ABSTRACT
Metastasis is the leading cause of death for colorectal cancer (CRC) patients, and the spreading tumor cells adhesion to endothelial cells is a critical step for extravasation and further distant metastasis. Previous studies have documented the important roles of gut microbiota-host interactions in the CRC malignancy, and Fusobacterium nucleatum (F. nucleatum) was reported to increase proliferation and invasive activities of CRC cells. However, the potential functions and underlying mechanisms of F. nucleatum in the interactions between CRC cells and endothelial cells and subsequent extravasation remain unclear. Here, we uncovered that F. nucleatum enhanced the adhesion of CRC cells to endothelial cells, promoted extravasation and metastasis by inducing ICAM1 expression. Mechanistically, we identified that F. nucleatum induced a new pattern recognition receptor ALPK1 to activate NF-κB pathway, resulting in the upregulation of ICAM1. Interestingly, the abundance of F. nucleatum in tumor tissues of CRC patients was positively associated with the expression levels of ALPK1 and ICAM1. Moreover, high expression of ALPK1 or ICAM1 was significantly associated with a shorter overall survival time of CRC patients. This study provides a new insight into the role of gut microbiota in engaging into the distant metastasis of CRC cells.
Introduction
Colorectal cancer (CRC) is the fourth most commonly diagnosed cancer worldwide and the second most frequent cause of death closely following lung cancer.Citation1 Cancer metastasis, a major clinical problem of human cancer, is responsible for most colorectal cancer patient mortality.Citation2 Thus, it is important to elucidate the underlying mechanisms of metastasis in CRC patients.
During metastasis, cancer cells leave the original tumor organ and migrate to the target metastasis organs through a process that involves intravasation, adhesion and extravasation, among which the adhesion to endothelial cells and trans-endothelial invasion of tumor cells are key steps in the metastatic process.Citation3,Citation4 Indeed, changes in the expression or functions of cell adhesion molecules have been implicated in all steps of tumor progression. Cell adhesion molecules belonging to the immunoglobulin superfamily commonly play critical and necessary roles in metastasis,Citation5–7 among which intercellular adhesion molecule 1 (ICAM1) is a well-known transmembrane glycoprotein involved in cell-cell direct interactions.Citation7 The interaction between ICAM1 and its specific ligand could facilitate the adhesion of cancer cells to the vascular endothelium and subsequently in the promotion of metastasis. Importantly, the expression of ICAM1 was positively correlated with cancer progression and metastasis.Citation8–10
Multiple studies have suggested a potential link between Fusobacterium nucleatum (F. nucleatum) and colorectal cancer.Citation11,Citation12 F. nucleatum is a Gram-negative anaerobic oncogenic bacterium enriched in human colorectal carcinoma compared with adjacent normal tissues.Citation13–15 The overabundance of F. nucleatum in CRC tissues has been reported to promote CRC cell proliferation, recurrence and chemoresistance.Citation16–18 However, the potential effects and underlying mechanisms of F. nucleatum in the interactions between CRC cells and endothelial cells and subsequent extravasation in CRC metastasis remain largely unknown.
NF-κB, initially established as an important transcription factor, has been demonstrated to participate in inflammation-associated colorectal cancer.Citation19 It is reported that F. nucleatum-mediated infection leads to the activation of NF-κB pathway via the suggested membrane receptor-TLR4/MYD88 cascade.Citation17 However, previous studies confirmed the invasive potential of F. nucleatum into CRC cells undergoing a non-obligate intracellular life stage, and it could be detected in CRC metastases of liver and lymph nodes,Citation20,Citation21 indicating that there may be another pathway to activate NF-κB in F. nucleatum-infected CRC cells. Interestingly, a recent study reported that alpha-kinase 1 (ALPK1), a member of the α-kinase family in cytoplasm, functions as a cytosolic innate immune receptor for bacterial adenosine diphosphate-heptose (ADP-Hep), which is present in all Gram-negative and some Gram-positive bacteria.Citation22,Citation23 It is reported that ALPK1 plays a crucial role in the activation of NF-κB signaling and subsequent production of pro-inflammatory cytokines and chemokines through the stimulation of its kinase domain.Citation24,Citation25 Therefore, we wondered the potential effects and mechanisms of ALPK1 in F. nucleatum-mediated CRC metastasis.
For the first time, our study proposed a F. nucleatum-induced cell-cell interaction model by which F. nucleatum treatment promoted CRC cells adhesion to endothelial cells, facilitated extravasation and metastasis through upregulating ICAM1 expression in CRC cells. Mechanistically, we uncovered that F. nucleatum upregulated the expression of ICAM1 by activating ALPK1/NF-κB pathway in CRC cells.
