ABSTRACT
Cardiovascular diseases (CVDs) still remain the leading concern of global health, accounting for approximately 17.9 million deaths in 2016. The pathogenetic mechanisms of CVDs are multifactorial and incompletely understood. Recent evidence has shown that alterations in the gut microbiome and its associated metabolites may influence the pathogenesis and progression of CVDs such as atherosclerosis, heart failure, hypertension, and arrhythmia, yet the underlying links are not fully elucidated. Owing to the progress in next-generation sequencing techniques and computational strategies, researchers now are available to explore the emerging links to the genomes, transcriptomes, proteomes, and metabolomes in parallel meta-omics approaches, presenting a panoramic vista of culture-independent microbial investigation. This review aims to outline the characteristics of meta-omics pipelines and provide a brief overview of current applications in CVDs studies which can be practical for addressing crucial knowledge gaps in this field, as well as to shed its light on cardiovascular risk biomarkers and therapeutic intervention in the near future.
Introduction
Cardiovascular diseases (CVDs) remain the leading cause of mortality and morbidity worldwide. It was estimated that 17.9 million people died from CVDs in 2016, representing approximately 30% of all global deaths.Citation1 Despite adherence to recommended therapies including high-dose statins and modification of lifestyle risk factors, curative effect is individual and varied, as well as significant residual risks remain.Citation2,Citation3 Additionally, although genetic predisposition contributes to the pathogenesis of CVDs, genetic variation could only explain 20% of the risk in disease progression.Citation4 Together, these observations suggest that the multiple mechanisms that participate in the pathophysiology of CVDs are incompletely understood, convincing data from biochemical, bioinformatical, epidemiological, and clinical studies will enhance our feasibility in CVDs prophylactic managements.
Within the last decade, research regarding the function and composition of the human gut microbiome has come to be a strong evolutionary force. The human gastrointestinal tract is populated by trillions of microorganisms including bacteria, viruses, fungi, and protozoa, which along with their genome, are collectively referred as the gut microbiome.Citation5 The gut microbiome is a complex community that interacts with host dynamic functions such as processing nutrients and modulation of the immune system which are essential to homeostasis maintenance.Citation6 An alteration in gut microbial composition and linked activity is known as gut dysbiosis,Citation7 and in-depth characterization of the human microbiome by meta-omics analysis spurred a paradigm shift in human health and disease without the need for culturing.Citation8 It has been demonstrated that microbial communities may be associated with the pathogenesis of several human diseases including obesity,Citation9,Citation10 inflammatory bowel disease,Citation11,Citation12 cancer,Citation13,Citation14 and rheumatic disease;Citation15,Citation16 moreover, increasing evidence has shown that changes in the gut microbiome and its associated metabolites may influence the pathogenesis and progression of CVDs such as atherosclerosis, heart failure, hypertension, and arrhythmia.Citation17,Citation18 However, the underlying mechanistic links are multifactorial and yet to be determined.
The purpose of this review is to provide a brief overview of recent progress in our understanding of gut microbiome in the development of CVDs and to outline meta-omics approaches that can be practical for addressing crucial knowledge gaps in this field. Together, this body of work supports the notion that the gut microbiome may shed its light on cardiovascular risk biomarkers and therapeutic intervention in the near future.
Gut meta-omics algorithms and current applications in CVDs
While isolating and culturing each individual microorganism is an intractable task, our capability to accurate characterization and quantification these microbial communities and perceive their impact on human biology from the collective nucleotide contents contained in a stool sample has been transformed by progress in technology, particularly next-generation sequencing (NGS) techniques and computational strategies. These advances have opened up a panoramic vista of untangling the emerging links to mapping the genomes, transcriptomes, proteomes, and metabolomes in parallel, presenting an exciting future field of investigation, especially considering the reality that gut microorganisms play a key role in plasticity and adaptation to environmental change and physiological stress. Here we summarized a set of pipelines for properly performing meta-omics studies (), which can provide information from different layers of the biosystem.
Figure 1. Meta-omics algorithm for investigating the host–microbiome interactions
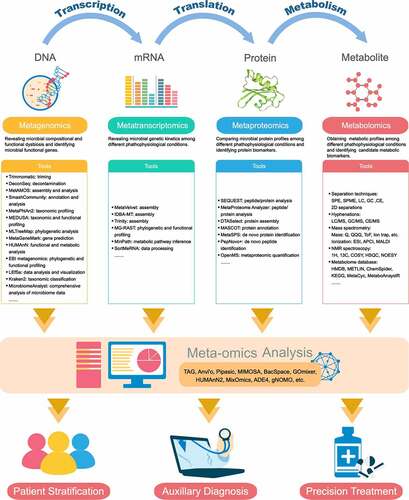
Metagenomics: revealing the diversity and function of the human gut microbiome
The composition of human-associated microbial communities has been well assessed by means of culture-independent sequencing technologies, for example by sequencing the 16S ribosomal ribonucleic acid (rRNA) genes to yield phylogenetic or taxonomic profiles on account of its applicability in prokaryotes and availability of considerable reference database, giving insight into non-cultivated microorganisms.Citation19 The amplicon-based (marker gene) technologies are limited in their phylogenetic ranges;Citation20 for example, 16S primarily targets in bacteria, whereas 18S in eukaryotes, and internal transcribed spacer (ITS) typically in fungi. Besides, these technologies also suffer from biases from copy-number variations,Citation21 amplification inefficacy,Citation22 and inconsistent characterization.Citation23,Citation24 More recently, genome-wide sequencing approaches, such as metagenomics, have further expanded the popularity and effectiveness for studying the microbiome. Metagenomics is the method of generating millions of short reads (mostly performed using Illumina platforms or Roche 454 pyrosequencing) that randomly sampled from all microbial genomes present in a defined environment or ecosystem, including viral and eukaryotic DNA. Based on adequate sequencing depth (the number of sequencing reads per sample), the taxonomic resolution to species or strain level by aligning reads to annotation reference genomes, and the assembly of whole microbial genomes from the overlapped DNA sequence reads (contigs) are possible.Citation25,Citation26 The complete genome of dominantly abundant microbes can also be reconstructed,Citation27,Citation28 providing a more comprehensive view of their biological potential. Adding a higher level of refinement, metagenomic operational taxonomic units (mOTUs) are clustered as groups of covariance single-copy marker genes, providing a powerful approach to classify and quantify known and currently unknown microorganisms at species-level resolution.Citation29 Additionally, microbial growth rates can be extrapolated from the metagenomic sequencing read coverage, revealing the rates at which bacteria undergo DNA replication in each sample, which could provide insights into pathology.Citation21
Gut microbial communities can be recognized not only as independent groups of microbes, but as collections of biochemical functions affecting the host physiology. Remarkably, the bioinformatics analysis of metagenomics sequences, so-called annotation, enables researchers to profile major metabolic pathways and functional modules carried by an entire community that of clinical interest at the gene level.Citation30 Biological interpretation of genome sequences is computationally annotated and evaluated by homology using reference databases such as COG,Citation31 KEGG,Citation32 or eggNOG.Citation33 Alternatively, raw sequence reads can be directly utilized to generate functional profiles (using pipelines such as EBI metagenomicsCitation34 and HUMAnNCitation30 or by mapping them to higher-quality catalogs of reference genes in the human gut microbiome).Citation35 It is important to emphasize that gene-scale functional profiling is still in its infancy; it provides permissive links between the microbial organismal compositions and metabolic potential, not the metabolic activity per se. However, these profiles are appropriate as inputs into more sophisticated metabolic activity networks and molecular mechanisms, allowing us to obtain more comprehensive information from annotated genomes, and making this integration more biologically meaningful.Citation36
The application of metagenomics has massive potentiality in revealing the interactions and mechanisms between the gut microbiome and diseases. However, it also has limitations and requires improvements. First, as metagenomics requires a much higher average genome size than 16S rDNA sequence analysis,Citation37 the involved costs and time are relatively far greater. Second, quality control protocols include human DNA contamination removing are essential to obtain high coverage required for metagenomics.Citation38 Third, different DNA extraction kits, as well as laboratory sampling and storage protocols, also have biases on the assessment of microbial configuration.Citation39 Comparing data across studies applying different bacterial DNA extraction methodologies is challenging.Citation40 Fourth, a proportion of data cannot be assigned due to the insufficient reference databases, particularly a lack of matches for eukaryotes and viruses, which impacts the extraction of organismal and functional information.Citation41 Thus, future advances in the study of the human gut microbiome using metagenomics are promising and more efforts are urgently needed.
Metatranscriptomics, metaproteomics and metabolomics: enormous complements to characterize microbial communities
Metagenomics provides us with an unprecedented tool that can be used to access the genetic potential and metabolic features of the microorganisms that exist in our biosphere. However, the abundances of metagenomically prevalent pathways inferred by the pipelines describe only the enzymes encoded by one or more microbial genomes, and the relationships to gene expression, protein abundance, and metabolite concentration may not be straightforward. With the rapidly growing area of metatranscriptomics,Citation42 metaproteomics,Citation43 and metabolomics,Citation44 experimental, computational, and conceptual connections between the meta-omics have started to put forward a better understanding of the dynamic gut ecosystem.
A metatranscriptomic assay routinely involves reverse transcription and cDNA sequencing of high-quality and sufficient amounts of RNA isolated from a microbiome sample. Such measurements of gene expression at the community level yield important information regarding gene expression patterns induced/repressed under specific conditions.Citation45 Additionally, identifying genomes with active transcription can inherently discriminate metabolically active microbes from dormant or dead microbes and extracellular DNA.Citation46 However, due to the ubiquitous presence of RNases in host-derived samples, it is also challenging to handle the laborious and complex procedures including sample collection, storage, and preparation when performing metatranscriptomic analysis.Citation47 In contrast with metagenomics and metatranscriptomics, metaproteomics can not only identify and quantify the expressed proteins present in a sample by the development of higher resolution mass spectrometers and quantitative proteomics techniques, but link the proteins to closely related species/strains of microorganisms via reference databases and the bioinformatic tools, which should in principle provide superior insight into taxon-specific functional activities and metabolic pathways, since not all transcripts are subsequently translated into proteins.Citation48 Metaproteomics is not yet a mature area and can suffer from a variety of drawbacks including insufficient universal guidelines and protocols for performing metaproteomic experiments and interpreting metaproteomic results.Citation49 In addition to the microbiome’s functional activity, a further goal of microbiome research is to measure the levels and flux of metabolites. Metabolomics is uniquely suited to capture these functional host–microbe interactions via mass spectroscopy (MS), nuclear magnetic resonance (NMR), or other analytical methods. Metabolomics research strategies can be conducted in a targeted or non-targeted manner. Targeted metabolomics can accurately quantify a set of known metabolites, while non-targeted methods can cover as many metabolites as possible.Citation50,Citation51 In fact, many gut metabolites may be of microbial origin or microbially modified,Citation52 typically result in a mutualistic relationship between microbe and host, highlighting the necessity to correlate specific microbes or genetic components with the specific molecules that, in turn, mediate human health phenotypes. Metabolomics shares similar weaknesses of incomplete databases, and remains intricate to distinguish host- and microbiome-origin metabolites and link the metabolites to specific taxa.Citation53
Despite these challenges, the future potential for multi-omics data integration represents also opportunities. Multi-omics approaches provide additional insights beyond the capacity of single omics studies, illuminating gene regulation and correlating the presence of microbes with metabolites. Correlation analysis and multivariate statistical methods are the most straightforward and commonly used methods for statistically studying the interconnected relationships,Citation54,Citation55 as well as identifying the key features in multi-omics data integration,Citation56 so that the functional phenotype of gut microorganisms can be inferred. The development of computational methods that complement experimental approaches will likely revolutionize our ability to integrate and interpret multi-omics data and help the research transition into the mechanism.Citation57 Within this context, multi-omics analysis of microbiome with efficiency and quality will become a sufficiently powerful tool to elucidate the ecologic roles of the human gut microbiome.
