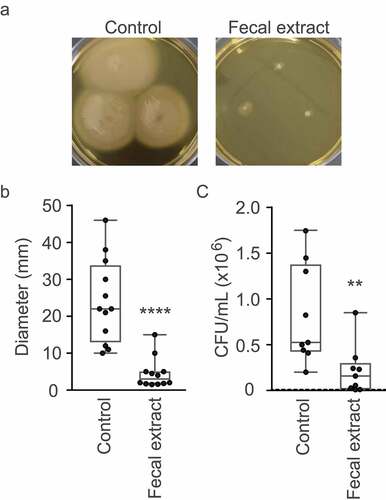
The RNAseq results described above suggested that the human gut metabolome contains molecules that may inhibit V. cholerae motility. To confirm this hypothesis, we performed phenotypic tests to assess swimming motility of V. cholerae when grown in the absence or presence of the fecal extract in BHI medium with 0.3% agar. As can be observed in and , the fecal extract severely inhibited V. cholerae motility, with a reduction of 5.3-fold in the diameter of motility halos in the presence of the fecal extract when compared to the control. This confirms that the modulation of motility and chemotaxis gene expression elicited by the fecal extract translated into a significantly reduced swimming capacity. We also investigated the effect of the fecal extract on mucin layer penetration in vitro. Motility is an important trait for enteric pathogens, as they must navigate the intestinal lumen to reach their preferred colonization niche. In the case of V. cholerae, it must traverse the thick mucus layer that covers the intestinal epithelium to reach and attach to host cells. Therefore, we measured the ability of V. cholerae cells to traverse a mucin column when exposed to the fecal extract. The number of bacteria recovered from the bottom of columns after 30 min of incubation was 15 times lower when V. cholerae was grown in the presence of the fecal extract (), thereby showing that bioactive molecules from the fecal metabolome significantly hamper the ability of V. cholerae to penetrate mucin.
Figure 5. Effect of the fecal extract on V. cholerae motility and mucin penetration
The human gut metabolome promotes V. cholerae biofilm formation
Planktonic and sessile bacterial behaviors are commonly subjected to shared regulatory control, where the expression of genetic requirements for one of these behaviors is usually associated with the repression of the other. Therefore, since the fecal extract strongly repressed V. cholerae motility, we investigated if the extract could affect V. cholerae biofilm formation. As shown in , V. cholerae growth in the presence of a fecal extract increased surface association and biofilm formation by 1.8-fold. However, total bacterial biomass (biofilm + planktonic) was slightly reduced by growth in the presence of the fecal extract (control: 0.843 ± 0.078; extract: 0.737 ± 0.089), indicating that the increase in biofilm formation observed was not due to an increase in growth.
Gut microbiota members produce bioactive molecules that modulate V. cholerae motility and mucin penetration
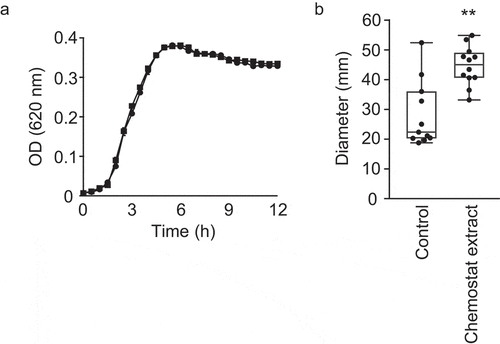
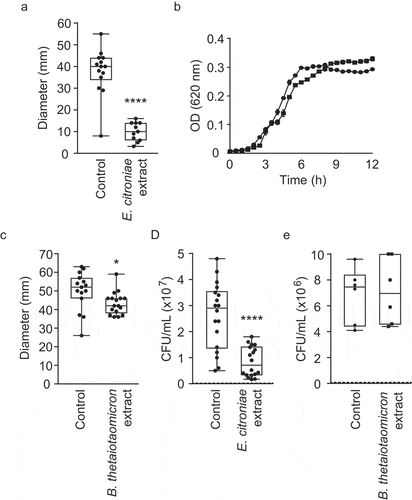
After determining that the human gut metabolome contains compounds that inhibit V. cholerae motility, we sought to establish whether the microbiota, and not the host, was responsible for this activity. Also, we sought to determine which component of the gut microbiota could be the source of the bioactive compound. For that purpose, we cultured a highly complex, defined microbial community containing 124 bacterial strains comprising 69 distinct species (Table S2) in a chemostat reactor designed to mimic the conditions encountered by the human gut microbiome.Citation33 After community stabilization, we extracted small molecules from this consortium and tested the effect of this extract on V. cholerae growth and swimming motility. V. cholerae growth was completely unaltered in the presence of this extract (). Interestingly, however, we found that the community extract caused a modest but statistically significant increase (1.6-fold) in V. cholerae swimming motility ().