Results
F. nucleatum promotes CRC cells adhesion to endothelial cells and facilitates extravasation by upregulating ICAM1.
When we study the role of gut microbiota in CRC cell malignancy, an interesting phenomenon during culturing CRC cells attracted our attention that F. nucleatum treatment, but not Escherichia coli (E. coli) or phosphate buffer saline (PBS) control, may increase cell-cell interaction ability of CRC cells. As we know, the spreading tumor cells adhesion to endothelial cells is a critical step for extravasation and further distant metastasis. It is possible that the F. nucleatum-induced adhesion ability of CRC cells may facilitate its interaction with endothelial cells and extravasation. To verify our hypothesis, we designed a tumor cell-endothelial cell interaction model. Human Umbilical Vein Endothelial Cells (HUVECs) were seeded in culture plates until full confluence. Then GFP-labeled HCT116 cells pretreated with F. nucleatum (F. nucleatum strain 25586), E. coli or PBS control were added onto the HUVECs monolayers. The unbound tumor cells were washed away after 15 minutes and the adherent HCT116 cells were analyzed ()). F. nucleatum treatment strongly enhanced the adhesion of HCT116 cells to HUVECs compared with E. coli or PBS control ()). Meanwhile, in concordance with our previous findings,Citation26 F. nucleatum treatment dramatically promoted cell migration of both HCT116 cells ()) and LoVo cells (Supplementary Figure 1a). In addition, another F. nucleatum strain 10953 was applied to assess whether this is a general property of F. nucleatum. F. nucleatum strain 10953 treatment, similar with F. nucleatum strain 25586, obviously promoted CRC cell-endothelial cell adhesion and CRC cells migration (Supplementary Figure 1b-c), suggesting that this may be a general property of F. nucleatum. Interestingly, when we employed a Gram-negative bacteria Akkermansia muciniphila (A. muciniphila)Citation27 and a Gram-positive bacteria Bifidobacterium adolescentis (B. adolescentis)Citation28 to identify the specific phenotype of F. nucleatum, we found that F. nucleatum treatment dramatically promoted CRC cells adhesion to endothelial cells, as well as the migration ability, but not A. muciniphila or B. adolescentis (Supplementary Figure 1d-e).
Figure 1. F. nucleatum promotes CRC cells adhesion to endothelial cells and facilitates extravasation by upregulating ICAM1. (a) Schematic illustration of adhesion assay in vitro. (b) GFP-labeled HCT116 cells pretreated with F. nucleatum, E. coli or PBS control for 6 hours were subjected to adhesion assay. Representative images of adherent cells were shown (left) and the adherent cells were quantified by counting in five fields (right). Scale bar, 100 μm. (c) HCT116 cells were pretreated with F. nucleatum, E. coli or PBS control for 6 hours and subjected to migration assay. The migrated cells were quantified at 18 hours by counting in five fields. Scale bar, 100 μm. (d) Schematic illustration of cancer cell extravasation model in vivo. (e) The colonized GFP-labeled HCT116 cells in mice lung tissues were quantified by immunofluorescence assay, in which lung sections were stained with a marker of vascular endothelial cells CD31 (red), and the nuclei were counterstained with DAPI (blue). Scale bar, 100 μm. (f) 256 genes with significant mRNA upregulation (fold change > 2, p.adj < 0.05) in RNA-sequencing, 880 genes in the GO class of cell-cell adhesion (GO: 0098609), and 133 genes in the KEGG pathway of cell adhesion molecules (hsa04514) with key functions were subjected to Venn diagram analysis. Venn diagram showing the overlapping genes. (g) Western blot analysis was performed to detect the protein levels of ICAM1 in HCT116 cells and LoVo cells treated with F. nucleatum, E. coli or PBS control. (h) Quantitative RT-PCR analysis of ICAM1 in PDX tumor tissues treated with F. nucleatum or PBS control. Data are shown as mean ± SD. ** P< .01, *** P < .001, **** P < .0001, Student’s t test (b, c, h), Mann-Whitney test (e).
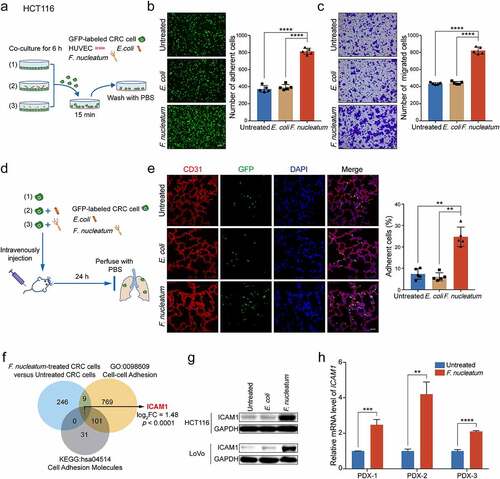
To confirm these in vitro results, we intravenously injected F. nucleatum-treated GFP-labeled HCT116 cells into SCID mice. The pulmonary blood vessels were perfused with PBS after 24 hours and the colonized HCT116 cells in lungs were analyzed ()). F. nucleatum treatment robustly increased the trans-endothelial invasion and colonization of HCT116 cells compared with E. coli or PBS treatment ()).