Sequencing measurements revealed altered gut microbial composition associated with CVDs
Epidemiological studies have revealed a series of associations between alterations in the organismal composition of the gut microbiome and cardiovascular traits. In , we summarize recent findings from clinical cohorts showing associations between gut microbes and CVDs in humans. From these studies, a discovery pipeline for identifying common and distinctive microbial players in CVDs emerges.
Table 1. Altered Gut Microbiome Composition Associated with CVDs in humans
Current evidence mainly shows the associations between gut microbiome configurations and susceptibility to CVDs, while the discrepant results of these studies are obvious due to the various factors that contribute to gut microbiome composition (e.g., subjects toward a region-specific genetic background and dietary habits; different sequencing strategies, etc.). A beneficial spin-off from these massive sequencing projects has been their deposition into online accessible databases, which has enabled researchers to leverage and reuse these data to create reference databases and conduct further studies. Importantly, several studies have also taken advantage of machine learning algorithms to either differentiate between health and different disease states or identify microbe-related features that predict clinical outcomes,Citation67,Citation70,Citation88 which would lend a hand to push the frontiers of translational microbiome research. Given that CAD like other common complex disorders develops over time and involves both genetics and environmental risk factors, full mechanistic insight will allow the generation of reliable prediction models based on coordinated sets of microbial data at multiple time points, especially when acute clinical outcomes (e.g., stable CAD vs. myocardial infarction) need to be modeled.Citation89
Nevertheless, the ability to extract precise insights of the interplay between the gut microbiome and CVDs from published research has so far been restricted due to lack of appropriate data in large enough cohorts, as well as flexibility, effectiveness, and robustness of data integration. As the gut microbiome is multiplex, dynamic, and spatially structured, as well as the fact that biological processes do not operate separately but manifest conjointly as molecular cascades and interact across multidimensional omics domains to affect CVDs etiology, it is progressively recognized that specialize in any explicit form of information solely provides limited knowledge into the underlying molecular mechanisms and CVDs phenotypes. Considering the great promise such studies hold for the treatment and prevention for CVDs, it is expected that parallel measurements on multi-omics will be rapidly collected during the next couple of years, and novel methods with standard pipelines for efficient data integration, modeling, visualization, and interpretation will be urgently needed to efficiently cope with this multi-dimensional data,Citation90 and transform into practicable precision medicine tools.
Gut-heart axis: the interplay between the microbiome and dietary-derived metabolites affects the pathogenesis of CVDs
The gut microbiome is known to aid in the harvest of nutrients from diet, with about half of the total genome of the gut microbiome being related to the metabolism of central carbon and amino acid as well as the biosynthesis of secondary metabolites.Citation91 Dietary components that are unutilized by the host can serve as growth nutrient substrates for the gut microbes, which in turn generate myriad molecules, many of which impact the host physiology,Citation92 functioning as an endocrine organ.Citation93 Through the development of metabolomics techniques and efforts in a series of experiments, there is increasing evidence that some of these metabolites contribute to CVDs pathology ().
Figure 2. Downstream effects of microbe-derived metabolites and their roles in CVDs
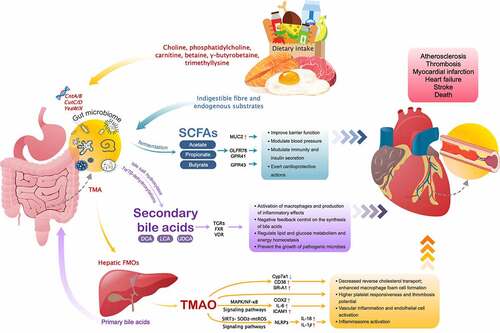
The microbe-dependent metabolite trimethylamine-N-oxide (TMAO)
TMAO was firstly reported by Wang et al. in a landmark paper, which has become the most compelling evidence of a causative link between the gut microbiome, oral intake of nutrient precursors, and CAD risk.Citation94 The conversion of TMAO in humans has been shown to arise via a multistep, metaorganismal, and microbe-dependent pathway commencing with the dietary precursors including choline,Citation94 phosphatidylcholine,Citation94,Citation95 carnitine,Citation18 betaine,Citation94 γ-butyrobetaine (γ-BB),Citation96 and trimethyllysine (TML),Citation97 which are most abundant in foods found in a western diet, including red meat, egg yolks, milk, and other animal products with high levels of saturated or unsaturated fat.Citation98,Citation99 Gut microbial metabolism of these precursors begins with specific microbial trimethylamine (TMA) lyases encoded by genes including choline-TMA lyase system (CutC/D) and the carnitine Rieske-type oxygenase/reductase system (CntA/B and YeaW/X), which humans do not possess, to generate TMA.Citation100 After absorption, the TMA produced is then transported to liver via the portal vein and readily oxidized by host hepatic FMOs (flavin-containing monooxygenases; mainly FMO3) into TMAO.Citation94
A role for TMAO in atherosclerosis has been implied in most, but not all, mechanistic and animal model studies.Citation18,Citation94,Citation101–103 TMAO can enhance cholesterol accumulation in macrophages by significantly increasing the mRNA and surface protein levels of CD36 receptor and scavenger receptor A1 (SR-A1), as well as the formation of macrophage-derived foam cells,Citation94 which are markers of atherosclerotic plaques.Citation104 Cholesterol 7alpha-hydroxylase (Cyp7a1) is one of the major enzymes in bile acid synthesis. TMAO can reduce Cyp7a1 expression, inhibit cholesterol transport, cause cholesterol accumulation and form cell formation, and ultimately result in atheromatous plaque formation.Citation18 Research evidence further suggests that feeding TMAO or either carnitine or choline to mice also inhibits in vivo cholesterol efflux in macrophages.Citation18 Moreover, another mechanism by which TMAO may increase the risk of cardiovascular events is mediated via platelet reactivity and thrombosis potential.Citation105–108 TMAO alters platelet intracellular Ca2+ signaling, heightening its responsiveness to submaximal stimulus-dependent activation (e.g., thrombin, ADP, collagen).Citation106 Inflammasome activation and vascular inflammation are critically involved in the progression of atherosclerosis and thrombotic complications.Citation109–111 Seldin et al. reported that acute injection of physiological levels of TMAO in mice induced vascular inflammation, including activation of mitogen-activated protein kinase (MAPK) and nuclear factor-kappa B (NF-κB) signaling pathways, upregulating the levels of cyclooxygenase 2 (COX2), IL-6, and intercellular adhesion molecule 1 (ICAM1), leading to subsequent leukocytes recruitment.Citation109 Similarly, investigations by Chen et al. have suggested TMAO can also promote the release of inflammatory cytokines IL-18 and IL-1β via priming and activation of the NLRP3 (nucleotide binding oligomerization domain–like receptor family pyrin domain-containing-3) inflammasome in vitro and in vivo through SIRT3-SOD2-mtROS (sirtuin-3-superoxide dismutase 2-mitochondrial reactive oxygen species) pathway to induce vascular inflammation.Citation110
Several epidemiological studies showed the prognostic value of plasma levels of TMAO that predicted future cardiovascular events including myocardial infarction, stroke, and all-cause mortality.Citation18,Citation95,Citation104,Citation112,Citation113 In addition, elevated levels of plasma TML, alone, or together with TMAO, have been shown to be associated with both near- and long-term major adverse cardiac events after acute coronary syndrome, independent of troponin T levels,Citation114 and circulating levels of γ-BB and its precursor TML may also act as cardiovascular risk markers in patients with carotid artery atherosclerosis.Citation115 Furthermore, several independent studies reported that TMAO may be a strong predictor of clinical outcomes in patients with HF.Citation116–118 A meta-analysis of eight studies also demonstrated that higher circulating TMAO concentration was positively associated with hypertension prevalence, which was dose-dependent.Citation119 Hence, with these inspiring results, although gut microbiome analyses are not yet ready for clinical use, there is a need to include TMAO and its precursors in future studies to delineate the mechanistic pathways involved in CVDs and explore potential therapeutic targets.
Bile Acids (BAs) serve as modulators of host metabolism
BAs are amphipathic molecules that are composed of a diverse array of structurally specific categories.Citation120 Primary BAs are initially synthesized from cholesterol in the host liver and stored in the gall bladder, then secreted into the intestinal (duodenum) lumen upon ingestion of a meal to assist the emulsification of fats and oils for digestion and absorption of lipid-soluble dietary nutrients.Citation121 Most of the primary BAs are actively absorbed and recycled back to the liver, while a small fraction escapes this enterohepatic circulation and enters the colon, where they are deconjugated into secondary BAs via bacterial bile salt hydrolase (BSH) activity identified in the bacterial genera including Bacteroides, Bifidobacterium, Lactobacillus, Clostridium, and Listeria,Citation121–123 as well as 7α/7β-dehydroxylations found only in anaerobic gut microbes,Citation124 providing microbes with a way in which to reduce the toxicity of bile acids.Citation125
The most well-discussed secondary BAs include deoxycholic acid (DCA), ursodeoxycholic acid (UDCA), and lithocholic acid (LCA), many of which have pleiotropic and hormonal functions through interaction with a variety of host receptors including farnesoid X receptor (FXR), specific G-protein–coupled receptors (GPCRs) like takeda G-protein–coupled receptor 5 (TGR5), and vitamin D receptor (VDR).Citation122,Citation126,Citation127 Activation of FXR and TGR5 regulates BAs synthesis, conjugation, transport, and detoxification, as well as glucose, lipid, and energy homeostasis.Citation128,Citation129 BAs also play an important role in regulating the gut microbiome structure by controlling gut bacteria overgrowth and maintaining intestinal barrier function via FXR signaling.Citation130 When bound to the TGR5 receptor, secondary BAs cause macrophage activation and subsequent production of inflammatory cytokines.Citation131 Additionally, studies have shown that BAs may control inflammation and fibrosis, maintain vascular integrity, and control aging and circadian rhythms.Citation132 These observations demonstrate the systematic regulatory role of BAs, providing a biochemical bridge for the gut microbiome to influence a range of host metabolic processes.
Perturbations to the dynamic interactions among dietary intake, gut microbiome, and specific BAs may thus contribute to cardiometabolic phenotypes and CVDs susceptibly. For example, altered BAs concentrations in plasma are correlated with insulin resistance in patients with type 2 diabetes mellitus,Citation133,Citation134 and increased relative abundances of Lactobacillus and Bifidobacterium in the gut may contribute to metabolic improvements via modulation of BAs signaling pathways when applying Acarbose treatments.Citation135 A recent cross-sectional study found an increased ratio of secondary to primary BAs in patients with chronic heart failure, which was associated with reduced overall survival in univariate analysis.Citation136 Microbial dysbiosis can lead to decreased BSH activity and impaired cholesterol elimination, thus, promoting atherosclerosis,Citation137 while a randomized placebo-controlled clinical trial showed that treatment with Lactobacillus reuteri, a bacteria with elevated BSH activity, significantly reduced low-density lipoprotein cholesterol (LDL-C) levels.Citation138 A better understanding of how perturbation in candidate BAs profiles is mechanistically linked to these processes of disease status or responses to therapy may enhance our views of CVDs pathogenesis in the future.