Figure 7. Impact of chemostat extract on V. cholerae growth and motility in vitro.
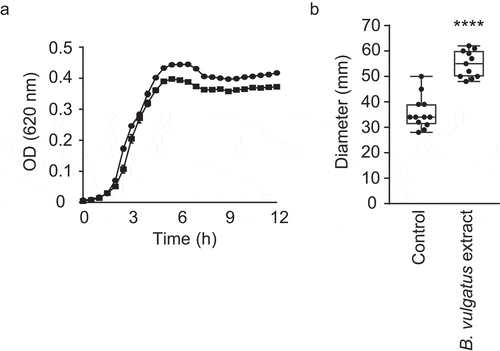
Given that a small-molecule extract from a complex microbial community induced V. cholerae motility, we sought to identify members of this community that could be involved in the induction of motility. Recently, You et al. showed that B. vulgatus has a critical role in protecting adult mice from V. cholerae infection, and that several microbiota-derived small molecules with growth-inhibitory activity against V. cholerae may be involved, supporting the notion that chemical interplay between the microbiota and V. cholerae is commonplace.Citation34 Although it is not the same strain as the one used by You and coworkers, the chemostat community described herein did contain a B. vulgatus strain. Therefore, we sought to determine if B. vulgatus produces small extracellular compounds that affect V. cholerae motility. Indeed, growth in the presence of a B. vulgatus extract caused a significant, 1.5-fold increase in V. cholerae motility, reminiscent of the increase caused by the extract from the complex community grown in the laboratory (1.6-fold; ). Again, V. cholerae growth was completely unaltered in the presence of this extract ().
Figure 8. Impact of small molecules produced by B. vulgatus on V. cholerae motility and growth
Although the presence of B. vulgatus in the defined community may explain its motility-enhancing effect, the negative impact of the human fecal extract on V. cholerae motility remained to be understood. Previously, we identified a specific member of the human gut microbiota, E. citroniae, that produces a bioactive compound when grown in pure culture that inhibits S. enterica host cell invasion gene expression.Citation11 Therefore, to test if E. citroniae could recapitulate the repression of V. cholerae motility observed with the fecal extract, we cultured one of the bioactive E. citroniae strains previously described (FM-V5-E) and tested the effect of secreted compounds on V. cholerae motility and growth. As can be seen in , E. citroniae metabolites led to potent inhibition (3.8-fold) of V. cholerae motility () but showed no significant effect on V. cholerae growth (). As a comparison, we tested extracts from similarly grown cultures of B. thetaiotaomicron (ATCC29741). This species was chosen because it is a highly-abundant, highly-prevalent member of the human gut microbiota.Citation35–37 Additionally, this species has been shown to produce small molecules with impact on enteric pathogen virulence gene expression.Citation13,Citation38 shows that, although statistically significant, the effect of a B. thetaiotaomicron extract on V. cholerae motility was negligible (1.2-fold). This phenotype was further confirmed by mucin penetration assays using both B. thetaiotaomicron and E. citroniae extracts. While B. thetaiotaomicron had virtually no impact on mucin penetration by V. cholerae, an E. citroniae extract inhibited mucin penetration 3.1-fold ( and ).
Figure 9. Impact of small molecules produced by E. citroniae and B. thetaiotaomicron on V. cholerae growth, swimming motility, and mucin penetration
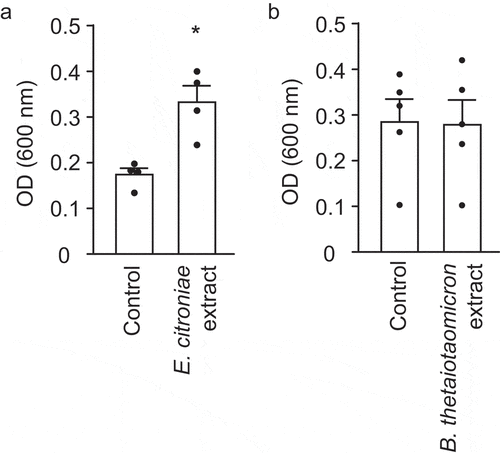
Biofilm formation analyses showed that B. thetaiotaomicron extracts do not alter the ability of V. cholerae to form biofilms on surfaces (), although E. citroniae extracts significantly (p< .05) increased biofilm formation by 1.9-fold (). However, total bacterial biomass (biofilm + planktonic) was unaltered by either extract (E. citroniae control: 0.751 ± 0.108; E. citroniae extract: 0.74 ± 0.043; B. thetaiotaomicron control: 0.763 ± 0.085; B. thetaiotaomicron extract: 0.753 ± 0.045;), indicating that the increase in biofilm formation was not due to an effect on growth.
