ABSTRACT
Most cirrhosis etiologies, such as alcohol, hepatitis C, and obesity, involve behavior that require the loss of inhibitory control. Once cirrhosis develops, patients can also develop cognitive impairment due to minimal hepatic encephalopathy (MHE). Both processes could have distinct imprints on the gut-liver-brain axis. Determine the impact of inhibitory control versus traditional cirrhosis-related cognitive performance on gut microbial composition and function. Outpatients with cirrhosis underwent two tests for MHE: inhibitory control test (MHEICT, computerized associated with response inhibition) and psychometric hepatic encephalopathy score (MHEPHES, paper-pencil HE-specific associated with subcortical impairment) along with stool collection for metagenomics. MHEICT/not, MHEPHES/not, and discordant (positive on one test but negative on the other) were analyzed for demographics, bacterial species, and gut-brain modules (GBM) using multi-variable analyses. Ninety-seven patients [47 (49%) MHEPHES, 76 (78%) MHEICT, 41 discordant] were enrolled. MHEPHES/not: Cirrhosis severity was worse in MHEPHES without differences in alpha/beta diversity on bacterial species or GBMs. Pathobionts (Enterobacteriaceae) and γ-amino-butryic acid (GABA) synthesis GBM were higher in MHEPHES. MHEICT/not: We found similar cirrhosis severity and metagenomic alpha/beta diversity in MHEICT versus not. However, alpha/beta diversity of GBMs were different in MHEICT versus No-MHE patients. Alistipes ihumii, Prevotella copri, and Eubacterium spp. were higher, while Enterococcus spp. were uniquely lower in MHEICT versus no-MHE and discordant comparisons. GBMs belonging to tryptophan, menaquinone, GABA, glutamate, and short-chain fatty acid synthesis were also unique to MHEICT. Gut microbial signature of impaired inhibitory control, which is associated with addictive disorders that can lead to cirrhosis, is distinct from cirrhosis-related cognitive impairment.
Introduction
Recent research suggests that an altered gut–brain axis is associated with several conditions, such as depression, anxiety, Parkinson’s disease, cirrhosis, and hepatic encephalopathy (HE)Citation.1 In cirrhosis, the impact on the brain is multifactorial and includes liver disease and multiple co-morbid conditions, as well as etiologies of the cirrhosis (e.g., alcohol, hepatitis C, obesity, and diabetes)Citation.2 Therefore, cirrhosis and HE are a microcosm of several factors that can impact the gut-brain axis, where gut microbial manipulation can be used as therapyCitation.3 However, cognitive impairment in cirrhosis can precede the confusional status of overt HECitation.3 This cognitive impairment or minimal HE (MHE) is an anamnestic form of mild cognitive impairment, which portends further complications and can impact survivalCitation.3 MHE can be measured using several strategies, such as paper-pencil or computerized testsCitation.4 The paper-pencil psychometric hepatic encephalopathy score (PHES), which includes five tests evaluates psychomotor speed, cognitive flexibility, and accuracy, while the inhibitory control test (ICT) evaluates working memory and inhibitory controlCitation.3 Since these tests interrogate separate parts of the brain, the gut contribution to individual test performance may increase our understanding of the gut-brain axis changes as cirrhosis progressesCitation.5 Prior 16SrRNA analyses have shown that microbial taxa differentially associate with specialized brain imaging changes and specific cognitive tests in MHE, but a deeper metagenomic evaluation of the microbiota that are associated with specific cognitive impairments are needed.Citation6–9
Our aim was to determine the linkage between bacterial metagenomic composition and function with specific cognitive tasks in patients with cirrhosis.
Methods:
Outpatients with cirrhosis and healthy controls were recruited from Virginia Commonwealth University and the Central Virginia Veterans Healthcare system after IRB approval. After informed consent, all subjects underwent PHES and ICT. All stool were collected in RNALater with DNA extraction using published techniques.
Clinical data for patients with cirrhosis pertaining to demographics, liver disease etiology, Model for End-stage Liver Disease (MELD) score, prior HE history, and current therapy with lactulose or rifaximin and cognitive analysis were collected.PHES details: This is a validated five test paper-pencil battery which tests visuo-motor coordination, psychomotor speed, and reaction timeCitation.5 It consists of the number connection test-A, number connection test B, digit symbol test, serial dotting test, and line tracing test (has two components: time and errors). Of these, a high raw score on digit symbol and low time for completion or errors in the remaining tests indicate normal cognition. Based on population control values, the standard deviations are calculated for each sub-test, and the total is added to give one valueCitation.3 A low score on the total PHES score indicates better performance.
ICT details:Citation10 ICT involves the presentation of several letters at 500-ms intervalsCitation.11 Interspersed within these letters are the letters X and Y. The subject is instructed to respond to every X and Y during the initial part of the training run, which establishes the prepotent response. In the latter part of the training run, the subject is instructed only to respond when X and Y are alternating (called targets) and inhibit responding when X and Y are not alternating (called lures). After the training run, 6 test runs, which last approximately 2 min each, are administered with a total of 40 lures, 212 targets, and 1728 random letters in between. At the end of the test, the lure and target response rates are automatically calculated. Lower lure response and higher target response indicate better performanceCitation.11 MHE on PHES and ICT were based on norms created for the Virginia populationCitation.12 We also determined concordant or discordant (negative on one and positive on the other versus vice-versa) performances on these tests.