Altered gut microbiome with reduced capacity for short chain fatty acids (SCFAs) production
The human gut microbiome is capable of fermenting carbohydrates in the form of dietary non-digestible fibers, leading to the production of SCFAs, mainly butyrate, propionate, and acetate.Citation139 SCFAs act as main energy source for colonocytes and play an important role in maintaining gut mucosal integrity by facilitating the tight junction assembly.Citation140 Gut dysbiosis can bring a reduction in SCFAs production, causing increased mucosal permeability and leakage of lipopolysaccharide (LPS) and other pro-inflammatory bacterial products such as circulating endotoxins into the systemic circulation, as well as allowing bacterial translocation.Citation139
A previous study has reported that SCFAs could stimulate MUC2 mucin expression in intestinal epithelial cells by regulating prostaglandins production, thus may improve barrier function.Citation141 Other mechanistic studies provided numerous lines of evidence show that SCFAs may serve as microbial mediators that contribute to the vasomotor tone and blood pressure via the olfactory receptor 78 (Olfr78) and G-protein–coupled receptors (GPRs) including GPR41 (also known as free fatty acid receptor 3, FFAR3) and GPR43 (also known as FFAR2).Citation142–145 Olfr78 is expressed in renal artery branches and the juxtaglomerular afferent arteriole, mediates renin secretion so that modulate vascular resistance, contributing to hypertension.Citation143 By contrast, GPR41 and GPR43, which are expressed within the vasculature, can exert vasodilator action.Citation142,Citation144 In addition, SCFAs can activate GPR41 or GPR43 leading to consequent production of chemokines and cytokines, which are necessary for adequate recruitment of leukocytes and activation of effector T lymphocytes during immune responses.Citation146 SCFAs also play a key role in glucose homeostasis by promoting the release of glucagon-like peptide-1 (GLP-1) via the activation of GPR43, resulting in increased insulin, decreased glucagon secretion, and suppressed appetite.Citation147,Citation148 Importantly, multiple studies demonstrate that GLP-1 can exert robust cardioprotective actions including attenuation of ischemic cardiac injury, preservation of ventricular function, as well as improved survival,Citation149–151 which holds great perspectives to favorably modify outcomes in patients with CVDs.
More recently, propionate has been shown to exert cardioprotective effects via attenuating adverse ventricular remodeling, atherosclerotic lesion area, and cardiovascular inflammation in a Treg-dependent manner.Citation152 Likewise, the susceptibility to cardiac ventricular arrhythmias was significantly lower in propionate-treated mice, indicating viable links between SCFAs and arrhythmia development.Citation152 Moreover, Tang et al. demonstrated that gut microbe-derived SCFAs would benefit cardiac repair, reduce the size of infarct area and improve outcomes after MI through modulation of immune composition in mice model.Citation153 In ApoE−/− mice, elevated levels of butyrate due to inoculation with the butyrate-producer Roseburia intestinalis led to ameliorated atherosclerosis.Citation154 Overall, the possible mechanisms behind SCFAs and CVDs are manifold and need to be elucidated.
A growing number of microbe-derived metabolites, including a variety of aromatic amino acid products like uremic toxins and indole, are thought to potentially favor disease pathogenesis via the aryl hydrocarbon receptor.Citation155,Citation156 Phenylacetylglutamine (PAGln), a phenylalanine-derived metabolite interacts with adrenergic receptors, was reported to induce hyperreactivity of platelet and be associated with incident risk for major adverse cardiac events in an independent cohort.Citation157 The emphasis is given on the complexities of translating these metabolic signatures into practical clinical biomarkers used to identify patients at risk of CVDs, particularly in terms of whether these changes can lead to new therapeutic strategies like dietary interventions and microbial adjustments.
Other potential microbial aspects in the development of CVDs
Oral microorganisms and cardiovascular risk
The oral cavity is also colonized with a highly heterogeneous microbial community,Citation158 oral infections occur frequently in humans and often cause caries, gingivitis, and periodontitis,Citation159 which can lead to edematous swelling and high bleeding tendency, facilitating the transmission of these periodontal pathogens into the bloodstream via the injured oral epithelium along with their by-products or inflammatory mediators (e.g., IL-1 or TNF-α).Citation160 Within this complexity, an ever-growing number of oral bacteria have been associated with CVDs, and particularly hypertensionCitation161 and atherosclerosis ().Citation162 Bacterial DNA of oral origin (e.g., Streptococcus mutans) and periodontopathogens such as Porphyromonas gingivalis and Treponema denticola has been frequently detected in atherosclerotic lesions suggesting that bacteria may be rapped at the site of the atherosclerotic lesion and act as potent inducers which may increase inflammatory activity.Citation163–166 The pathogen-associated molecular patterns (PAMPs, e.g., LPS or peptidoglycan), found commonly on many different pathogens, can bind to a limited number of pattern-recognition receptors (PRRs) expressed on both macrophage and endothelial cells, and through subsequent intracellular signaling trigger the inflammatory response,Citation167,Citation168 leading to a more vulnerable plaque, thereby increasing the risk of CVDs. Some of these receptors include toll-like receptors (TLRs), nucleotide-binding oligomerization domains (NODs), CD14, complement receptor-3, lectins, and scavenger receptors.Citation169–171 In addition to recognizing PAMPs, these receptors are also involved in the uptake of oxidized LDL by macrophages, which play a causative role in exacerbating atherosclerosis.Citation172
Figure 3. Relationship between atherosclerosis and oral diseases
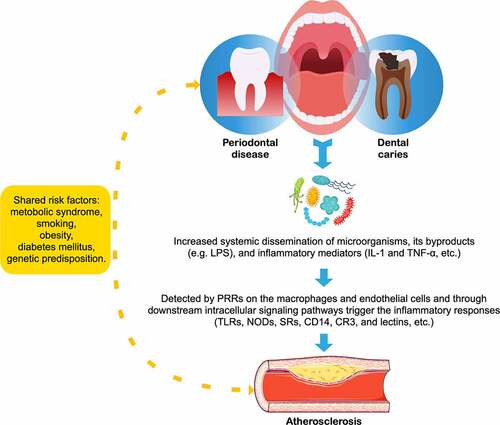
Oral diseases and CVDs share a large set of overlapping risk factors, such as smoking, diabetes mellitus, age, obesity, and poor socioeconomic conditions,Citation173 as well as common genetic susceptibility variantsCitation174 and biomarkers,Citation175 suggesting a possible common pathophysiology of periodontitis and CVDs. Previous observational studies have supported an association between periodontitis and atherosclerosis independent of known confounders; however, the evidence did not support a causative relationship.Citation176 Recently, it was inspiring that a population-based cohort study containing 247,696 participants in Korea demonstrated oral hygiene improvements such as frequent tooth brushing and regular dental visits for professional cleaning were associated with 9% and 14% significantly lower risk of cardiovascular events after multivariable adjustment.Citation177 Considering as a highly feasible and daily manageable intervention to prevent CVDs, in the near future, randomized controlled trials would require a large population, an adequate follow-up length, and an ethically acceptable control group to elucidate the effects of oral hygiene behaviors on attenuating the cardiovascular risk originating from periodontal disease and dental caries.
Host ecological acclimation and adaptation powered by gut microbiome
The gut microbiome and its hosts are no longer heralded as autonomous entities, but rather as holobionts. As such, their collective genomes forge a hologenome, and these concepts afford a holistic view of biological complexity.Citation178 Therefore, while human evolution undoubtedly shaped the gut microbiome via changes in factors such as host genetic background, dietary habits, and social behavior during the admixture and introgression processes of human ancestors,Citation179–181 we believe genetic interactions between the gut microbiome and hosts might play an essential role in increasing a host’s acclimation capacity (individual level) and adaptation capacity (population level), even modulating disease severity.Citation182 One of the examples in humans suggested that the gut bacterium Bacteroides plebeius in Japanese individuals has acquired genes encoding carbohydrate active enzymes, which are absent in the human genome, from the seaweed-associated bacterium Zobellia galactanivorans in non-sterile food (i.e., sushi).Citation183 Additionally, the differences in digestion ability due to allelic variations of human genes LCT (encoding lactase) and AMY1 (encoding salivary amylase) could be modified by Bifidobacterium (a lactose-metabolizing genus),Citation184 Ruminococcus (a resistant-starch fermentation genus),Citation185 or other functionally redundant microbes, thus may mitigate energetic trade-offs and contribute to host-adaptive evolution. Similar patterns were also observed when humans encounter environmental fluctuations,Citation186 and some commensal bacteria can even determine the phenotype of hosts carrying risk alleles for inflammatory disease.Citation182
Obviously, these reciprocal interactions facilitate rapid adjustments in host phenotypes across much shorter timescales compared with the classic models of selective pressure on the human genome. A better understanding of the relationship between the host genome and gut microbiome in the context of adaptive evolution will add another dimension to our knowledge as we moved with our microbes through time and space. Therefore, intertwined processes with host genetics must be considered when exploring the potential of the gut microbiome to influence CVDs pathogenesis. Currently, no study has tested the hypothesis and its biological relevance in an eco-evolutionary trajectory, investigating host genome–microbiome interactions in a wider range of human populations will help researchers go beyond collections of anecdotes to form the foundation of a theory that takes microbiome contributions to host physiology and disease progression into account in a formal framework. Microbial transplantations may help to demonstrate the effect of the gut microbiome on host phenomes, combining with metagenomics and other meta-omics approaches to reveal mechanisms underlying genotype-phenotype variation involving simultaneous pathological changes in diverse levels that contribute to disease risk. Furthermore, as microbial gene repertoires are highly malleable, it offers not only the metagenomic plasticity that expedites host acclimation and adaptation, but exciting prospects for novel therapies.
Conclusions and future perspectives
Although our understanding of how the gut microbiome impacts the pathogenesis of CVDs is still rudimentary, the rate at which new discoveries are emerging is impressive. As outlined, there is overwhelming evidence that microbe-related processes, in general, are linked to multiple CVD-relevant phenotypes, including but not limited to atherosclerosis, blood pressure, platelet reactivity and thrombosis potential, cardiac rhythm, lipid metabolism, and vascular inflammation, which provides us with exciting areas of investigation to translate it into microbiome-based clinical or pharmaceutical practice. Unsurprisingly, inhibitors of TMA formation that targeting microbial TMA lyases have been developed, which could reduce TMAO levels and reverse atherosclerosis in mice models.Citation187 Moreover, bile acid activation of FXR can inhibit NF-κB pathway, thereby ameliorating inflammation and improving myocardial function;Citation188 TGR5 agonists can exert a cardioprotective function and improve myocardial response to physiologic, inotropic, and hemodynamic stress in mice,Citation189 and thus may consider their roles in the treatment of heart failure in the future. Whereas dietary interventions, prebiotics and probiotics, antibiotics, and fecal microbiota transplantation (FMT) are all potential candidates showing the ability to favorably alter gut microbial profiles and its functional output in some human studies, results are highly variable, and findings from animal models have not yet been transformed to evidence of clinical efficacy, and large, prospective interventional studies will be needed to validate these gut microbiome-targeted therapeutics.Citation190
In light of recent findings of adherence to a Mediterranean diet or discontinuation of dietary red meat consumption is negatively associated with TMAO levels,Citation98,Citation191 and high-fiber diet or acetate supplementary being beneficial to reducing blood pressure and cardiac fibrosis,Citation192 dietary interventions could be a logical and accessible next step for targeting the gut-heart axis. However, one challenge with particular relevance to microbiome research is the tremendous inter-individual variations in the gut microbiome configuration,Citation193 and the following host metabolism occasioned by dietary interventions was also person-specific.Citation194,Citation195 Hence, personalized diet recommendations based on an individual’s microbiome, particular responders to a specific food, as well as personal health information including physiologic parameters and family history would become a new rational design for prevention and treatment of disease, and the exploration of precision medicine will progress immensely.