Figure 10. Effect of E. citroniae and B. thetaiotaomicron extracts on V. cholerae biofilm formation
Statistical analyses
The results are reported as means with their respective standard errors. The results were analyzed using two-tailed Student´s t-test in GraphPad Prism 6. Differences were considered significant if the p value was less than 0.05.
Discussion
Bacteria rely heavily on small molecules to interact with their environment. Such compounds function as cues to inform cells of the conditions encountered in their surroundings. Also, they can perform dedicated signaling functions, and the production of and response to these signals – small compounds of diverse chemical properties – is used by most bacteria to regulate their behavior and that of neighboring cells.Citation39,Citation40 Interestingly, these compounds are also used for interkingdom communication, and examples of microbe-host interactions that occur through small molecules and signaling mechanisms continue to be reported.Citation41–44 Recently, Uchimura et al. demonstrated that there is an extensive penetration of a broad range of bacterial small molecules into host tissues, suggesting that the effects of microbial compounds on host physiology are likely to be much larger than currently appreciated.Citation7
Using metabolomics, we have previously shown that the human gut is teeming with small molecules, and that many of the molecules detected may represent new compounds.Citation45 Some of the metabolites produced by members of the gut microbiota have biological activity, exerting strong effects on pathogen virulence gene expression. Previously, we reported that small molecules present in organic extracts of human feces display antivirulence activity against S. enterica.Citation11 Interestingly, metabolites produced by strains of E. citroniae, a member of the gut microbiota, caused potent inhibition of invasion gene expression by this enteric pathogen. Other studies have shown that B. thetaiotaomicron, a predominant member of the human gut microbiota, can inhibit Shiga toxin production by enterohemorrhagic E. coli (EHEC) O157:H7.Citation13,Citation38 The Bacteroides genus represents one of the most important members of the human gut microbiota, and accounts for 10 to 30% of the total bacterial population.Citation35 These and other studies demonstrate that small molecule-dependent interactions between members of the human microbiota and invading pathogens may be prevalent, and that such interactions could be involved in the role played by the gut microbiota in protecting the host against enteric infections.
Here, we hypothesized that small molecules derived from the gut microbiota may have significant effects on V. cholerae. We began investigating this by analyzing the effect of small molecules from organic extracts of human feces on V. cholerae growth. The microbiota can directly inhibit the growth of diarrheal pathogens through inhibitory structures that depend on cell contact, such as the type VI secretory system or by secreting bacteriocins (antimicrobial peptides) or small molecules, such as secondary bile acids.Citation46–48 Our results show that small molecules extracted from feces of a healthy donor have a modest effect on V. cholerae growth (maximum of 36.6% growth reduction), suggesting that molecules with biological activity against this pathogen are present in the fecal metabolome, although the mechanism for this effect on growth is still unknown. This effect is somewhat similar to what was observed with S. enterica.Citation11 Interestingly, Silva et al. have previously studied the effect of fecal samples as well as bacterial strains cultured from a fecal sample on V. cholerae growth, and identified Peptostreptococcus sp. and Lactobacillus sp. that secreted unidentified metabolites that inhibited the growth of V. cholerae in vitro and in vivo.Citation49 Therefore, it is possible that the mild effect observed in our growth curves could be due to a microbiota-derived inhibitory compound, although further studies are required to support this hypothesis.