Stool Collection and analysis details: Metagenomic DNA from fecal samples was extracted using the MO BIO PowerFecal DNA Isolation Kit (Qiagen) and stored in our repository at −80°C until the metagenomics analysisCitation.13 Samples were processed in an automated, high throughput manner using the QiaCube DNA/RNA Purification System (Qiagen) with bead beating in 0.1 mm glass bead plates. Isolated DNA was quantified and normalized using the Quant-iT Picogreen dsDNA Assay Kit. Shotgun metagenomic libraries were prepared with a procedure adapted from the Nextera Library Prep Kit (Illumina). Libraries were subsequently pooled and assessed using the Agilent Bioanalyzer. Sequencing was performed on either an Illumina NextSeq 550 (1 x 150 bp, NextSeq 500/550 High Output v2 kit) or an Illumina NovaSeq 6000 (1 x 100 bp, NovaSeq 6000 S2 Reagent Kit). Metagenomic analysis: Reads were processed and annotated using the BoosterShot in-house pipelineCitation.13 Bcl files were converted to fastq format using bcl2fastq (Illumina). Cutadapt Citation14 was used for adapter and quality (final Q-score > 20) trimming. Reads shorter than 50 bp were filtered out using cutadapt, and all reads were trimmed to 100 bp prior to downstream alignment and annotation. Quality sequences were then aligned at 97% identity to a curated database (Venti) containing all representative genomes in RefSeq Citation15 for bacteria and additional manually curated strains using the BURST optimal alignerCitation.16 Ties in alignment were broken by minimizing the overall number of unique Operational Taxonomic Units (OTUs). For taxonomic assignment, each input sequence was assigned the lowest common ancestor, which was consistent across at least 80% of all reference sequences tied for best hit. Counts were normalized to the species-level average genome length. OTUs accounting for less than one millionth of all species-level genomic markers were discarded, as well as those with either less than 0.01% of their unique genome or less than 1% of the whole genome covered by reads in any sample. The Shannon index, Chao1 index and observed OTU alpha diversity metrics were calculated from count tables rarefied to 40,000 reads per sample using the veganCitation17 R package.
We first analyzed patients who were PHES positive compared to PHES negative in the entire population and then similar analysis for those who were ICT positive and negative. We then analyzed those who were discordant and those who were MHEICT positive versus those who negative on ICT and similarly for MHEPHES (Fig S1). We performed MAAslin2 analysis of patients including age, gender, diabetes, PPI use, prior HE, lactulose and rifaximin use, psychoactive medications, depression and anxiety with the bacterial species comparing patients MHEICT versus not, similarly for PHES and for discordant patientsCitation.18 Alpha diversity (richness, Shannon, and Simpson), and beta-diversity (PERMANOVA and PCoA) were performed using BiomMinerCitation.19 Finally, similar analyses were performed based on gut-brain module (GBM) between MHEICT/not, MHEPHES/not and those who were discordantCitation.8 GBMs assembly database is a metabolic reconstruction framework specific for translating shotgun metagenomic data into microbial neuroactive metabolic potential was constructed based on extensive literature and database (MetaCyc) reviewCitation.8 A set of 56 GBMs was assembled, each corresponding to a process of synthesis or degradation of a neuroactive compound by the members of the gut microbiota. Module structure follows the Kyoto Encyclopedia of Genes and Genomes (KEGG) database syntax as previously constructed for the gut microbial metabolic food chainCitation.9 GBM presence in the metagenome is defined with a detection threshold of at least 66% coverage to provide tolerance to miss-annotations, and missing data are incomplete(draft) genomes. GBM abundances were derived from an orthologue abundance table using Omixer-RPM version 1.0 (https://github.com/raeslab/omixer-rpm) by matching calculated KEGG IDs with GBMs database curated KEGG IDs.
Results:
Demographic comparisons:
Ninety-seven patients with cirrhosis were included (). Approximately, half of the patients had MHEPHES diagnosed per norms. These patients were more likely to be advanced in their cirrhosis, with a higher proportion of men with alcohol-related etiology and PPI use compared to those negative on PHES. On the other hand, no significant changes in demographics and cirrhosis characteristics were found in MHEICT versus no-MHE. The results of MHE were concordant in 56 patients (15 patients negative on both and 41 positive on both). However, discordance was seen in the remaining 41 patients (35 patients MHEICT but not on PHES and 6 MHEPHES positive and negative on ICT, Figure S1).
Table 1. Details of Patients with Minimal Hepatic Encephalopathy on Psychometric Hepatic Encephalopathy Score (PHES) and on Inhibitory Control test (ICT) (N = 97)
Bacterial species comparisons:
MHEPHES versus not:. No changes in alpha/beta-diversity (PERMANOVA p = .23) were seen between PHES positive/negative patients (). On MAAsLin2 top 30 variables with MHE-PHES, MELD score, several Lactobacillus spp. and species belonging to Enterobacteriaceae (Klebsiella, Klyuvera, Pectobacterium) but not Enterococcus spp. were higher. Several members of the Prevotellaceae family (Prevotella ruminicola), short-chain fatty acid (SCFA) producers such as Ruminococcaceae spp. and Clostridium aerotolerans, and species belonging to Desulfovibrio and Bacteroides were not associated with MHEPHES.