To understand the underlying mechanisms of F. nucleatum-induced intercellular adhesion ability of CRC cells, we performed RNA-sequencing in HCT116 cells with or without F. nucleatum treatment (GSE171611). As shown in ), we identified gene ICAM1 overlapping among F. nucleatum-upregulated genes (log2 fold change > 1, p.adj < 0.05), the GO class of cell-cell adhesion and the KEGG pathway of cell adhesion molecules. To confirm the profiling results, we detected the protein expression of ICAM1 in F. nucleatum-treated CRC cells and observed that ICAM1 was indeed significantly increased in F. nucleatum-treated HCT116 cells and LoVo cells ()). Importantly, we established patient-derived xenograft (PDX) model with tumor tissues generated from CRC patients. All of the PDX tumor tissues from three patients treated with F. nucleatum ex vivo showed a significant upregulation of ICAM1 levels ()).
ICAM1 is involved in F. nucleatum-induced CRC cells adhesion to endothelial cells and migration in vitro.
To further understand the functions of ICAM1 in CRC progression, we silenced ICAM1 in HCT116 cells using two specific siRNAs (-b)). Knockdown of ICAM1 dramatically inhibited the adhesion of HCT116 cells to HUVECs ()). Meanwhile, as shown in ), ICAM1 silencing showed remarkable tumor migration suppressive effects. Similar results were also observed in LoVo cells (Supplementary Figure 2a-c).
Figure 2. ICAM1 is involved in F. nucleatum-induced CRC cells adhesion to endothelial cells and migration in vitro. (a-b) Quantitative RT-PCR (a) and Western blot analysis (b) of ICAM1 were performed in HCT116 cells transfected with two siRNAs targeting ICAM1 or control siRNAs. (c-d) Adhesion assay (c) and migration assay (d) of HCT116 cells transfected with the indicated siRNAs. The migrated cells were observed at 24 hours. Scale bar, 100 μm. (e-f) Quantitative RT-PCR (e) and Western blot analysis (f) were performed in HCT116 cells. They were transfected with the indicated siRNAs, and then co-cultured with F. nucleatum or PBS. (g-h) HCT116 cells transfected with the indicated siRNAs were co-cultured with F. nucleatum, and subjected to adhesion assay (g) and migration assay (h). The migrated cells were observed at 18 hours. Scale bar, 100 μm. Data are shown as mean ± SD. *** P < .001, **** P < .0001, by Student’s t test.
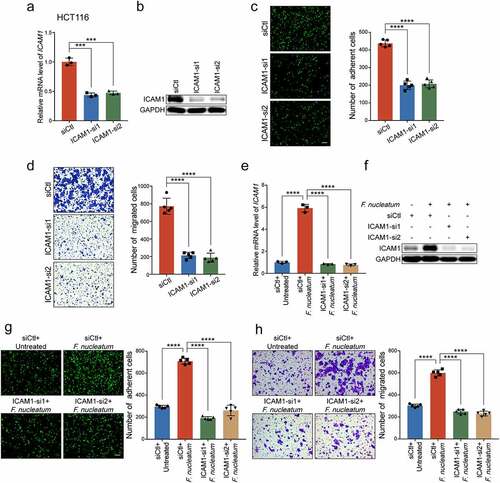
To further determine whether F. nucleatum exerted oncogenic functions in an ICAM1 dependent manner, we performed ICAM1 loss of function assays in CRC cells. The upregulated mRNA and protein levels of ICAM1 in F. nucleatum-treated CRC cells were significantly decreased when we silenced ICAM1 in HCT116 cells (-f)) and LoVo cells (Supplementary Figure 2d-e). As expected, ICAM1 knockdown abrogated the promotion of F. nucleatum-mediated HCT116 cells adhesion to endothelial cells ()). Moreover, silencing of ICAM1 significantly reversed the increased cell migration ability induced by F. nucleatum (), Supplementary Figure 2f), highlighting the important role of ICAM1 in sustaining the functions mediated by F. nucleatum. Taken together, we suggest that ICAM1 is responsible for CRC cells adhesion to endothelial cells and migration induced by F. nucleatum in vitro.
ICAM1 is involved in F. nucleatum-mediated CRC cells extravasation and metastasis in vivo.