Transcriptome analyses revealed a pleiotropic effect of small molecules from human feces on V. cholerae gene expression, with almost 25% of the genome being differentially expressed in the presence of the fecal extract. Although the fecal metabolome is rich in chemical diversity, the magnitude of this effect on V. cholerae was unexpected, given that our similar study using S. enterica revealed differential expression of approximately 5% of the genome.Citation11 RNAseq results demonstrated an activation of the sulfate assimilation pathway in the presence of the fecal extract. Small-scale reduction of sulfate by this route is used by several organisms, not just those classified as sulfate reducers, being important for the synthesis of sulfur-containing biomolecules.Citation50 In a study by Sigalevich et al., it was observed that the transition from anaerobic to aerobic conditions affects the rate of sulfate reduction in Desulfovibrio oxyclinae.Citation51 Although V. cholerae is not a sulfate-reducing microorganism, we observed a significant increase in the expression of genes related to the assimilation of sulfate in the presence of the fecal extract. As this occurred during growth in an atmosphere rich in oxygen and under agitation, we hypothesize that some metabolites present in the extract may induce the activation of a pathway normally associated with an anaerobic environment, as the one seen in the gut.
As we have previously shown, some microbiota species produce molecules that can dampen virulence attributes of enteric pathogens. When S. enterica is grown in the presence of small molecules extracted from human feces, the expression of genes coding for one of its type-3 secretion systems, which are essential for its ability to invade host cells and colonize the intestine, is reduced. The ability to invade human epithelial cells in vitro is also decreased in the presence of a human fecal extract.Citation11 A key component of the early phase of intestinal colonization by V. cholerae is motility, as well as the associated chemotaxis.Citation52 In this work, we show that 44% of genes involved in V. cholerae motility were downregulated in the presence of the fecal extract. Understanding the precise role of chemotaxis in V. cholerae colonization is complicated by the presence of three chemotaxis operons (cluster I, II and III) and at least 45 methyl-accepting chemotaxis proteins, MCPs.Citation53 Only one of the chemotaxis operons (cluster II) is required for standard chemotaxis.Citation54 The function of the other two chemotaxis operons is not presently known. In this work, genes belonging to the three clusters were affected by the fecal extract, with inhibition, although weak, of cluster II gene expression. Many intestinal pathogens rely on chemotaxis, amongst many other factors, to aid in their virulence strategies, and chemotaxis may increase pathogen fitness and virulence in the gut.Citation55 Phenotypic analysis to verify if the downregulation of V. cholerae motility genes observed affected its swimming ability showed that the fecal extract drastically reduces motility in vitro.
In a previous study, Klose and Mekalanos (1998) showed that although V. cholerae has five flagellins – FlaA, FlaB, FlaC, FlaD, and FlaE – only FlaA is essential for motility; a strain containing a mutation in flaA does not produce a flagellum and is therefore nonmotile.Citation56 By RNAseq analysis we observed that flaA was repressed in the presence of the fecal extract, which may be one of the reasons for the reduction of V. cholerae motility in the presence of this extract. In addition, other flagellar genes important for bacterial locomotion were also downregulated, such as the motor genes motX, motA, and motB.
The impact of small molecules produced by specific components of the gut microbiota on V. cholerae motility was also analyzed. Our results showed that small molecules from different bacterial species have different effects on V. cholerae motility. While B. thetaiotaomicron extracts did not elicit significant effects on V. cholerae, B. vulgatus extracts significantly increased motility. You et al. recently described a B. vulgatus strain from the murine microbiota that displayed a protective effect on host susceptibility against V. cholerae infection.Citation34 As the authors showed, B. vulgatus suppressed V. cholerae colonization of the adult mouse intestine, and they attributed this effect to the production of inhibitory molecules. In our work, we did not observe an effect of B. vulgatus on V. cholerae growth. However, swimming motility was increased in the presence of the extract, a phenotype that was also observed when V. cholerae was cultured in the presence of an extract from a complex microbial community cultured in vitro.
The induction of V. cholerae motility by extracts from the 124-strain defined chemostat community was unexpected. We have previously shown that in vitro complex communities are able to recapitulate the impact of the human fecal metabolome on S. enterica virulence gene expression.Citation11 Therefore, we set out to reconstitute a complex and diverse in vitro community to test its effect on V. cholerae motility, expecting that a phenotype similar to the one obtained with fecal extracts would be observed. Aside from the fact that S. enterica and V. cholerae will likely have different regulatory mechanisms involved in the response to microbiome-derived small molecules, one of the differences between the two experiments is that in the case of S. enterica chemostats were inoculated with human feces, whereas isolated strains were used to assemble the defined community described herein. Therefore, it is likely that the former communities were a better representation of those found in the human gut. In that sense, it is possible that the community described herein lacked strains with the ability to repress V. cholerae motility. Although the defined community tested did contain an E. citroniae strain, this is not the same strain previously shown to have activity against S. enterica and shown here to inhibit V. cholerae motility (FM-V5-E). We chose not to use FM-V5-E in the chemostat community because during the establishment of complex microbial communities different strains co-adapt to maintain community structure and function. Therefore, we assembled a community of species and strains isolated from the same donor. As previously shown, communities assembled using strains from different donors can lose some of its members and generate a metabolic output that is significantly different from communities assembled using strains from the same donor.Citation57 Therefore, inserting FM-V5-E in a community composed of strains from a different donor would have unpredictable effects.