Figure 1. Bacterial species comparison between patients with MHEPHES (n = 47) versus not (n = 50)1A: Alpha diversity analyses did not show any differences between groups 1B: Cleveland plot derived from DESeq2 comparison 1 C: PCoA showing no significant separation between groups (PERMANOVA not significant)
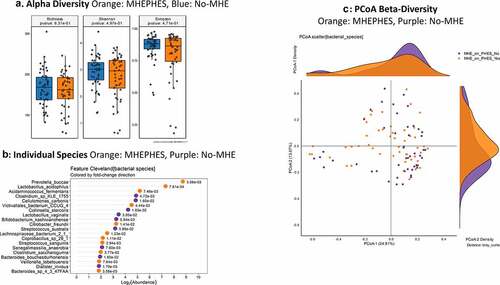
As a whole on MAasLin2, higher age and MELD score, and prior HE, lactulose and rifaximin use were associated with MHEPHES. Several species belonging to symbionts such as Akkermansia, as well as potential SCFA producers belonging to Lachnospiraceae and Ruminococcaceae were higher in those without MHE, while the reverse was seen with potential pathobionts belonging to Proteobacteria and urease-producing Streptococcus species ( and Table S1).
Table 2. MAAsLin2 Top 30 Bacterial Species and Clinical Variables associated with MHE on Individual tests
MHEICT versus not: Similar to PHES, no changes in alpha-diversity was seen and PERMANOVA of borderline significance (p = .08; ) was seen for beta-diversity. On MAAsLin2, most bacterial species in the top 30 were negatively linked to MHEICT with the majority of these belonging to Enterococcus, Veillonella, Clostridia and Ruminococcacae spp. Dakarella massiliensis was the only microbe linked to MHE-ICT in the top 30 ( and Table S2). Remaining microbes associated with MHEICT were Alistipes ihumii, Megasphaera massiliensis, Prevotella copri, Eubacterium spp., and Bifidobacterium adolescentis.
Table 3. MAAsLin2 Top 30 Bacterial Species and Clinical Variables
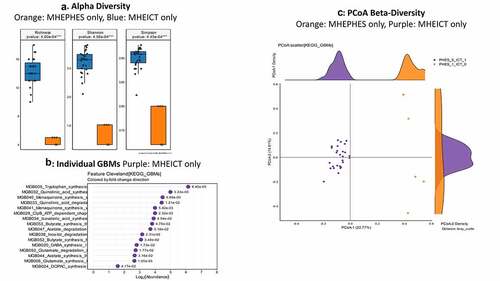
Discordant: No changes in alpha/beta-diversity were seen in MHEICT only versus MHEPHES only patients (). On MAAsLin2, several Enterococcus, Streptococcus and pathobiont gram-negative species were higher in MHEPHES only patients while Prevotella copri, Eggerthela and Alistipes spp. were higher in MHEICT only patients in the top 30 ( and Table S3). MHEICT only versus No MHEICT: For the 35 patients impaired on ICT versus 21 with normal performance, we found lower Enterococcus spp. and higher Prevotella spp., Dakarella massiliensis and potential autochthonous species in those with MHEICT only (). MHEPHES only versus No MHEPHES: When we compared the 6 patients only impaired on PHES to the 50 patients who had normal performance on PHES, we found higher Enterococcus, Streptococcus spp. in MHEPHES only patients. Also, higher cirrhosis severity (higher MELD score, Prior HE, lactulose and rifaximin use) were higher in those with MHEPHES only patients compared to No MHEPHES patients ().
Gut-brain modules:
MHEPHES versus not: No differences in GBM alpha/beta-diversity was regardless of MHEPHES/not. MHEPHES patients had a higher abundance of GBMs related to GABA and glutamate synthesis, nitric oxide and propionate degradation on Metatstats, while those lower were related to butyrate, isovalerate, menaquinone and DOPAC synthesis, and degradation of quinolonic acid, NO and glutamate ( and ). MHEICT versus not: GBM alpha-diversity was higher in MHEICT versus No MHEICT, which also were separated on PCoA (PERMANOVA, p = .001). GBMs that were higher in MHEICT versus No MHEICT were focused on quinolonic acid, menaquinone, GABA, DOPAC and SCFA pathways, while the opposite was seen for GHB degradation ( and ). Discordant: In patients who were MHEICT but not PHES, there was higher GBM alpha-diversity. PERMANOVA (p = .001) also showed a clear separation between MHEICT-NoPHES and MHEPHES-NoICT ( and ). GABA synthesis III was the only GBM uniquely higher in MHEPHES. In MHEICT, DOPAC synthesis, SCFAs (Isovalerate synthesis-I KADH pathway, Butyrate synthesis-I), menaquinone synthesis, tryptophan and quinolinic acid synthesis, inositol and glutamate degradation and ClpB-ATP-dependent chaperone protein were higher.