Given the complexity and heterogeneity of the gut microbiome, as well as the state-of-the-art technological advances, a meta-omics approach holds the promise as a path toward unraveling host–microbiome interactions and assessing causality in clinical settings. However, the current application of meta-omics integrations in the field of CVDs has so far been limited. Obviously, one of the main reasons for this is inadequate research design and lack of standardized methods. The parallel measurements on multiple omics layers and proper preprocessing are important for removing outliers and non-biological variations and increasing the comparability between data from different studies. In the coming future, standardized meta-omics pipelines and updated bioinformatic methods with high-resolution of microbial taxa characterization at the strain level can effectively capture a holistic view of pathogenic mechanisms, and ultimately achieve more comprehensive phenotyping of our physiology to select composite diagnostic or prognostic biomarkers for risk stratification and precision medicine, realizing the full potential of this field.
Author Contributions
All authors were involved in the conception of this review; Jing Xu wrote the manuscript and drew the figures; Yuejin Yang checked and reviewed the manuscript.
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP. Heart disease and stroke statistics-2020 update: a report from the American Heart Association. Circulation. 2020;141(9):e139–25. PubMed PMID: 31992061. doi:10.1161/cir.0000000000000757. Epub 2020/01/30.
- Young DR, Hivert MF, Alhassan S, Camhi SM, Ferguson JF, Katzmarzyk PT. Sedentary behavior and cardiovascular morbidity and mortality: a science advisory from the American Heart Association. Circulation. 2016; 134(13):e262–79. Epub 2016/08/17. PubMed PMID: 27528691. doi: 10.1161/cir.0000000000000440.
- Ferguson JF, Allayee H, Gerszten RE, Ideraabdullah F, Kris-Etherton PM, Ordovás JM. Nutrigenomics, the microbiome, and gene-environment interactions: new directions in cardiovascular disease research, prevention, and treatment: a scientific statement from the American Heart Association. Circ Cardiovasc Genet. 2016;9(3):291–313. PubMed PMID: 27095829. doi:10.1161/hcg.0000000000000030. Epub 2016/04/21.
- Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376(9750):1393–1400. PubMed PMID: 20971364; PubMed Central PMCID: PMCPMC2965351. doi:10.1016/s0140-6736(10)61267-6. Epub 2010/10/26.
- Loscalzo J. Gut microbiota, the genome, and diet in atherogenesis. N Engl J Med. 2013;368(17):1647–1649. PubMed PMID: 23614591. doi:10.1056/NEJMe1302154. Epub 2013/04/26.
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. PubMed PMID: 22424233; PubMed Central PMCID: PMCPMC5050011. doi:10.1016/j.cell.2012.01.035. Epub 2012/03/20.
- Weiss GA, Hennet T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol Life Sci. 2017;74(16):2959–2977. PubMed PMID: 28352996. doi:10.1007/s00018-017-2509-x. Epub 2017/03/30.
- Zhang X, Li L, Butcher J, Stintzi A, Figeys D. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome. 2019;7(1):154. PubMed PMID: 31810497; PubMed Central PMCID: PMCPMC6898977. doi:10.1186/s40168-019-0767-6. Epub 2019/12/08.
- Maruvada P, Leone V, Kaplan LM, Chang EB. The human microbiome and obesity: moving beyond associations. Cell Host Microbe. 2017;22(5):589–599. PubMed PMID: 29120742. doi:10.1016/j.chom.2017.10.005. Epub 2017/11/10.
- Miyamoto J, Igarashi M, Watanabe K, Karaki SI, Mukouyama H, Kishino S. Gut microbiota confers host resistance to obesity by metabolizing dietary polyunsaturated fatty acids. Nat Commun. 2019;10(1):4007. PubMed PMID: 31488836; PubMed Central PMCID: PMCPMC6728375. doi:10.1038/s41467-019-11978-0. Epub 2019/09/07.
- McIlroy J, Ianiro G, Mukhopadhya I, Hansen R, Hold GL. Review article: the gut microbiome in inflammatory bowel disease-avenues for microbial management. Aliment Pharmacol Ther. 2018;47(1):26–42. PubMed PMID: 29034981. doi:10.1111/apt.14384. Epub 2017/10/17.
- Sitkin S, Demyanova E, Vakhitov T, Pokrotnieks J. Altered sphingolipid metabolism and its interaction with the intestinal microbiome is another key to the pathogenesis of inflammatory bowel disease. Inflamm Bowel Dis. 2019;25(12):e157–e8. PubMed PMID: 31560761. doi:10.1093/ibd/izz228. Epub 2019/09/29.
- Rajagopala SV, Vashee S, Oldfield LM, Suzuki Y, Venter JC, Telenti A. The human microbiome and cancer. Cancer Prev Res (Phila). 2017;10(4):226–234. PubMed PMID: 28096237. doi:10.1158/1940-6207.Capr-16-0249. Epub 2017/01/18.
- Liang W, Yang Y, Wang H, Wang H, Yu X, Lu Y. Gut microbiota shifts in patients with gastric cancer in perioperative period. Medicine (Baltimore). 2019;98(35):e16626. PubMed PMID: 31464899; PubMed Central PMCID: PMCPMC6736490. doi:10.1097/md.0000000000016626. Epub 2019/08/30.
- Yeoh N, Burton JP, Suppiah P, Reid G, Stebbings S. The role of the microbiome in rheumatic diseases. Curr Rheumatol Rep. 2013;15(3):314. PubMed PMID: 23378145. doi:10.1007/s11926-012-0314-y. Epub 2013/02/05.
- Ehp VD, Del Chierico F, Malattia C, Russo A, Pires Marafon D, Ter Haar NM. Microbiome analytics of the gut microbiota in patients with juvenile idiopathic arthritis: a longitudinal observational cohort study. Arthritis Rheumatol. 2019;71(6):1000–1010. PubMed PMID: 30592383; PubMed Central PMCID: PMCPMC6593809. doi:10.1002/art.40827. Epub 2018/12/29.
- Ascher S, Reinhardt C. The gut microbiota: an emerging risk factor for cardiovascular and cerebrovascular disease. Eur J Immunol. 2018;48(4):564–575. PubMed PMID: 29230812. doi:10.1002/eji.201646879. Epub 2017/12/13.
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–585. PubMed PMID: 23563705; PubMed Central PMCID: PMCPMC3650111. doi:10.1038/nm.3145. Epub 2013/04/09.
- Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–422. PubMed PMID: 29795328. doi:10.1038/s41579-018-0029-9. Epub 2018/05/26.
- Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14(6):405–416. PubMed PMID: 24854590; PubMed Central PMCID: PMCPMC4332855. doi:10.1038/nri3684. Epub 2014/05/24.
- Korem T, Zeevi D, Suez J, Weinberger A, Avnit-Sagi T, Pompan-Lotan M. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science. 2015;349(6252):1101–1106. PubMed PMID: 26229116; PubMed Central PMCID: PMCPMC5087275. doi:10.1126/science.aac4812. Epub 2015/08/01.
- Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010;4(5):642–647. PubMed PMID: 20090784. doi:10.1038/ismej.2009.153. Epub 2010/01/22.
- Claesson MJ, Wang Q, O’Sullivan O, Greene-Diniz R, Cole JR, Ross RP. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38(22):e200. PubMed PMID: 20880993; PubMed Central PMCID: PMCPMC3001100. doi:10.1093/nar/gkq873. Epub 2010/10/01.
- Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ. Opinion: re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3(9):733–739. PubMed PMID: 16138101. doi:10.1038/nrmicro1236. Epub 2005/09/03.
- Scholz M, Ward DV, Pasolli E, Tolio T, Zolfo M, Asnicar F. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods. 2016;13(5):435–438. PubMed PMID: 26999001. doi:10.1038/nmeth.3802. Epub 2016/03/22.
- Mukherjee S, Seshadri R, Varghese NJ, Eloe-Fadrosh EA, Meier-Kolthoff JP, Göker M. 1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life. Nat Biotechnol. 2017;35(7):676–683. PubMed PMID: 28604660. doi:10.1038/nbt.3886. Epub 2017/06/13.
- Pope PB, Smith W, Denman SE, Tringe SG, Barry K, Hugenholtz P. Isolation of succinivibrionaceae implicated in low methane emissions from tammar wallabies. Science. 2011;333(6042):646–648. PubMed PMID: 21719642. doi:10.1126/science.1205760. Epub 2011/07/02.
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428(6978):37–43. PubMed PMID: 14961025. doi:10.1038/nature02340. Epub 2004/02/13.
- Sunagawa S, Mende DR, Zeller G, Izquierdo-Carrasco F, Berger SA, Kultima JR. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods. 2013;10(12):1196–1199. PubMed PMID: 24141494. doi:10.1038/nmeth.2693. Epub 2013/10/22.
- Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8(6):e1002358. PubMed PMID: 22719234; PubMed Central PMCID: PMCPMC3374609. doi:10.1371/journal.pcbi.1002358. Epub 2012/06/22.
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28(1):33–36. PubMed PMID: 10592175; PubMed Central PMCID: PMCPMC102395. doi:10.1093/nar/28.1.33. Epub 1999/12/11.
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42(D1):D199–205. PubMed PMID: 24214961; PubMed Central PMCID: PMCPMC3965122. doi:10.1093/nar/gkt1076. Epub 2013/11/12.
- Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014;42(Database issue):D231–9. PubMed PMID: 24297252; PubMed Central PMCID: PMCPMC3964997. doi:10.1093/nar/gkt1253. Epub 2013/12/04.
- Hunter S, Corbett M, Denise H, Fraser M, Gonzalez-Beltran A, Hunter C. EBI metagenomics – a new resource for the analysis and archiving of metagenomic data. Nucleic Acids Res. 2014;42(Database issue):D600–6. PubMed PMID: 24165880; PubMed Central PMCID: PMCPMC3965009. doi:10.1093/nar/gkt961. Epub 2013/10/30.
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32(8):834–841. PubMed PMID: 24997786. doi:10.1038/nbt.2942. Epub 2014/07/07.
- Chan SHJ, Cai J, Wang L, Simons-Senftle MN, Maranas CD. Standardizing biomass reactions and ensuring complete mass balance in genome-scale metabolic models. Bioinformatics. 2017;33(22):3603–3609. PubMed PMID: 29036557. doi:10.1093/bioinformatics/btx453. Epub 2017/10/17.
- Raes J, Korbel JO, Lercher MJ, Von Mering C, Bork P. Prediction of effective genome size in metagenomic samples. Genome Biol. 2007;8(1):R10. PubMed PMID: 17224063; PubMed Central PMCID: PMCPMC1839125. doi:10.1186/gb-2007-8-1-r10. Epub 2007/01/17.
- Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486(7402):215–221. Epub 2012/06/16. doi: 10.1038/nature11209. PubMed PMID: 22699610; PubMed Central PMCID: PMCPMC3377744.
- Kennedy NA, Walker AW, Berry SH, Duncan SH, Farquarson FM, Louis P. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS One. 2014;9(2):e88982. PubMed PMID: 24586470; PubMed Central PMCID: PMCPMC3933346. doi:10.1371/journal.pone.0088982. Epub 2014/03/04.