Another potential reason for the discrepancy between results obtained with fecal versus chemostat extracts is that the chemostat community may contain strains (B. vulgatus strains and others) whose motility-inducing effects counteract the inhibitory effect observed with E. citroniae. Also, although complex and diverse, it is important to stress that the microbial community grown in the chemostat was composed of 69 different species selected from a collection of strains previously isolated from a healthy donor. Therefore, they do not fully represent the microbial diversity found in fecal samples. Further supporting this notion, we showed that, although fecal extracts moderately inhibited V. cholerae growth, chemostat extracts have virtually no effect on growth. Therefore, it is clear that the microbial and chemical composition of these two matrices (fecal vs. chemostat extracts) is significantly different. The difference observed in their ability to inhibit V. cholerae growth likely comes from the fact that fecal extracts will inevitably display higher chemical complexity. For instance, chemostat extracts probably lack various host factors that are likely present in fecal extracts, such as antimicrobial peptides. Thus, the absence of certain strains and compounds in the chemostat community can have unpredictable effects on the net biological activity of the chemostat extracts. Altogether, these data suggest that the effect of the fecal extract on V. cholerae motility observed is likely to represent the combined effect of multiple bioactive compounds whose ‘messages’ are somehow integrated into V. cholerae regulatory circuits. Nevertheless, we were able to detect a bacterial species that, when grown in pure culture, could recapitulate the effect of the fecal extract. Although E. citroniae is a minor member of the human gut microbiota, it has strong antivirulence activity against S. enterica. Whether the same molecule exerts the observed effects on S. enterica and V. cholerae remains to be determined. It is also important to point out that E. citroniae may not be the exact bacterial species responsible for the bioactivity of the fecal extract. It is possible, and very likely, that other strains present in the human gut microbiota will have motility-inhibiting activities, although this remains to be determined. Nevertheless, our results using E. citroniae establish that members of the microbiota can inhibit V. cholerae motility independently of host factors and will facilitate the purification and identification of the bioactive compound.
Bacterial chemotaxis and motility are often required for bacterial pathogenesis, as these processes allow cells to reach their niche, detect nutrients and sense cues from other bacteria or the host.Citation58 Thus, bioactive molecules that can inhibit these processes may hold therapeutic promise, since they may prevent the establishment of infections. Studies with V. cholerae have shown that, to cause infection, cells need to swim through the intestinal lumen, traverse the mucus layer, and then colonize epithelial cells.Citation32 The ability of V. cholerae to penetrate mucin is impaired in the presence of fecal and E. citroniae extracts, suggesting that small molecules present in these extracts may affect host colonization. Hindrance of V. cholerae virulence by these small molecules may allow the use of such molecules in the development of therapeutics. As an alternative, strains that produce the bioactive compound could be used as probiotics to lower burden of V. cholerae infection and therefore prevent or treat cholera. Such strains could be either gut microbiota isolates from healthy subjects or engineered strains with desirable properties. Interestingly, antivirulence activity of the probiotic fermented beverage Kombucha against V. cholerae has been shown. In this case, small molecules present in the beverage were shown to inhibit motility, alter motility gene expression and inhibit mucin penetration in vitro.Citation59 The idea of developing new therapeutic alternatives for cholera based on the gut microbiota is particularly enticing in the context of this work, since the V. cholerae strain used herein is resistant to multiple antibiotics, and a non-antibiotic therapeutic intervention would be widely useful during infections with drug-resistant organisms. However, further studies will be required to address the therapeutic value of such compounds, and to reveal the details of interspecies interactions in the human microbiota and how they contribute to the balance between host colonization resistance and susceptibility.