Table 4. Comparison of Gut-Brain Modules Different in Patients According to Cognitive Strategy used
Figure 2. Gut brain module comparison between patients with MHEPHES (n = 47) versus not (n = 50) 1A: Alpha diversity analyses did not show any differences between groups 1B: Cleveland plot derived from Metastats comparison 1 C: PCoA showing no significant separation between groups (PERMANOVA not significant)
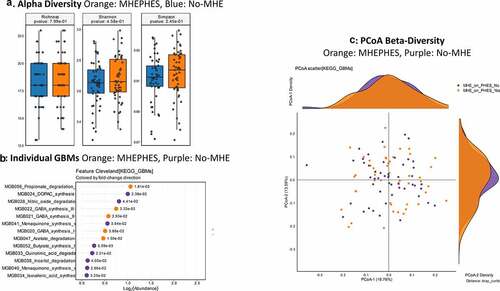
Figure 3. Bacterial species comparison between patients with MHEICT (n = 76) versus not (n = 21) 1A: Alpha diversity analyses did not show any differences between groups 1B: Cleveland plot derived from DESeq2 comparison 1 C: PCoA showing trend toward a significant separation between groups (PERMANOVA p = .08)
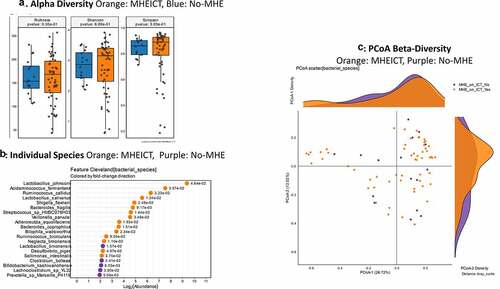
Figure 4. Gut brain module comparison between patients with MHEICT (n = 76) versus not (n = 21)1A: Alpha diversity analyses showed significantly higher diversity in the MHEICT group compared to no-MHE 1B: Cleveland plot derived from Metastats comparison 1 C: PCoA showing a significant separation between groups
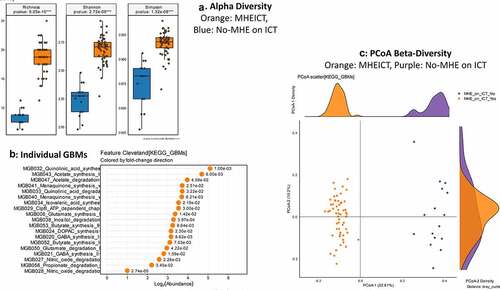
Figure 5. Bacterial species comparison between patients with MHEICT-only (n = 35) versus MHEPHES only (n = 6)1A: Alpha diversity analyses did not show any differences between groups 1B: Cleveland plot derived from DESeq2 comparison 1 C: PCoA showing no significant separation between groups
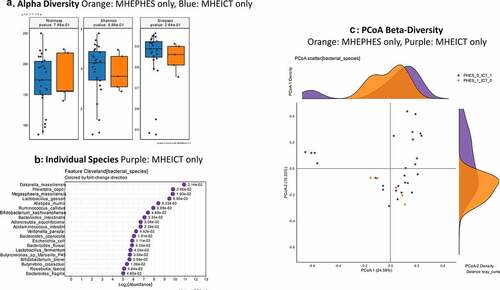
Figure 6. Gut brain module comparison between patients with MHEICT-only (n = 35) versus MHEPHES only (n = 6) 1A: Alpha diversity analyses showed significantly higher diversity in the MHEICT only group compared to MHEPHES only group 1B: Cleveland plot derived from Metastats comparison 1 C: PCoA showing a significant separation between groups
Discussion:
In this study, we found that gut-brain axis changes in cirrhosis differed based on impairment on a measure of inhibitory control, a major determinant of several high-risk impulsive human behaviors which can result in self-harm. These include alcohol misuse, food addiction, and illicit drug use, which can lead to liver disease directly or through hepatitis C. With the further progression of liver disease, an inflammatory milieu and hyperammonemia can impair cognitive performance on visuospatial, psychomotor, and cognitive flexibility-related functions. Therefore, the cognitive impairment in cirrhosis could be a result of cirrhosis itself, the etiology of the cirrhosis or both. Gut-brain axis alterations in cirrhosis have been studied by several groups and favorable manipulation using laxatives, antibiotics, and fecal microbiota transplant have led to beneficial cognitive outcomes.Citation20–24 A deeper characterization of functional microbial modules and their linkage with cognitive dysfunction in cirrhosis is relevant to define potential targets that can be extended beyond cirrhosis. As previously reported, there was relatively poor concordance between MHEICT versus MHEPHESCitation.25 Regardless, we did not find alpha or beta-diversity changes between groups with/without both forms of MHE. This similarity is at odds with the demographic and cirrhosis profile since MHEPHES patients had more advanced liver disease compared to unimpaired patients while MHEICT patients did not. This points toward a relationship between the microbiota and cognitive function that could be independent of the liver disease.Citation26–31
In addition, the PHES emphasizes psychomotor speed, which on a neuronal level reflects the integration between primary motor function (i.e., dopaminergic-based subcortical-cortical motor circuit) and non-motor function (i.e., cognition and emotion)Citation.32 It is a 5-test battery, which is mostly sub-cortical and is usually specific for cirrhosis. On the other hand, ICT evaluates working memory storage and an individual’s ability to override, or inhibit, a habitual behavioral responseCitation33 that depends on prefrontal cortex integrity. In addition to inhibitory control, good ICT performance requires strong working memory skills. Working memory (WM) involves the temporary storage, and subsequent manipulation of data. Given the PHES’s emphasis is on simple psychomotor speed (which depends on subcortical structures), compared to the more complex, and cognitively demanding task in ICT that emphasizes higher cortical processing, it is not surprising that more patients were impaired on ICT.