- Wesolowska-Andersen A, Bahl MI, Carvalho V, Kristiansen K, Sicheritz-Pontén T, Gupta R. Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome. 2014;2(1):19. PubMed PMID: 24949196; PubMed Central PMCID: PMCPMC4063427. doi:10.1186/2049-2618-2-19. Epub 2014/06/21.
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. PubMed PMID: 20203603; PubMed Central PMCID: PMCPMC3779803. doi:10.1038/nature08821. Epub 2010/03/06.
- Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11(6):1358–1375. PubMed PMID: 19207571. doi:10.1111/j.1462-2920.2008.01863.x. Epub 2009/02/12.
- Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3(2):179–189. PubMed PMID: 18971961. doi:10.1038/ismej.2008.108. Epub 2008/10/31.
- Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3(5):431–438. PubMed PMID: 15821725. doi:10.1038/nrmicro1152. Epub 2005/04/12.
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. PubMed PMID: 24336217; PubMed Central PMCID: PMCPMC3957428. doi:10.1038/nature12820. Epub 2013/12/18.
- Heintz-Buschart A, May P, Laczny CC, Lebrun LA, Bellora C, Krishna A. Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nat Microbiol. 2016;2(1):16180. PubMed PMID: 27723761. doi:10.1038/nmicrobiol.2016.180. Epub 2016/10/11.
- Bikel S, Valdez-Lara A, Cornejo-Granados F, Rico K, Canizales-Quinteros S, Soberón X. Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome. Comput Struct Biotechnol J. 2015;13:390–401. doi:10.1016/j.csbj.2015.06.001. PubMed PMID: 26137199; PubMed Central PMCID: PMCPMC4484546. Epub 2015/07/03.
- Zhang X, Chen W, Ning Z, Mayne J, Mack D, Stintzi A. Deep metaproteomics approach for the study of human microbiomes. Anal Chem. 2017;89(17):9407–9415. PubMed PMID: 28749657. doi:10.1021/acs.analchem.7b02224. Epub 2017/07/28.
- Zhang X, Figeys D. Perspective and guidelines for metaproteomics in microbiome studies. J Proteome Res. 2019;18(6):2370–2380. PubMed PMID: 31009573. doi:10.1021/acs.jproteome.9b00054. Epub 2019/04/23.
- Karu N, Deng L, Slae M, Guo AC, Sajed T, Huynh H. A review on human fecal metabolomics: methods, applications and the human fecal metabolome database. Anal Chim Acta. 2018;1030:1–24. doi:10.1016/j.aca.2018.05.031. PubMed PMID: 30032758. Epub 2018/07/24.
- Lamichhane S, Sen P, Dickens AM, Orešič M, Bertram HC. Gut metabolome meets microbiome: a methodological perspective to understand the relationship between host and microbe. Methods. 2018;149:3–12. doi:10.1016/j.ymeth.2018.04.029. PubMed PMID: 29715508. Epub 2018/05/02.
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106(10):3698–3703. PubMed PMID: 19234110; PubMed Central PMCID: PMCPMC2656143. doi:10.1073/pnas.0812874106. Epub 2009/02/24.
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. PubMed PMID: 22674330. doi:10.1126/science.1223813. Epub 2012/06/08.
- Franzosa EA, Hsu T, Sirota-Madi A, Shafquat A, Abu-Ali G, Morgan XC. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol. 2015;13(6):360–372. PubMed PMID: 25915636; PubMed Central PMCID: PMCPMC4800835. doi:10.1038/nrmicro3451. Epub 2015/04/29.
- Blanco-Míguez A, Fdez-Riverola F, Sánchez B, Lourenço A. Resources and tools for the high-throughput, multi-omic study of intestinal microbiota. Brief Bioinform. 2019;20(3):1032–1056. PubMed PMID: 29186315. doi:10.1093/bib/bbx156. Epub 2017/12/01.
- Weiss S, Van Treuren W, Lozupone C, Faust K, Friedman J, Deng Y. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 2016;10(7):1669–1681. PubMed PMID: 26905627; PubMed Central PMCID: PMCPMC4918442. doi:10.1038/ismej.2015.235. Epub 2016/02/26.
- McHardy IH, Goudarzi M, Tong M, Ruegger PM, Schwager E, Weger JR. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome. 2013;1(1):17. PubMed PMID: 24450808; PubMed Central PMCID: PMCPMC3971612. doi:10.1186/2049-2618-1-17. Epub 2014/01/24.
- Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. 2011;108 Suppl 1 (Suppl 1):4592–4598. Epub 2010/10/13. doi: 10.1073/pnas.1011383107. PubMed PMID: 20937873; PubMed Central PMCID: PMCPMC3063583
- Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. Epub 2012/12/06. doi: 10.1038/ncomms2266. PubMed PMID: 23212374; PubMed Central PMCID: PMCPMC3538954 competing financial interests.
- Yin J, Liao SX, He Y, Wang S, Xia GH, Liu FT. Dysbiosis of gut microbiota with reduced Trimethylamine-N-Oxide level in patients with large-artery atherosclerotic stroke or transient ischemic attack. J Am Heart Assoc. 2015;4(11). Epub 2015/11/26. doi: 10.1161/jaha.115.002699. PubMed PMID: 26597155; PubMed Central PMCID: PMCPMC4845212.
- Feng Q, Liu Z, Zhong S, Li R, Xia H, Jie Z. Integrated metabolomics and metagenomics analysis of plasma and urine identified microbial metabolites associated with coronary heart disease. Sci Rep. 2016;6:22525. doi:10.1038/srep22525. PubMed PMID: 26932197; PubMed Central PMCID: PMCPMC4773756. Epub 2016/03/05.
- Emoto T, Yamashita T, Sasaki N, Hirota Y, Hayashi T, So A. Analysis of gut microbiota in coronary artery disease patients: a possible link between gut microbiota and coronary artery disease. J Atheroscler Thromb. 2016;23(8):908–921. PubMed PMID: 26947598; PubMed Central PMCID: PMCPMC7399299. doi:10.5551/jat.32672. Epub 2016/03/08.
- Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8(1):845. PubMed PMID: 29018189; PubMed Central PMCID: PMCPMC5635030. doi:10.1038/s41467-017-00900-1. Epub 2017/10/12.
- Gózd-Barszczewska A, Kozioł-Montewka M, Barszczewski P, Młodzińska A, Humińska K. Gut microbiome as a biomarker of cardiometabolic disorders. Ann Agric Environ Med. 2017;24(3):416–422. PubMed PMID: 28954482. doi:10.26444/aaem/75456. Epub 2017/09/29.
- Yoshida N, Emoto T, Yamashita T, Watanabe H, Hayashi T, Tabata T. Bacteroides vulgatus and bacteroides dorei reduce gut microbial Lipopolysaccharide production and inhibit atherosclerosis. Circulation. 2018;138(22):2486–2498. doi:10.1161/CIRCULATIONAHA.118.033714. PubMed PMID: 30571343.
- Zhu Q, Gao R, Zhang Y, Pan D, Zhu Y, Zhang X. Dysbiosis signatures of gut microbiota in coronary artery disease. Physiol Genomics. 2018;50(10):893–903. PubMed PMID: 30192713. doi:10.1152/physiolgenomics.00070.2018. Epub 2018/09/08.
- Liu H, Chen X, Hu X, Niu H, Tian R, Wang H. Alterations in the gut microbiome and metabolism with coronary artery disease severity. Microbiome. 2019;7(1):68. PubMed PMID: 31027508; PubMed Central PMCID: PMCPMC6486680. doi:10.1186/s40168-019-0683-9. Epub 2019/04/28.
- Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM. Gut dysbiosis is linked to hypertension. Hypertension. 2015;65(6):1331–1340. PubMed PMID: 25870193; PubMed Central PMCID: PMCPMC4433416. doi:10.1161/hypertensionaha.115.05315. Epub 2015/04/15.
- Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, Dekker Nitert M. Increased systolic and diastolic blood pressure is associated with altered gut microbiota composition and Butyrate production in early pregnancy. Hypertension. 2016;68(4):974–981. PubMed PMID: 27528065. doi:10.1161/hypertensionaha.116.07910. Epub 2016/08/17.
- Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5(1):14. doi:10.1186/s40168-016-0222-x. PubMed PMID: 28143587
- Yan Q, Gu Y, Li X, Yang W, Jia L, Chen C. Alterations of the gut microbiome in hypertension. Front Cell Infect Microbiol. 2017;7:381. doi:10.3389/fcimb.2017.00381. PubMed PMID: 28884091; PubMed Central PMCID: PMCPMC5573791. Epub 2017/09/09.
- Kim S, Goel R, Kumar A, Qi Y, Lobaton G, Hosaka K. Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin Sci (Lond). 2018;132(6):701–718. PubMed PMID: 29507058; PubMed Central PMCID: PMCPMC5955695. doi:10.1042/cs20180087. Epub 2018/03/07.
- Menni C, Lin C, Cecelja M, Mangino M, Matey-Hernandez ML, Keehn L. Gut microbial diversity is associated with lower arterial stiffness in women. Eur Heart J. 2018;39(25):2390–2397. doi:10.1093/eurheartj/ehy226. PubMed PMID: 29750272
- Sun S, Lulla A, Sioda M, Winglee K, Wu MC, Jacobs DR Jr. Gut microbiota composition and blood pressure. Hypertension. 2019;73(5):998–1006. PubMed PMID: 30905192; PubMed Central PMCID: PMCPMC6458072. doi:10.1161/hypertensionaha.118.12109. Epub 2019/03/25.
- Huart J, Leenders J, Taminiau B, Descy J, Saint-Remy A, Daube G. Gut microbiota and fecal levels of short-chain fatty acids differ upon 24-hour blood pressure levels in men. Hypertension. 2019;74(4):1005–1013. PubMed PMID: 31352822. doi:10.1161/hypertensionaha.118.12588. Epub 2019/07/30.
- Verhaar BJH, Collard D, Prodan A, Levels JHM, Zwinderman AH, Bäckhed F. Associations between gut microbiota, faecal short-chain fatty acids, and blood pressure across ethnic groups: the HELIUS study. Eur Heart J. 2020;41(44):4259–4267. PubMed PMID: 32869053; PubMed Central PMCID: PMCPMC7724641. doi:10.1093/eurheartj/ehaa704. Epub 2020/09/02.
- Sandek A, Bauditz J, Swidsinski A, Buhner S, Weber-Eibel J, Von Haehling S. Altered intestinal function in patients with chronic heart failure. J Am Coll Cardiol. 2007;50(16):1561–1569. PubMed PMID: 17936155. doi:10.1016/j.jacc.2007.07.016. Epub 2007/10/16.
- Sandek A, Swidsinski A, Schroedl W, Watson A, Valentova M, Herrmann R. Intestinal blood flow in patients with chronic heart failure: a link with bacterial growth, gastrointestinal symptoms, and cachexia. J Am Coll Cardiol. 2014;64(11):1092–1102. PubMed PMID: 25212642. doi:10.1016/j.jacc.2014.06.1179. Epub 2014/09/13.
- Pasini E, Aquilani R, Testa C, Baiardi P, Angioletti S, Boschi F. Pathogenic gut flora in patients with chronic heart failure. JACC Heart Fail. 2016;4(3):220–227. PubMed PMID: 26682791. doi:10.1016/j.jchf.2015.10.009. Epub 2015/12/20.
- Luedde M, Winkler T, Heinsen FA, Rühlemann MC, Spehlmann ME, Bajrovic A. Heart failure is associated with depletion of core intestinal microbiota. ESC Heart Fail. 2017;4(3):282–290. PubMed PMID: 28772054; PubMed Central PMCID: PMCPMC5542738. doi:10.1002/ehf2.12155. Epub 2017/08/05.