Supplemental Material
Download Zip (1.5 MB)Acknowledgments
We thank the anonymous reviewers for their careful assessment of the manuscript and constructive criticism. We thank Ana Carolina Vicente and the Bacteria Culture Collection of Environment and Health at Fundação Oswaldo Cruz for providing the V. cholerae strain used in this study. We are also grateful to Plataforma de Sequenciamento de Ácidos Nucleicos de Nova Geração–RPT01J and Plataforma de Bioinformática–RPT04A (Rede de Plataformas Tecnológicas FIOCRUZ). This work was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001, Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) as well as the Fundação Oswaldo Cruz Inova Fiocruz/VPPCB Program.
Disclosure statement
Author E. A.-V. is co-founder and CSO of NuBiyota, a company seeking to create ‘microbial ecosystem therapeutics’ for the treatment of disease in humans.
Supplementary Material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Possemiers S, Grootaert C, Vermeiren J, Gross G, Marzorati M, Verstraete W, De Wiele T. The intestinal environment in health and disease – recent insights on the potential of intestinal bacteria to influence human health. Curr Pharm Des. 2009;15(18):2051–19. doi:10.2174/138161209788489159.
- Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152(1–2):39–50. doi:10.1016/j.cell.2012.10.052.
- Kell D. Metabolomics and systems biology: making sense of the soup. Curr Opin Microbiol. 2004;7(3):296–307. doi:10.1016/j.mib.2004.04.012.
- Han J, Danell RM, Patel JR, Gumerov DR, Scarlett CO, Speir JP, Parker CE, Rusyn I, Zeisel S, Borchers CH. Towards high-throughput metabolomics using ultrahigh-field Fourier transform ion cyclotron resonance mass spectrometry. Metabolomics. 2008;4(2):128–140. doi:10.1007/s11306-008-0104-8.
- Beebe K, Sampey B, Watkins SM, Milburn M, Eckhart AD. Understanding the apothecaries within: the necessity of a systematic approach for defining the chemical output of the human microbiome. Clin Transl Sci. 2014;7(1):74–81. doi:10.1111/cts.12131.
- Han J, Antunes LCM, Finlay BB, Borchers CH. Metabolomics: towards understanding host–microbe interactions. Future Microbiol. 2010;5(2):153–161. doi:10.2217/fmb.09.132.
- Uchimura Y, Fuhrer T, Li H, Lawson MA, Zimmermann M, Yilmaz B, Zindel J, Ronchi F, Sorribas M, Hapfelmeier S, Ganal-Vonarburg SC, de Agüero MG, McCoy KD, Sauer U, Macpherson AJ. Antibodies set boundaries limiting microbial metabolite penetration and the resultant mammalian host response. Immunity. 2018;49(3):545–559. doi:10.1016/j.immuni.2018.08.004.
- Postler TS, Ghosh S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab. 2017;26(1):110–130. doi:10.1016/j.cmet.2017.05.008.
- Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, Felin J, Perkins R, Borén J, Orešič M, Bäckhed F. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2010;51(5):1101–1112. doi:10.1194/jlr.M002774.
- Donia MS, Fischbach MA. Small molecules from the human microbiota. Science (80-). 2015;349(6246):1254766. doi:10.1126/science.1254766.
- Antunes LCM, McDonald JAK, Schroeter K, Carlucci C, Ferreira RBR, Wang M, Yurist-Doutsch S, Hira G, Jacobson K, Davies J, Allen-Vercoe E, Finlay BB. Antivirulence activity of the human gut metabolome. MBio. 2014;5(4):e01183–14. doi:10.1128/mBio.01183-14.
- Peixoto RJM, Alves ES, Wang M, Ferreira RBR, Granato A, Han J, Gill H, Jacobson K, Lobo LA, Domingues RMCP, Borchers CH, Davies JE, Finlay BB, Antunes LCM. Repression of Salmonella host cell invasion by aromatic small molecules from the human fecal metabolome. Appl Environ Microbiol. 2017;83(19):e01148–17. doi:10.1128/AEM.01148-17.
- De Sablet T, Chassard C, Bernalier-Donadille A, Vareille M, Gobert AP, Martin C. Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2009;77(2):783–790. doi:10.1128/IAI.01048-08.
- Theriot CM, Young VB. Microbial and metabolic interactions between the gastrointestinal tract and Clostridium difficile infection. Gut Microbes. 2014;5(1):86–95. doi:10.4161/gmic.27131.