However, regardless of testing strategy, SCFA producers and symbionts tended to be lower in those with MHE even on multi-variable analyses. On the other hand, potential gram-negative pathobionts belonging to Enterobacteriaceae, as well as Lactobacillus and Veillonella spp. were higher in both MHE groups. These taxa have been associated with cirrhosis progression, as well as the production of GABA, which can promulgate cognitive impairmentCitation.34 This was further reflected in the GBMs with those centered around the synthesis of GABA, and the degradation of propionate and NO (through NO dioxygenase) were higher in MHE regardless of modality. GABA is a major inhibitory neurotransmitter, which is associated with cirrhosis progression and HE. GABA is produced by several different microbiota including Lactobacillus, Escherichia, and Bifidobacterium, which are elevated in advancing cirrhosis and in MHE patients.Citation35 GABA synthesis I–II are over-represented in E. coli and K. aerogenes spp. and involve putrescine to GABA conversion via glutamate or 2-oxoglutarateCitation.36 Glutamate is a major excitatory neurotransmitter while GABA is inhibitory; pathways overexpressing conversion to GABA are likely related to cognitive impairment. NO dioxygenase degradation usually protects against nitrosative stress through the expression of flavoHgb in bacteria and fungi,Citation37 however, the overexpression can lead to oxidative stress, which has been found in HECitation.38 Moreover, the major inducer, NO, is suppressed in patients with cirrhosis and portal hypertensionCitation.39 Propionate is a major SCFA which can affect the gut barrier function as well as promote immune surveillance, neuronal health and integrity of the blood–brain barrierCitation.40 Therefore, the association of MHE with enhanced propionate degradation is not surprising.
While MHE on PHES was associated with advancing cirrhosis severity, MHEICT was not. Therefore, the discordance between Enterobacteriaceae and Enterococcus spp. based on mode for MHE diagnosis is interesting because prior studies have indicated that both the taxa increase with advancing cirrhosis. Since this pattern of higher Enterobacteriaceae members in both MHE groups but reduction in Enterococcus abundance in MHEICT persisted despite multi-variable adjustment, it could reflect an underlying difference in gut-brain axis alteration independent of cirrhosis severity. The mechanism is unclear but serotonin, which is a product of Enterococcus spp.Citation34 promotes inhibitory control and lowers impulsivity, which could be contributory to lower Enterococcus spp. in MHEICT.Citation41,Citation42 This higher Enterococcus spp. in MHEPHES and lower in MHEICT was further confirmed when subgroups impaired on only one test were compared to those that were unimpaired. In addition, similar patterns to that seen when the entire group of cognitively impaired patients were compared to the rest were also seen when exclusively impaired patients were analyzed. This included a greater role of cirrhosis severity in MHEPHES and higher Prevotella and Dakarella spp. in MHEICT. Moreover, since the prevalence of psychoactive medications and diagnoses were similar across groups, this is unlikely to be an epiphenomenon of medication use. Certain Enterococcus spp. are also able to synthesize dopamine, which is associated with lower impulsivity; therefore, lower Enterococcus may be contributory to poor response inhibition.Citation43–45 This was further corroborated by higher DOPAC, a dopamine degradation metabolite, in MHEICT using GBM analysis.
Species associated with MHEICT but not PHES were Prevotella spp.,Dakarella massiliensis, Megasphaera massiliensis, and Alistipes ihumii. Two taxa, Prevotella copri and Dakarella massiliensis (member of Sutterellaceae) are associated with inflammation, altered glycemic status, and advancing liver disease, respectively.Citation46,Citation47 However, the other two species, Megasphaera massiliensis is a butyrate and medium chain FA producer Citation48 and Alistipes spp., associated with protection from disease progression in HECitation.49 This could reflect the similar cirrhosis severity between groups with/without MHEICT rather than MHEPHES. This was further extended by findings that clinical variables such as MELD score, HE, lactulose and rifaximin use were only significantly independent of bacteria in MHEPHES multi-variable analysis but not in MHEICT. This indicates that the microbial outputs could be more related to ICT rather than the cirrhosis severity, while PHES performance is the reverse. While these may be partly due to the relatively preserved hepatic function in MHEICT patients versus MHEPHES ones compared to unimpaired, these taxa were significant on MAASLin2 despite controlling for these clinical factors.