- Kamo T, Akazawa H, Suda W, Saga-Kamo A, Shimizu Y, Yagi H. Dysbiosis and compositional alterations with aging in the gut microbiota of patients with heart failure. PLoS One. 2017;12(3):e0174099. PubMed PMID: 28328981; PubMed Central PMCID: PMCPMC5362204 Ltd., Daiichi Sankyo Co., Ltd., Shionogi & Co., Ltd., Nippon Boehringer Ingelheim Co., Ltd., Bristol-Myers Squibb K.K., MSD K.K., Sanofi K.K., and Sumitomo Dainippon Pharma Co., Ltd. There are no patents, products in development, or marketed products to declare. doi:10.1371/journal.pone.0174099. Epub 2017/03/23.
- Kummen M, Mayerhofer CCK, Vestad B, Broch K, Awoyemi A, Storm-Larsen C. Gut microbiota signature in heart failure defined from profiling of 2 independent cohorts. J Am Coll Cardiol. 2018;71(10):1184–1186. PubMed PMID: 29519360. doi:10.1016/j.jacc.2017.12.057. Epub 2018/03/10.
- Cui X, Ye L, Li J, Jin L, Wang W, Li S. Metagenomic and metabolomic analyses unveil dysbiosis of gut microbiota in chronic heart failure patients. Sci Rep. 2018;8(1):635. PubMed PMID: 29330424; PubMed Central PMCID: PMCPMC5766622. doi:10.1038/s41598-017-18756-2. Epub 2018/01/14.
- Zuo K, Yin X, Li K, Zhang J, Wang P, Jiao J. Different types of atrial fibrillation share patterns of gut microbiota dysbiosis. mSphere. 2020;5(2). Epub 2020/03/20. doi: 10.1128/mSphere.00071-20. PubMed PMID: 32188747; PubMed Central PMCID: PMCPMC7082137.
- Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JA. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ Res. 2015;117(9):817–824. PubMed PMID: 26358192; PubMed Central PMCID: PMCPMC4596485. doi:10.1161/circresaha.115.306807. Epub 2015/09/12.
- Kelly TN, Bazzano LA, Ajami NJ, He H, Zhao J, Petrosino JF. Gut microbiome associates with lifetime cardiovascular disease risk profile among Bogalusa heart study participants. Circ Res. 2016;119(8):956–964. PubMed PMID: 27507222; PubMed Central PMCID: PMCPMC5045790. doi:10.1161/circresaha.116.309219. Epub 2016/08/11.
- Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. PubMed PMID: 29489753. doi:10.1038/nature25973. Epub 2018/03/01.
- Jie Z, Xia H, Zhong S-L, Feng Q, Li S, Liang S. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8(1):845. doi:10.1038/s41467-017-00900-1. PubMed PMID: 29018189.
- Arneson D, Shu L, Tsai B, Barrere-Cain R, Sun C, Yang X. Multidimensional integrative genomics approaches to dissecting cardiovascular disease. Front Cardiovasc Med. 2017;4:8. doi:10.3389/fcvm.2017.00008. PubMed PMID: 28289683; PubMed Central PMCID: PMCPMC5327355. Epub 2017/03/16.
- Huang S, Chaudhary K, Garmire LX. More is better: recent progress in multi-omics data integration methods. Front Genet. 2017;8:84. doi:10.3389/fgene.2017.00084. PubMed PMID: 28670325; PubMed Central PMCID: PMCPMC5472696. Epub 2017/07/04.
- Griffin JL, Wang X, Stanley E. Does our gut microbiome predict cardiovascular risk? A review of the evidence from metabolomics. Circ Cardiovasc Genet. 2015;8(1):187–191. PubMed PMID: 25691688; PubMed Central PMCID: PMCPMC4333723. doi:10.1161/circgenetics.114.000219. Epub 2015/02/19.
- Xu J, Yang Y. Implications of gut microbiome on coronary artery disease. Cardiovasc Diagn Ther. 2020;10(4):869–880. PubMed PMID: 32968642; PubMed Central PMCID: PMCPMC7487393. doi:10.21037/cdt-20-428. Epub 2020/09/25.
- Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535(7610):56–64. PubMed PMID: 27383980; PubMed Central PMCID: PMCPMC5991619. doi:10.1038/nature18846. Epub 2016/07/08.
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi:10.1038/nature09922. PubMed PMID: 21475195
- Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–1584. PubMed PMID: 23614584; PubMed Central PMCID: PMCPMC3701945. doi:10.1056/NEJMoa1109400. Epub 2013/04/26.
- Koeth RA, Levison BS, Culley MK, Buffa JA, Wang Z, Gregory JC. γ-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab. 2014;20(5):799–812. PubMed PMID: 25440057; PubMed Central PMCID: PMCPMC4255476. doi:10.1016/j.cmet.2014.10.006. Epub 2014/12/03.
- Li XS, Wang Z, Cajka T, Buffa JA, Nemet I, Hurd AG. Untargeted metabolomics identifies trimethyllysine, a TMAO-producing nutrient precursor, as a predictor of incident cardiovascular disease risk. JCI Insight. 2018;3(6). Epub 2018/03/23. doi: 10.1172/jci.insight.99096. PubMed PMID: 29563342; PubMed Central PMCID: PMCPMC5926943.
- Wang Z, Bergeron N, Levison BS, Li XS, Chiu S, Jia X. Impact of chronic dietary red meat, white meat, or non-meat protein on trimethylamine N-oxide metabolism and renal excretion in healthy men and women. Eur Heart J. 2019;40(7):583–594. PubMed PMID: 30535398; PubMed Central PMCID: PMCPMC6374688. doi:10.1093/eurheartj/ehy799. Epub 2018/12/12.
- Zhu W, Wang Z, Tang WHW, Hazen SL. Gut microbe-generated Trimethylamine N-Oxide from Dietary Choline is Prothrombotic in subjects. Circulation. 2017;135(17):1671–1673. PubMed PMID: 28438808; PubMed Central PMCID: PMCPMC5460631. doi:10.1161/circulationaha.116.025338. Epub 2017/04/26.
- Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci U S A. 2012;109(52):21307–21312. PubMed PMID: 23151509; PubMed Central PMCID: PMCPMC3535645. doi:10.1073/pnas.1215689109. Epub 2012/11/16.
- Ding L, Chang M, Guo Y, Zhang L, Xue C, Yanagita T. Trimethylamine-N-oxide (TMAO)-induced atherosclerosis is associated with bile acid metabolism. Lipids Health Dis. 2018;17(1):286. PubMed PMID: 30567573; PubMed Central PMCID: PMCPMC6300890. doi:10.1186/s12944-018-0939-6. Epub 2018/12/21.
- Aldana-Hernández P, Leonard KA, Zhao YY, Curtis JM, Field CJ, Jacobs RL. Dietary Choline or Trimethylamine N-oxide supplementation does not influence Atherosclerosis development in Ldlr-/- and Apoe-/- male mice. J Nutr. 2020;150(2):249–255. PubMed PMID: 31529091. doi:10.1093/jn/nxz214. Epub 2019/09/19.
- Xue J, Zhou D, Poulsen O, Imamura T, Hsiao YH, Smith TH. Intermittent hypoxia and hypercapnia accelerate atherosclerosis, partially via trimethylamine-oxide. Am J Respir Cell Mol Biol. 2017;57(5):581–588. PubMed PMID: 28678519; PubMed Central PMCID: PMCPMC5705907. doi:10.1165/rcmb.2017-0086OC. Epub 2017/07/06.
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. PubMed PMID: 21475195; PubMed Central PMCID: PMCPMC3086762. doi:10.1038/nature09922. Epub 2011/04/09.
- Skye SM, Zhu W, Romano KA, Guo CJ, Wang Z, Jia X. Microbial transplantation with human gut commensals containing CutC is sufficient to transmit enhanced platelet reactivity and thrombosis potential. Circ Res. 2018;123(10):1164–1176. PubMed PMID: 30359185; PubMed Central PMCID: PMCPMC6223262. doi:10.1161/circresaha.118.313142. Epub 2018/10/26.
- Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165(1):111–124. PubMed PMID: 26972052; PubMed Central PMCID: PMCPMC4862743. doi:10.1016/j.cell.2016.02.011. Epub 2016/03/15.
- Zhu W, Buffa JA, Wang Z, Warrier M, Schugar R, Shih DM. Flavin monooxygenase 3, the host hepatic enzyme in the metaorganismal trimethylamine N-oxide-generating pathway, modulates platelet responsiveness and thrombosis risk. J Thromb Haemost. 2018;16(9):1857–1872. PubMed PMID: 29981269; PubMed Central PMCID: PMCPMC6156942. doi:10.1111/jth.14234. Epub 2018/07/08.
- Roberts AB, Gu X, Buffa JA, Hurd AG, Wang Z, Zhu W. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat Med. 2018;24(9):1407–1417. PubMed PMID: 30082863; PubMed Central PMCID: PMCPMC6129214. doi:10.1038/s41591-018-0128-1. Epub 2018/08/08.
- Seldin MM, Meng Y, Qi H, Zhu W, Wang Z, Hazen SL. Trimethylamine N-Oxide promotes vascular inflammation through signaling of Mitogen-Activated Protein Kinase and Nuclear Factor-κB. J Am Heart Assoc. 2016;5(2). Epub 2016/02/24. doi: 10.1161/jaha.115.002767. PubMed PMID: 26903003; PubMed Central PMCID: PMCPMC4802459.
- Chen ML, Zhu XH, Ran L, Lang HD, Yi L, Mi MT. Trimethylamine-N-Oxide induces vascular inflammation by activating the NLRP3 inflammasome through the SIRT3-SOD2-mtROS signaling pathway. J Am Heart Assoc. 2017;6(9). Epub 2017/09/06. doi: 10.1161/jaha.117.006347. PubMed PMID: 28871042; PubMed Central PMCID: PMCPMC5634285.
- Zhang X, Li Y, Yang P, Liu X, Lu L, Chen Y. Trimethylamine-N-Oxide promotes vascular calcification through activation of NLRP3 (Nucleotide-binding domain, Leucine-Rich-containing Family, Pyrin domain-containing-3) inflammasome and NF-κB (Nuclear Factor κB) signals. Arterioscler Thromb Vasc Biol. 2020;40(3):751–765. PubMed PMID: 31941382. doi:10.1161/atvbaha.119.313414. Epub 2020/01/17.
- Tang WH, Wang Z, Kennedy DJ, Wu Y, Buffa JA, Agatisa-Boyle B. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res. 2015;116(3):448–455. PubMed PMID: 25599331; PubMed Central PMCID: PMCPMC4312512. doi:10.1161/circresaha.116.305360. Epub 2015/01/20.
- Li XS, Obeid S, Klingenberg R, Gencer B, Mach F, Räber L. Gut microbiota-dependent trimethylamine N-oxide in acute coronary syndromes: a prognostic marker for incident cardiovascular events beyond traditional risk factors. Eur Heart J. 2017;38(11):814–824. Epub 2017/01/13. doi: 10.1093/eurheartj/ehw582. PubMed PMID: 28077467; PubMed Central PMCID: PMCPMC5837488.