- Carlson-Banning KM, Sperandio V. Enterohemorrhagic Escherichia coli outwits hosts through sensing small molecules. Curr Opin Microbiol. 2018;41:83–88. doi:10.1016/j.mib.2017.12.002.
- Silva AJ, Benitez JA. Vibrio cholerae biofilms and cholera pathogenesis. PLoS Negl Trop Dis. 2016;10(2):e0004330. doi:10.1371/journal.pntd.0004330.
- Cobaxin M, Martínez H, Ayala G, Holmgren J, Å S, Sánchez J. Cholera toxin expression by El Tor Vibrio cholerae in shallow culture growth conditions. Microb Pathog. 2014;66:5–13. doi:10.1016/j.micpath.2013.11.002.
- Louis P, O’Byrne CP. Life in the gut: microbial responses to stress in the gastrointestinal tract. Sci Prog. 2010;93(1):7–36. doi:10.3184/003685009X12605525292307.
- Kim S, Covington A, Pamer EG. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol Rev. 2017;279(1):90–105. doi:10.1111/imr.12563.
- Ott SJ, Waetzig GH, Rehman A, Moltzau-Anderson J, Bharti R, Grasis JA, Cassidy L, Tholey A, Fickenscher H, Seegert D, Rosenstiel P, Schreiber S. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology. 2017;152(4):799–811. doi:10.1053/j.gastro.2016.11.010.
- Marin MA, Fonseca EL, Andrade BN, Cabral AC, Vicente ACP. Worldwide occurrence of integrative conjugative element encoding multidrug resistance determinants in epidemic Vibrio cholerae O1. PLoS One. 2014;9(9):e108728. doi:10.1371/journal.pone.0108728.
- Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, Brown EM, Schroeter K, Allen-Vercoe E. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013;1(1):3. doi:10.1186/2049-2618-1-3.
- McDonaldJAK, Schroeter K, Fuentes S, Heikamp-dejong I, Khursigara CM, De Vos WM, Allen-Vercoe E. Evaluation of microbial community reproducibility, stability and composition in a human distal gut chemostat model. J Microbiol Methods. 2013;95(2):167–174. doi:10.1016/j.mimet.2013.08.008.
- Glatthardt T, Campos JCM, Chamon RC, de Sá Coimbra TF, Rocha GDA, de Melo MAF, Parente TE, Lobo LA, Antunes LCM, dos Santos KRN, Ferreira RBR. Small molecules produced by commensal Staphylococcus epidermidis disrupt formation of biofilms by Staphylococcus aureus. Appl Environ Microbiol. 2020; 86:e02539–19
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–359. doi:10.1038/nmeth.1923.
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi:10.1038/nbt.1621.
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. doi:10.1038/nbt.2450.
- Goff L, Trapnell C, Kelley D. cummeRbund: analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. R Packag. version 2.30.0.2020.
- Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, von Mering C, Bork P. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47(D1):D309–14. doi:10.1093/nar/gky1085.
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, von Mering C. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–13. doi:10.1093/nar/gky1131.
- Liu Z, Miyashiro T, Tsou A, Hsiao A, Goulian M, Zhu J. Mucosal penetration primes Vibrio cholerae for host colonization by repressing quorum sensing. Proc Natl Acad Sci. 2008;105(28):9769–9774. doi:10.1073/pnas.0802241105.
- Yen S, Jak M, Schroeter K, Oliphant K, Sokolenko S, Blondeel EJM, Allen-Vercoe E, Aucoin MG. Metabolomic analysis of human fecal microbiota: a comparison of feces-derived communities and defined mixed communities. J Proteome Res. 2015;14(3):1472–1482. doi:10.1021/pr5011247.
- You JS, Yong JH, Kim GH, Moon S, Nam KT, Ryu JH, Yoon MY, Yoon SS. Commensal-derived metabolites govern Vibrio cholerae pathogenesis in host intestine. Microbiome. 2019;7(1):132. doi:10.1186/s40168-019-0746-y.
- Hold GL, Pryde SE, Russell VJ, Furrie E, Flint HJ. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol Ecol. 2002;39(1):33–39. doi:10.1111/j.1574-6941.2002.tb00904.x.
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. The long-term stability of the human gut microbiota. Science (80-). 2013;341(6141):1237439. doi:10.1126/science.1237439.
- Zitomersky NL, Coyne MJ, Comstock LE. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect Immun. 2011;79(5):2012–2020. doi:10.1128/IAI.01348-10.