The only GBM higher in MHEPHES was GABA production through the shunt, which is higher in lactic acid-producing bacteria,Citation50 which reflect worsened disease in cirrhosis. Despite the relatively similarity in cirrhosis severity, MHEICT patients had several uniquely higher GBM abundances than MHEPHES. Even more so than the bacterial species, several GBMs were lower in MHEPHES and higher in MHEICT that skewed toward several important processes. These included SCFA/branched-chain SCFA production, inositol, and glutamate degradation, DOPAC, tryptophan, menaquinone, and quinolinic acid synthesis and ClpB-ATP-dependent chaperone protein. Glutamate degradation through an NAD-linked dehydrogenase is ubiquitous and is ammoniagenicCitation.9 Quinolinic acid degradation from aspartate is involved in the formation of NAD that is required for the above-mentioned glutamate degradationCitation.51 Two pathways requiring chorismate resulting in tryptophan and menaquinone generation were higher in MHEICT, both of which are associated with neuroactive potential. Menaquinone is critical for electronic transport chain integrity and can be anti-oxidant in the brain.Citation52,Citation53 DOPAC is a degradation product of dopamine, which along with isovaleric acid, an SCFA, is associated with lower depression in the general population. Both isovalerate and DOPAC pathways were higher in MHEICT but not PHES patients. DOPAC production through isoflavinoids (quercetin) can be beneficial from a free radical scavenger perspective and is found in some human fecal bacteria, such as Clostridium perfringens and Bacteroides fragilis, but not Escherichia coli or Lactobacillus acidophilus. This fits the profile of cirrhosis progression associated with higher Lactobacillus and Enterobacteriaceae members and MHEPHES in contrast to ICT. Inositol degradation may be potentially injurious due to reduction in membrane stabilizing phosphatidylinositol and brain osmotic protector myoinositol and could potentiate ICT-related cognitive impairment. ClpB chaperone protein caseinolytic protease B (ClpB), which is found in Rikenellaceae and Clostridiaceae and are negatively related to obesityCitation.54
These findings underline the association of metagenomic structural and functional changes in gut microbiota with differing cognitive profiles in patients with cirrhosis. Understanding cirrhosis as a culmination of etiologies and concomitant comorbid conditions is important to interpret these results and potentially extend them beyond this population. It is striking that despite minimal changes in cirrhosis severity between groups impaired on ICT, there was a major change in GBMs that was unique to this impairment compared to the traditional PHES modality while PHES changes largely followed the underlying cirrhosis itself. Unlike in the general population, we did not find a major impact of depression, anxiety, or other psychoactive medication use on either MHEICT or MHEPHES. This could be due to the major impact of cirrhosis on the microbiota that would reduce the relative influence of these medications or conditions. These unique changes to ICT could be due to reduced inhibitory control that is inherent in addictive disorders or substance abuse disorders that often precede cirrhosis. ICT-related changes focused on aromatic amino acid and SCFA metabolism suggest that microbiota involved in these pathways could specifically be targeted to improve outcomes. In prior studies, fecal microbiota transplant has been used to beneficially improve cognitive function in cirrhosis, and reduced craving toward alcohol.Citation55–57 However, those studies were focused on one donor for all recipients. These results could form the basis for developing new therapeutic options focused on microbiota that are associated with inhibitory control changes that could be extended beyond cirrhosis.
Our study is limited by the relatively modest sample size and the large proportion who were positive on MHEICT. However, we analyzed discordance and found similar changes regardless of PHES impairment. We excluded those with active substance abuse to avoid confounding and had a relatively narrow age range with mostly men. Future studies across genders need to be performed.
We conclude that impairment on inhibitory control has a distinct metagenomic and GBM signature in the gut microbiota of patients with cirrhosis, which is independent of degree of cirrhosis severity, mood disorders, or psychoactive medications. This is different from microbial changes found with traditional psychometric hepatic encephalopathy score impairment that largely follows cirrhosis severity. Since impaired inhibitory control forms a major basis of addictive disorders, the microbial changes that are unique to this present an opportunity to design trials focused on manipulating these specific microbial taxa.
High scores on ICT targets and digit symbol indicate better performance, while low scores on all others, including composite PHES score, indicate better performance, MHE on PHES or ICT is adjusted for age, gender, and educational performance; the raw scores are presented above. Both indicates Hepatitis and alcohol.
Variables that are positively linked to MHE are in bold font, rest are associated with the absence of MHELOG2FC: Log 2-fold change.
Financial support:
This was partly supported from VA Merit Review 2I0CX00176, NCATS R21TR003095 and R21TR002024AHRQ grant RO1HS025412 and the McGuire Research Institute.
Supplemental Material
Download Zip (98.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on thepublisher’s website.
Additional information
Funding
References
- Cryan JF, O’Riordan KJ, Cowan CSM, et al. The Microbiota-Gut-Brain Axis. Physiol Rev. 2019;99:1877–16.
- Davis BC, Bajaj JS. Effects of Alcohol on the Brain in Cirrhosis: beyond Hepatic Encephalopathy. Alcohol Clin Exp Res. 2018;42(4):660–667. doi:10.1111/acer.13605.
- Vilstrup H, Amodio P, Bajaj J, et al. Hepatic encephalopathy in chronic liver disease: 2014 Practice Guideline by the American Association for the Study of Liver Diseases and the European Association for the Study of the Liver. Hepatology. 2014;60(2):715–735. doi:10.1002/hep.27210.
- Acharya C, Bajaj JS. Current Management of Hepatic Encephalopathy. Am J Gastroenterol. 2018;113(11):1600–1612.
- Weissenborn K, Ennen JC, Schomerus H, Ruckert N, Hecker H. Neuropsychological characterization of hepatic encephalopathy. J Hepatol. 2001;34:768–773.
- Bajaj JS, Fagan A, White MB, et al. Specific Gut and Salivary Microbiota Patterns Are Linked With Different Cognitive Testing Strategies in Minimal Hepatic Encephalopathy. Am J Gastroenterol. 2019;114(7):1080–1090. doi:10.14309/ajg.0000000000000102.
- Ahluwalia V, Betrapally NS, Hylemon PB et al. Impaired Gut-Liver-Brain Axis in Patients with Cirrhosis. Sci Rep. 2016;6(1):26800. doi:10.1038/srep26800.
- Valles-Colomer M, Falony G, Darzi Y et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol. 2019;4(4):623–632. doi:10.1038/s41564-018-0337-x.
- Vieira-Silva S, Falony G, Darzi Y et al. Species-function relationships shape ecological properties of the human gut microbiome. Nat Microbiol. 2016;1(8):16088. doi:10.1038/nmicrobiol.2016.88.