- Li XS, Obeid S, Wang Z, Hazen BJ, Li L, Wu Y. Trimethyllysine, a trimethylamine N-oxide precursor, provides near- and long-term prognostic value in patients presenting with acute coronary syndromes. Eur Heart J. 2019;40(32):2700–2709. doi:10.1093/eurheartj/ehz259. PubMed PMID: 31049589
- Skagen K, Trøseid M, Ueland T, Holm S, Abbas A, Gregersen I. The Carnitine-butyrobetaine-trimethylamine-N-oxide pathway and its association with cardiovascular mortality in patients with carotid atherosclerosis. Atherosclerosis. 2016;247:64–69. doi:10.1016/j.atherosclerosis.2016.01.033. PubMed PMID: 26868510. Epub 2016/02/13.
- Tang WH, Wang Z, Fan Y, Levison B, Hazen JE, Donahue LM. Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-N-oxide in patients with heart failure: refining the gut hypothesis. J Am Coll Cardiol. 2014;64(18):1908–1914. PubMed PMID: 25444145; PubMed Central PMCID: PMCPMC4254529. doi:10.1016/j.jacc.2014.02.617. Epub 2014/12/03.
- Trøseid M, Ueland T, Hov JR, Svardal A, Gregersen I, Dahl CP. Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J Intern Med. 2015;277(6):717–726. PubMed PMID: 25382824. doi:10.1111/joim.12328. Epub 2014/11/11.
- Suzuki T, Heaney LM, Bhandari SS, Jones DJ, Ng LL. Trimethylamine N-oxide and prognosis in acute heart failure. Heart. 2016;102(11):841–848. PubMed PMID: 26869641. doi:10.1136/heartjnl-2015-308826. Epub 2016/02/13.
- Ge X, Zheng L, Zhuang R, Yu P, Xu Z, Liu G. The Gut microbial metabolite Trimethylamine N-Oxide and hypertension risk: a systematic review and dose-response meta-analysis. Adv Nutr. 2020;11(1):66–76. PubMed PMID: 31269204; PubMed Central PMCID: PMCPMC7442397. doi:10.1093/advances/nmz064. Epub 2019/07/04.
- Lau K, Srivatsav V, Rizwan A, Nashed A, Liu R, Shen R. Bridging the gap between gut microbial dysbiosis and cardiovascular diseases. Nutrients. 2017;9(8). Epub 2017/08/11. doi: 10.3390/nu9080859. PubMed PMID: 28796176; PubMed Central PMCID: PMCPMC5579652.
- Wang Z, Zhao Y. Gut microbiota derived metabolites in cardiovascular health and disease. Protein Cell. 2018;9(5):416–431. PubMed PMID: 29725935; PubMed Central PMCID: PMCPMC5960473. doi:10.1007/s13238-018-0549-0. Epub 2018/05/05.
- Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016;7(1):22–39. PubMed PMID: 26939849; PubMed Central PMCID: PMCPMC4856454. doi:10.1080/19490976.2015.1127483. Epub 2016/03/05.
- Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008;105(36):13580–13585. PubMed PMID: 18757757; PubMed Central PMCID: PMCPMC2533232. doi:10.1073/pnas.0804437105. Epub 2008/09/02.
- Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. PubMed PMID: 16299351. doi:10.1194/jlr.R500013-JLR200. Epub 2005/11/22.
- Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. 2018;57(1):1–24. PubMed PMID: 28393285; PubMed Central PMCID: PMCPMC5847071. doi:10.1007/s00394-017-1445-8. Epub 2017/04/11.
- Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res. 2009;50(8):1509–1520. PubMed PMID: 19346331; PubMed Central PMCID: PMCPMC2724047. doi:10.1194/jlr.R900007-JLR200. Epub 2009/04/07.
- Keitel V, Stindt J, Häussinger D. Bile Acid-activated receptors: GPBAR1 (TGR5) and other G Protein-coupled receptors. Handb Exp Pharmacol. 2019;256:19–49. doi:10.1007/164_2019_230. PubMed PMID: 31302759. Epub 2019/07/16.
- Kuipers F, Bloks VW, Groen AK. Beyond intestinal soap–bile acids in metabolic control. Nat Rev Endocrinol. 2014;10(8):488–498. PubMed PMID: 24821328. doi:10.1038/nrendo.2014.60. Epub 2014/05/14.
- Li T, Chiang JY. Bile acids as metabolic regulators. Curr Opin Gastroenterol. 2015;31(2):159–165. PubMed PMID: 25584736; PubMed Central PMCID: PMCPMC4332523. doi:10.1097/mog.0000000000000156. Epub 2015/01/15.
- Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103(10):3920–3925. PubMed PMID: 16473946; PubMed Central PMCID: PMCPMC1450165. doi:10.1073/pnas.0509592103. Epub 2006/02/14.
- Joyce SA, Gahan CG. Disease-associated changes in Bile Acid profiles and links to altered gut microbiota. Dig Dis. 2017;35(3):169–177. PubMed PMID: 28249284. doi:10.1159/000450907. Epub 2017/03/02.
- Adorini L, Schoonjans K, Friedman SL. Megatrends in bile acid receptor research. Hepatol Commun. 2017;1(8):831–835. PubMed PMID: 29404496; PubMed Central PMCID: PMCPMC5678912. doi:10.1002/hep4.1085. Epub 2018/02/07.
- Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E. Human insulin resistance is associated with increased plasma levels of 12α-hydroxylated bile acids. Diabetes. 2013;62(12):4184–4191. PubMed PMID: 23884887; PubMed Central PMCID: PMCPMC3837033. doi:10.2337/db13-0639. Epub 2013/07/26.
- Choucair I, Nemet I, Li L, Cole MA, Skye SM, Kirsop JD. Quantification of bile acids: a mass spectrometry platform for studying gut microbe connection to metabolic diseases. J Lipid Res. 2020;61(2):159–177. PubMed PMID: 31818878; PubMed Central PMCID: PMCPMC6997600. doi:10.1194/jlr.RA119000311. Epub 2019/12/11.
- Gu Y, Wang X, Li J, Zhang Y, Zhong H, Liu R. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat Commun. 2017;8(1):1785. PubMed PMID: 29176714; PubMed Central PMCID: PMCPMC5702614. doi:10.1038/s41467-017-01682-2. Epub 2017/11/28.
- Mayerhofer CCK, Ueland T, Broch K, Vincent RP, Cross GF, Dahl CP. Increased secondary/primary Bile Acid ratio in Chronic heart failure. J Card Fail. 2017;23(9):666–671. PubMed PMID: 28688889. doi:10.1016/j.cardfail.2017.06.007. Epub 2017/07/10.
- Ryan PM, Stanton C, Caplice NM. Bile acids at the cross-roads of gut microbiome-host cardiometabolic interactions. Diabetol Metab Syndr. 2017;9(1):102. PubMed PMID: 29299069; PubMed Central PMCID: PMCPMC5745752. doi:10.1186/s13098-017-0299-9. Epub 2018/01/05.
- Jones ML, Martoni CJ, Prakash S. Cholesterol lowering and inhibition of sterol absorption by Lactobacillus reuteri NCIMB 30242: a randomized controlled trial. Eur J Clin Nutr. 2012;66(11):1234–1241. PubMed PMID: 22990854. doi:10.1038/ejcn.2012.126. Epub 2012/09/20.
- Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7(3):189–200. PubMed PMID: 26963409; PubMed Central PMCID: PMCPMC4939913. doi:10.1080/19490976.2015.1134082. Epub 2016/03/11.
- Trøseid M, Andersen G, Broch K, Hov JR. The gut microbiome in coronary artery disease and heart failure: current knowledge and future directions. EBioMedicine. 2020;52:102649. doi:10.1016/j.ebiom.2020.102649. PubMed PMID: 32062353; PubMed Central PMCID: PMCPMC7016372. Epub 2020/02/18.
- Willemsen LE, Koetsier MA, Van Deventer SJ, Van Tol EA. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut. 2003;52(10):1442–1447. PubMed PMID: 12970137; PubMed Central PMCID: PMCPMC1773837. doi:10.1136/gut.52.10.1442. Epub 2003/09/13.
- Pluznick J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes. 2014;5(2):202–207. PubMed PMID: 24429443; PubMed Central PMCID: PMCPMC4063845. doi:10.4161/gmic.27492. Epub 2014/01/17.
- Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A. 2013;110(11):4410–4415. PubMed PMID: 23401498; PubMed Central PMCID: PMCPMC3600440. doi:10.1073/pnas.1215927110. Epub 2013/02/13.
- Natarajan N, Hori D, Flavahan S, Steppan J, Flavahan NA, Berkowitz DE. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G protein-coupled receptor 41. Physiol Genomics. 2016;48(11):826–834. PubMed PMID: 27664183; PubMed Central PMCID: PMCPMC6223570. doi:10.1152/physiolgenomics.00089.2016. Epub 2016/09/25.
- Hur KY, Lee MS. Gut microbiota and metabolic disorders. Diabetes Metab J. 2015;39(3):198–203. PubMed PMID: 26124989; PubMed Central PMCID: PMCPMC4483604. doi:10.4093/dmj.2015.39.3.198. Epub 2015/07/01.
- Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145(2):396–406. PubMed PMID: 23665276. doi:10.1053/j.gastro.2013.04.056. e1-10. Epub 2013/05/15.
- Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61(2):364–371. PubMed PMID: 22190648; PubMed Central PMCID: PMCPMC3266401. doi:10.2337/db11-1019. Epub 2011/12/23.
- Psichas A, Sleeth ML, Murphy KG, Brooks L, Bewick GA, Hanyaloglu AC. The short chain fatty acid propionate stimulates GLP-1 and PYY secretion via free fatty acid receptor 2 in rodents. Int J Obes (Lond). 2015;39(3):424–429. PubMed PMID: 25109781; PubMed Central PMCID: PMCPMC4356745. doi:10.1038/ijo.2014.153. Epub 2014/08/12.
- Drucker DJ. The cardiovascular biology of Glucagon-like Peptide-1. Cell Metab. 2016;24(1):15–30. PubMed PMID: 27345422. doi:10.1016/j.cmet.2016.06.009. Epub 2016/06/28.
- Lønborg J, Vejlstrup N, Kelbæk H, Bøtker HE, Kim WY, Mathiasen AB. Exenatide reduces reperfusion injury in patients with ST-segment elevation myocardial infarction. Eur Heart J. 2012;33(12):1491–1499. PubMed PMID: 21920963. doi:10.1093/eurheartj/ehr309. Epub 2011/09/17.
- Ussher JR, Baggio LL, Campbell JE, Mulvihill EE, Kim M, Kabir MG. Inactivation of the cardiomyocyte glucagon-like peptide-1 receptor (GLP-1R) unmasks cardiomyocyte-independent GLP-1R-mediated cardioprotection. Mol Metab. 2014;3(5):507–517. PubMed PMID: 25061556; PubMed Central PMCID: PMCPMC4099509. doi:10.1016/j.molmet.2014.04.009. Epub 2014/07/26.
- Bartolomaeus H, Balogh A, Yakoub M, Homann S, Markó L, Höges S. Short-chain fatty acid propionate protects from hypertensive cardiovascular damage. Circulation. 2019;139(11):1407–1421. PubMed PMID: 30586752; PubMed Central PMCID: PMCPMC6416008. doi:10.1161/circulationaha.118.036652. Epub 2018/12/28.
- Tang TWH, Chen HC, Chen CY, Yen CYT, Lin CJ, Prajnamitra RP. Loss of gut microbiota alters Immune system composition and Cripples Postinfarction Cardiac repair. Circulation. 2019;139(5):647–659. PubMed PMID: 30586712. doi:10.1161/circulationaha.118.035235. Epub 2018/12/28.