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science (80-). 2005;308(5728):1635–1638. doi:10.1126/science.1110591.
- Antunes LCM, Ferreira RBR, Buckner MMC, Finlay BB. Quorum sensing in bacterial virulence. Microbiology. 2010;156(8):2271–2282. doi:10.1099/mic.0.038794-0.
- Antunes LCM, Ferreira RBR. Intercellular communication in bacteria. Crit Rev Microbiol. 2009;35(2):69–80. doi:10.1080/10408410902733946.
- McCarville JL, Chen GY, Cuevas VD, Troha K, Ayres JS. Microbiota metabolites in health and disease. Annu Rev Immunol. 2020;38(1):147–170. doi:10.1146/annurev-immunol-071219-125715.
- Bosi A, Banfi D, Bistoletti M, Giaroni C, Baj A. Tryptophan metabolites along the microbiota-gut-brain axis: an interkingdom communication system influencing the gut in health and disease. Int J Tryptophan Res. 2020;13:1–25. doi:10.1177/1178646920928984.
- Kumar A, Russell RM, Pifer R, Menezes-Garcia Z, Cuesta S, Narayanan S, MacMillan JB, Sperandio V. The serotonin neurotransmitter modulates virulence of enteric pathogens. Cell Host Microbe. 2020;28(1):41–53. doi:10.1016/j.chom.2020.05.004.
- Moreira CG, Sperandio V. The epinephrine/norepinephrine/autoinducer-3 interkingdom signaling system in Escherichia coli O157:H7. Adv Exp Med Biol. 2016;874:247–261.
- Antunes LCM, Han J, Ferreira RBR, Lolić P, Borchers CH, Finlay BB. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother. 2011;55(4):1494–1503. doi:10.1128/AAC.01664-10.
- Cg B,Buffie CG , Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–208. doi:10.1038/nature13828.
- Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16(2):227–236. doi:10.1016/j.chom.2014.07.007.
- Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526(7575):719–722. doi:10.1038/nature15524.
- Silva SH, Vieira EC, Dias RS, Nicoli JR. Antagonism against Vibrio cholerae by diffusible substances produced by bacterial components of the human faecal microbiota. J Med Microbiol. 2001;50(2):161–164. doi:10.1099/0022-1317-50-2-161.
- Rückert C. Sulfate reduction in microorganisms — recent advances and biotechnological applications. Curr Opin Microbiol. 2016;33:140–146. doi:10.1016/j.mib.2016.07.007.
- Sigalevich P, Meshorer E, Helman Y, Cohen Y. Transition from anaerobic to aerobic growth conditions for the sulfate-reducing bacterium Desulfovibrio oxyclinae results in flocculation. Appl Environ Microbiol. 2000;66(11):5005–5012. doi:10.1128/AEM.66.11.5005-5012.2000.
- Butler SM, Camilli A. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc Natl Acad Sci. 2004;101(14):5018–5023. doi:10.1073/pnas.0308052101.
- Peterson KM, Gellings PS. Multiple intraintestinal signals coordinate the regulation of Vibrio cholerae virulence determinants. Pathog Dis. 2018;76(1):126. doi:10.1093/femspd/ftx126.
- Boin MA, Austin MJ, Häse CC. Chemotaxis in Vibrio cholerae. FEMS Microbiol Lett. 2004;239(1):1–8. doi:10.1016/j.femsle.2004.08.039.
- Matilla MA, Krell T. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol Rev. 2018;42:40–67.
- Klose KE, Mekalanos JJ. Differential regulation of multiple flagellins in Vibrio cholerae. J Bacteriol. 1998;180(2):303–316. doi:10.1128/JB.180.2.303-316.1998.
- Oliphant K, Parreira VR, Cochrane K, Allen-Vercoe E. Drivers of human gut microbial community assembly: coadaptation, determinism and stochasticity. Isme J. 2019;13(12):3080–3092. doi:10.1038/s41396-019-0498-5.
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5(12):1024–1037. doi:10.1038/nrm1524.
- Bhattacharya D, Sinha R, Mukherjee P, Howlader DR, Nag D, Sarkar S, Koley H, Withey JH, Gachhui R. Anti-virulence activity of polyphenolic fraction isolated from kombucha against Vibrio cholerae. Microb Pathog. 2020;140:103927. doi:10.1016/j.micpath.2019.103927.