- Garavan H, Ross TJ, Stein EA. Right hemispheric dominance of inhibitory control: an event-related functional MRI study. Proc Natl Acad Sci U S A. 1999;96(14):8301–8306. doi:10.1073/pnas.96.14.8301.
- Bajaj JS, Hafeezullah M, Franco J et al. Inhibitory control test for the diagnosis of minimal hepatic encephalopathy. Gastroenterology. 2008;135(5):1591–600 e1. doi:10.1053/j.gastro.2008.07.021.
- Allampati S, Duarte-Rojo A, Thacker LR et al. Diagnosis of Minimal Hepatic Encephalopathy Using Stroop EncephalApp: a Multicenter US-Based, Norm-Based Study. Am J Gastroenterol. 2016;111(1):78–86. doi:10.1038/ajg.2015.377.
- Bajaj JS, Sikaroodi M, Shamsaddini A, et al. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut. 2020;70(6):1162–1173.
- Martin M Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. https://doi.org/10.14806/ej.17.1.200.
- O’Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–45. doi:10.1093/nar/gkv1189.
- Al-Ghalith G, Knights D. BURST enables optimal exhaustive DNA alignment for big data. 2017. https://doi.org/10.1101/2020.09.08.287128
- Oksanen J, Blanchet FG, Friendly M, et al. vegan: community Ecology Package. R package version. 2019.
- [Accessed 20 May 2021]. https://huttenhower.sph.harvard.edu/maaslin/
- Shamsaddini A, Dadkhah K, Gillevet PM. BiomMiner: an advanced exploratory microbiome analysis and visualization pipeline. PLoS One. 2020;15(6):e0234860. doi:10.1371/journal.pone.0234860.
- Bajaj JS, Kassam Z, Fagan A, et al. Fecal Microbiota Transplant from a Rational Stool Donor Improves Hepatic Encephalopathy: a Randomized Clinical Trial. Hepatology. 2017;66(6):1727–1738. doi:10.1002/hep.29306.
- Bajaj JS, Saeian K, Christensen KM, et al. Probiotic yogurt for the treatment of minimal hepatic encephalopathy. Am J Gastroenterol. 2008;103(7):1707–1715. doi:10.1111/j.1572-0241.2008.01861.x.
- Bass NM, Mullen KD, Sanyal A, et al. Rifaximin treatment in hepatic encephalopathy. N Engl J Med. 2010;362(12):1071–1081. doi:10.1056/NEJMoa0907893.
- Prasad S, Dhiman RK, Duseja A, Chawla YK, Sharma A, Agarwal R. Lactulose improves cognitive functions and health-related quality of life in patients with cirrhosis who have minimal hepatic encephalopathy. Hepatology. 2007;45(3):549–559. doi:10.1002/hep.21533.
- Hatton G, Shawcross DL. Is treating the gut microbiome the key to achieving better outcomes in cirrhosis? Expert Rev Gastroenterol Hepatol. 2019;13(1):1–2. doi:10.1080/17474124.2019.1543587.
- Goldbecker A, Weissenborn K, Hamidi Shahrezaei G, et al. Comparison of the most favoured methods for the diagnosis of hepatic encephalopathy in liver transplantation candidates. Gut. 2013;62(10):1497–1504. doi:10.1136/gutjnl-2012-303262.
- Rehm J, Gmel GE Sr., Gmel G, et al. The relationship between different dimensions of alcohol use and the burden of disease-an update. Addiction. 2017;112(6):968–1001. doi:10.1111/add.13757.
- Solinas A, Piras MR, Deplano A. Cognitive dysfunction and hepatitis C virus infection. World J Hepatol. 2015;7(7):922–925. doi:10.4254/wjh.v7.i7.922.
- Ponziani FR, Putignani L, Paroni Sterbini F, et al. Influence of hepatitis C virus eradication with direct-acting antivirals on the gut microbiota in patients with cirrhosis. Aliment Pharmacol Ther. 2018;48(11–12):1301–1311. doi:10.1111/apt.15004.
- Perez-Matute P, Iniguez M, Villanueva-Millan MJ, et al. Short-term effects of direct-acting antiviral agents on inflammation and gut microbiota in hepatitis C-infected patients. Eur J Intern Med. 2019;67:47–58. doi:10.1016/j.ejim.2019.06.005.
- Ticinesi A, Tana C, Nouvenne A, Prati B, Lauretani F, Meschi T. Gut microbiota, cognitive frailty and dementia in older individuals: a systematic review. Clin Interv Aging. 2018;13:1497–1511. doi:10.2147/CIA.S139163.
- Bajaj JS. Alcohol, liver disease and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16(4):235–246. doi:10.1038/s41575-018-0099-1.
- Northoff G, Hirjak D, Wolf RC, Magioncalda P, Martino M. All roads lead to the motor cortex: psychomotor mechanisms and their biochemical modulation in psychiatric disorders. Mol Psychiatry. 2021;26(1):92–102. doi:10.1038/s41380-020-0814-5.
- Verbruggen F, Best M, Bowditch WA, Stevens T, McLaren IP. The inhibitory control reflex. Neuropsychologia. 2014;65:263–278. doi:10.1016/j.neuropsychologia.2014.08.014.
- TG Dinan, RM Stilling, Stanton C,et al. Collective unconscious: how gut microbes shape human behavior. J Psychiatr Res. 2015;63:1–9. doi:10.1016/j.jpsychires.2015.02.021.