- Kasahara K, Krautkramer KA, Org E, Romano KA, Kerby RL, Vivas EI. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat Microbiol. 2018;3(12):1461–1471. PubMed PMID: 30397344; PubMed Central PMCID: PMCPMC6280189. doi:10.1038/s41564-018-0272-x. Epub 2018/11/07.
- Sallée M, Dou L, Cerini C, Poitevin S, Brunet P, Burtey S. The aryl hydrocarbon receptor-activating effect of uremic toxins from tryptophan metabolism: a new concept to understand cardiovascular complications of chronic kidney disease. Toxins (Basel). 2014;6(3):934–949. PubMed PMID: 24599232; PubMed Central PMCID: PMCPMC3968369. doi:10.3390/toxins6030934. Epub 2014/03/07.
- Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med. 2016;22(6):586–597. PubMed PMID: 27158906; PubMed Central PMCID: PMCPMC4899206. doi:10.1038/nm.4106. Epub 2016/05/10.
- Nemet I, Saha PP, Gupta N, Zhu W, Romano KA, Skye SM. A cardiovascular disease-linked gut microbial metabolite acts via Adrenergic receptors. Cell. 2020;180(5). doi:10.1016/j.cell.2020.02.016. PubMed PMID: 32142679.
- Xu X, He J, Xue J, Wang Y, Li K, Zhang K. Oral cavity contains distinct niches with dynamic microbial communities. Environ Microbiol. 2015;17(3):699–710. PubMed PMID: 24800728. doi:10.1111/1462-2920.12502. Epub 2014/05/08.
- Aarabi G, Heydecke G, Seedorf U. Roles of oral infections in the pathomechanism of Atherosclerosis. Int J Mol Sci. 2018;19(7). Epub 2018/07/11. doi: 10.3390/ijms19071978. PubMed PMID: 29986441; PubMed Central PMCID: PMCPMC6073301.
- Eberhard J, Grote K, Luchtefeld M, Heuer W, Schuett H, Divchev D. Experimental gingivitis induces systemic inflammatory markers in young healthy individuals: a single-subject interventional study. PLoS One. 2013;8(2):e55265. PubMed PMID: 23408963; PubMed Central PMCID: PMCPMC3567060. doi:10.1371/journal.pone.0055265. Epub 2013/02/15.
- Martin-Cabezas R, Seelam N, Petit C, Agossa K, Gaertner S, Tenenbaum H. Association between periodontitis and arterial hypertension: a systematic review and meta-analysis. Am Heart J. 2016;180:98–112. doi:10.1016/j.ahj.2016.07.018. PubMed PMID: 27659888. Epub 2016/09/24.
- Chen H, Jiang W. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front Microbiol. 2014;5:508. doi:10.3389/fmicb.2014.00508. PubMed PMID: 25352835; PubMed Central PMCID: PMCPMC4195358. Epub 2014/10/30.
- Marcelino SL, Gaetti-Jardim E Jr., Nakano V, Canônico LA, Nunes FD, Lotufo RF. Presence of periodontopathic bacteria in coronary arteries from patients with chronic periodontitis. Anaerobe. 2010;16(6):629–632. PubMed PMID: 20816998. doi:10.1016/j.anaerobe.2010.08.007. Epub 2010/09/08.
- Fernandes CP, Oliveira FA, Silva PG, Alves AP, Mota MR, Montenegro RC. Molecular analysis of oral bacteria in dental biofilm and atherosclerotic plaques of patients with vascular disease. Int J Cardiol. 2014;174(3):710–712. PubMed PMID: 24820755. doi:10.1016/j.ijcard.2014.04.201. Epub 2014/05/14.
- Cavrini F, Sambri V, Moter A, Servidio D, Marangoni A, Montebugnoli L. Molecular detection of Treponema denticola and Porphyromonas gingivalis in carotid and aortic atheromatous plaques by FISH: report of two cases. J Med Microbiol. 2005;54(Pt 1):93–96. PubMed PMID: 15591262. doi:10.1099/jmm.0.45845-0. Epub 2004/12/14.
- Okuda K, Ishihara K, Nakagawa T, Hirayama A, Inayama Y, Okuda K. Detection of Treponema denticola in atherosclerotic lesions. J Clin Microbiol. 2001;39(3):1114–1117. PubMed PMID: 11230436; PubMed Central PMCID: PMCPMC87882. doi:10.1128/jcm.39.3.1114-1117.2001. Epub 2001/03/07.
- Aderem A. Phagocytosis and the inflammatory response. J Infect Dis. 2003;187(Suppl s2):S340–5. PubMed PMID: 12792849. doi:10.1086/374747. Epub 2003/06/07.
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. PubMed PMID: 16497588. doi:10.1016/j.cell.2006.02.015. Epub 2006/02/25.
- Areschoug T, Gordon S. Pattern recognition receptors and their role in innate immunity: focus on microbial protein ligands. Contrib Microbiol. 2008;15:45–60. doi:10.1159/000135685. PubMed PMID: 18511855. Epub 2008/05/31.
- Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124(4):823–835. PubMed PMID: 16497591. doi:10.1016/j.cell.2006.02.016. Epub 2006/02/25.
- Peiser L, Mukhopadhyay S, Gordon S. Scavenger receptors in innate immunity. Curr Opin Immunol. 2002;14(1):123–128. PubMed PMID: 11790542. doi:10.1016/s0952-7915(01)00307-7. Epub 2002/01/16.
- Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cell. 2001;104(4):503–516. PubMed PMID: 11239408. doi:10.1016/s0092-8674(01)00238-0. Epub 2001/03/10.
- Schaefer AS, Richter GM, Groessner-Schreiber B, Noack B, Nothnagel M, El Mokhtari NE. Identification of a shared genetic susceptibility locus for coronary heart disease and periodontitis. PLoS Genet. 2009;5(2):e1000378. PubMed PMID: 19214202; PubMed Central PMCID: PMCPMC2632758. doi:10.1371/journal.pgen.1000378. Epub 2009/02/14.
- Schaefer AS, Bochenek G, Jochens A, Ellinghaus D, Dommisch H, Güzeldemir-Akçakanat E. Genetic evidence for PLASMINOGEN as a shared genetic risk factor of coronary artery disease and periodontitis. Circ Cardiovasc Genet. 2015;8(1):159–167. PubMed PMID: 25466412. doi:10.1161/circgenetics.114.000554. Epub 2014/12/04.
- Li X, Tse HF, Jin LJ. Novel endothelial biomarkers: implications for periodontal disease and CVD. J Dent Res. 2011;90(9):1062–1069. PubMed PMID: 21321068. doi:10.1177/0022034510397194. Epub 2011/02/16.
- Lockhart PB, Bolger AF, Papapanou PN, Osinbowale O, Trevisan M, Levison ME. Periodontal disease and atherosclerotic vascular disease: does the evidence support an independent association?: a scientific statement from the American Heart Association. Circulation. 2012;125(20):2520–2544. PubMed PMID: 22514251. doi:10.1161/CIR.0b013e31825719f3. Epub 2012/04/20.
- Park SY, Kim SH, Kang SH, Yoon CH, Lee HJ, Yun PY. Improved oral hygiene care attenuates the cardiovascular risk of oral health disease: a population-based study from Korea. Eur Heart J. 2019;40(14):1138–1145. PubMed PMID: 30561631. doi:10.1093/eurheartj/ehy836. Epub 2018/12/19.
- Bordenstein SR, Theis KR. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 2015;13(8):e1002226. PubMed PMID: 26284777; PubMed Central PMCID: PMCPMC4540581. doi:10.1371/journal.pbio.1002226. Epub 2015/08/19.
- Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16(1):191. PubMed PMID: 26374288; PubMed Central PMCID: PMCPMC4570153. doi:10.1186/s13059-015-0759-1. Epub 2015/09/17.
- Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332(6032):970–974. PubMed PMID: 21596990; PubMed Central PMCID: PMCPMC3303602. doi:10.1126/science.1198719. Epub 2011/05/21.
- Moeller AH, Foerster S, Wilson ML, Pusey AE, Hahn BH, Ochman H. Social behavior shapes the chimpanzee pan-microbiome. Sci Adv. 2016;2(1):e1500997. PubMed PMID: 26824072; PubMed Central PMCID: PMCPMC4730854. doi:10.1126/sciadv.1500997. Epub 2016/01/30.
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141(7):1135–1145. PubMed PMID: 20602997; PubMed Central PMCID: PMCPMC2908380. doi:10.1016/j.cell.2010.05.009. Epub 2010/07/07.
- Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464(7290):908–912. PubMed PMID: 20376150. doi:10.1038/nature08937. Epub 2010/04/09.
- Szilagyi A. Adaptation to lactose in lactase non persistent people: effects on intolerance and the relationship between dairy food consumption and evaluation of diseases. Nutrients. 2015;7(8):6751–6779. PubMed PMID: 26287234; PubMed Central PMCID: PMCPMC4555148. doi:10.3390/nu7085309. Epub 2015/08/20.
- Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6(8):1535–1543. PubMed PMID: 22343308; PubMed Central PMCID: PMCPMC3400402. doi:10.1038/ismej.2012.4. Epub 2012/02/22.
- Suzuki TA, Ley RE. The role of the microbiota in human genetic adaptation. Science. 2020;370(6521). Epub 2020/12/05. doi: 10.1126/science.aaz6827. PubMed PMID: 33273073.
- Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E. Non-lethal inhibition of gut microbial Trimethylamine production for the treatment of Atherosclerosis. Cell. 2015;163(7):1585–1595. PubMed PMID: 26687352; PubMed Central PMCID: PMCPMC4871610. doi:10.1016/j.cell.2015.11.055. Epub 2015/12/22.
- Tang WHW, Li DY, Hazen SL. Dietary metabolism, the gut microbiome, and heart failure. Nat Rev Cardiol. 2019;16(3):137–154. PubMed PMID: 30410105; PubMed Central PMCID: PMCPMC6377322. doi:10.1038/s41569-018-0108-7. Epub 2018/11/10.
- Eblimit Z, Thevananther S, Karpen SJ, Taegtmeyer H, Moore DD, Adorini L. TGR5 activation induces cytoprotective changes in the heart and improves myocardial adaptability to physiologic, inotropic, and pressure-induced stress in mice. Cardiovasc Ther. 2018;36(5):e12462. PubMed PMID: 30070769; PubMed Central PMCID: PMCPMC6800140. doi:10.1111/1755-5922.12462. Epub 2018/08/03.
- Durack J, Lynch SV. The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med. 2019;216(1):20–40. PubMed PMID: 30322864; PubMed Central PMCID: PMCPMC6314516. doi:10.1084/jem.20180448. Epub 2018/10/17.
- De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65(11):1812–1821. PubMed PMID: 26416813. doi:10.1136/gutjnl-2015-309957. Epub 2015/09/30.
- Marques FZ, Nelson E, Chu PY, Horlock D, Fiedler A, Ziemann M. High-fiber diet and Acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation. 2017;135(10):964–977. PubMed PMID: 27927713. doi:10.1161/circulationaha.116.024545. Epub 2016/12/09.
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. Epub 2012/06/16. doi: 10.1038/nature11234. PubMed PMID: 22699609; PubMed Central PMCID: PMCPMC3564958.
- Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–1094. PubMed PMID: 26590418. doi:10.1016/j.cell.2015.11.001. Epub 2015/11/23.
- Kolodziejczyk AA, Zheng D, Elinav E. Diet-microbiota interactions and personalized nutrition. Nat Rev Microbiol. 2019;17(12):742–753. PubMed PMID: 31541197. doi:10.1038/s41579-019-0256-8. Epub 2019/09/22.