- Strandwitz P, Kim KH, Terekhova D, et al. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol. 2019;4(3):396–403. doi:10.1038/s41564-018-0307-3.
- Feehily C, Karatzas KA. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J Appl Microbiol. 2013;114(1):11–24. doi:10.1111/j.1365-2672.2012.05434.x.
- Forrester MT, Foster MW. Protection from nitrosative stress: a central role for microbial flavohemoglobin. Free Radic Biol Med. 2012;52(9):1620–1633. doi:10.1016/j.freeradbiomed.2012.01.028.
- Bosoi CR, Rose CF. Oxidative stress: a systemic factor implicated in the pathogenesis of hepatic encephalopathy. Metab Brain Dis. 2013;28(2):175–178. doi:10.1007/s11011-012-9351-5.
- Shah V, Kamath PS. Nitric oxide in liver transplantation: pathobiology and clinical implications. Liver Transpl. 2003;9(1):1–11. doi:10.1053/jlts.2003.36244.
- Silva YP, Bernardi A, Frozza RL. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front Endocrinol (Lausanne). 2020;11:25. doi:10.3389/fendo.2020.00025.
- Enge S, Fleischhauer M, Gartner A, et al. Brain-Derived Neurotrophic Factor (Val66Met) and Serotonin Transporter (5-HTTLPR) Polymorphisms Modulate Plasticity in Inhibitory Control Performance Over Time but Independent of Inhibitory Control Training. Front Hum Neurosci. 2016;10:370. doi:10.3389/fnhum.2016.00370.
- Robinson OJ, Roiser JP. The Role of Serotonin in Aversive Inhibition: behavioural, Cognitive and Neural Perspectives. Psychopathology Review. 2016;3(1):29–40. doi:10.5127/pr.034013.
- Villageliu D, Lyte M. Dopamine production in Enterococcus faecium: a microbial endocrinology-based mechanism for the selection of probiotics based on neurochemical-producing potential. PLoS One. 2018;13(11):e0207038. doi:10.1371/journal.pone.0207038.
- Jentsch JD, Pennington ZT. Reward, interrupted: inhibitory control and its relevance to addictions. Neuropharmacology. 2014;76(Pt):B:479–86. doi:10.1016/j.neuropharm.2013.05.022.
- Winstanley CA, Olausson P, Taylor JR, Jentsch JD. Insight into the relationship between impulsivity and substance abuse from studies using animal models. Alcohol Clin Exp Res. 2010;34:1306–1318.
- Franke T, Deppenmeier U. Physiology and central carbon metabolism of the gut bacterium Prevotella copri. Mol Microbiol. 2018;109(4):528–540. doi:10.1111/mmi.14058.
- Bajaj JS, Kakiyama G, Cox IJ, et al. Alterations in gut microbial function following liver transplant. Liver Transpl. 2018;24(6):752–761. doi:10.1002/lt.25046.
- Sharma M, Li Y, Stoll ML, Tollefsbol TO. The Epigenetic Connection Between the Gut Microbiome in Obesity and Diabetes. Front Genet. 2019;10:1329. doi:10.3389/fgene.2019.01329.
- Parker BJ, Wearsch PA, Veloo ACM, The Genus R-PA. Alistipes: gut Bacteria With Emerging Implications to Inflammation, Cancer, and Mental Health. Front Immunol. 2020;11:906. doi:10.3389/fimmu.2020.00906.
- Feehily C, O’Byrne CP, Karatzas KA. Functional gamma-Aminobutyrate Shunt in Listeria monocytogenes: role in acid tolerance and succinate biosynthesis. Appl Environ Microbiol. 2013;79(1):74–80. doi:10.1128/AEM.02184-12.
- Lima WC, Varani AM, Menck CF. NAD biosynthesis evolution in bacteria: lateral gene transfer of kynurenine pathway in Xanthomonadales and Flavobacteriales. Mol Biol Evol. 2009;26(2):399–406. doi:10.1093/molbev/msn261.
- Boersch M, Rudrawar S, Grant G, Zunk M. Menaquinone biosynthesis inhibition: a review of advancements toward a new antibiotic mechanism. RSC Advances. 2018;8(10):5099–5105. doi:10.1039/C7RA12950E.
- Farhadi Moghadam B, Fereidoni M. Neuroprotective effect of menaquinone-4 (MK-4) on transient global cerebral ischemia/reperfusion injury in rat. PLoS One. 2020;15(3):e0229769. doi:10.1371/journal.pone.0229769.
- Arnoriaga-Rodriguez M, Mayneris-Perxachs J, Burokas A, et al. Gut bacterial ClpB-like gene function is associated with decreased body weight and a characteristic microbiota profile. Microbiome. 2020;8(1):59. doi:10.1186/s40168-020-00837-6.
- Bajaj JS, Kassam Z, Fagan A, et al. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: a randomized clinical trial. Hepatology. 2017;66:1727–1738.
- Bajaj JS, Salzman NH, Acharya C, et al. Fecal Microbial Transplant Capsules are Safe in Hepatic Encephalopathy: a Phase 1, Randomized, Placebo-Controlled Trial. Hepatology. 2019;70(5):1690–1703.
- Bajaj JS, Gavis EA, Fagan A, et al. A Randomized Clinical Trial of Fecal Microbiota Transplant for Alcohol Use Disorder. Hepatology. 2020;73:S59–S60. doi:10.1016/S0168-8278(20)30660-7.