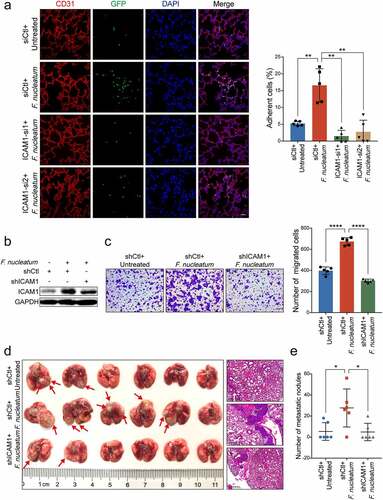
To further assess the role of ICAM1 in F. nucleatum-mediated trans-endothelial invasion and colonization in vivo, we depleted ICAM1 expression in GFP-labeled HCT116 cells and treated cells with F. nucleatum. The cancer cell extravasation assay showed that F. nucleatum dramatically promoted HCT116 cells extravasation, while knockdown of ICAM1 obviously reversed the trans-endothelial invasion and colonization ()). In addition, we established stable ICAM1-knockdown HCT116 cells by lentivirus-based shRNAs ()). In concordance with our previous findings, knockdown of ICAM1 significantly diminished the cell migration ability induced by F. nucleatum ()). We intravenously inoculated mice with the indicated cells to measure pulmonary metastases. Data from the gross image and H&E staining of lungs showed that F. nucleatum significantly increased the number of metastatic lesions, whereas knockdown of ICAM1 in HCT116 cells notably diminished F. nucleatum-induced lung metastasis (-e)). Taken together, these data suggest that F. nucleatum promotes extravasation and metastasis of CRC cells via the upregulation of ICAM1 in vivo.
Figure 3. ICAM1 is involved in F. nucleatum-mediated CRC cells extravasation and metastasis in vivo. (a) GFP-labeled HCT116 cancer cells transfected with two siRNAs targeting ICAM1 were co-cultured with F. nucleatum or PBS, and subjected to extravasation model in vivo. The colonized HCT116 cells in lung tissues were measured by immunofluorescence assay. Scale bar, 100 μm. (b-c) HCT116 cells stably infected with lentivirus-based ICAM1 shRNAs or control shRNAs were co-cultured with F. nucleatum or PBS control, and subjected to Western blot analysis (b) and migration assay (c). The migrated cells were quantified at 18 hours. Scale bar, 100 μm. (d-e) The ICAM1-knockdown HCT116 cells pretreated with F. nucleatum or PBS were tail-vein injected into nude mice to develop pulmonary metastases. Representative gross lungs and H&E stained lung sections were shown (d). Arrows indicated metastatic nodules. Pulmonary metastatic nodules per mice were quantified (e). Data are shown as mean ± SD. * P < .05, ** P < .01, **** P < .0001, Mann-Whitney test (a, e), Student’s t test (c).
F. nucleatum upregulates ICAM1 expression through the activation of NF-κB signaling pathway.
We next tried to investigate the underlying mechanisms for F. nucleatum upregulating the expression of ICAM1. KEGG pathway analysis and Gene Set Enrichment Analysis (GSEA) of the RNA-sequencing results suggested that NF-κB signaling pathway was activated by F. nucleatum infection (-b)). Western blot analysis confirmed that the phosphorylation levels of NF-κB subunit p65 was substantially upregulated, as well as the phosphorylation levels of NF-κB inhibitor IκBα in HCT116 cells ()) and LoVo cells (Supplementary Figure 3a) when treated with F. nucleatum, but not E. coli or PBS. Immunofluorescence assay also showed an increase in the nuclear localization of NF-κB p65 in cells with F. nucleatum treatment (), Supplementary Figure 3b). Collectively, these data indicate that NF-κB signaling is activated in CRC cells by F. nucleatum.
Figure 4. F.nucleatum upregulates ICAM1 expression through the activation of NF-κB signaling pathway. (a) KEGG pathway analysis of a total number of 503 genes upregulated with significant difference in RNA-sequencing. (b) GSEA showed the differentially expressed gene cluster related to NF-κB signaling pathway in HCT116 cells with or without F. nucleatum treatment. (c) Western blot analysis of phospho-IκBα, NF-κB subunit p65 and phospho-p65 in HCT116 cells treated with F. nucleatum, E. coli or PBS control. (d) Immunofluorescence assay of NF-κB p65 distribution in the indicated HCT116 cells. Cells were stained with specific antibody against p65 (green), and the nuclei were counterstained with DAPI (blue). Scale bar, 20 μm. (e-f) HCT116 cells were co-cultured with F. nucleatum or PBS control, and then treated with NF-κB inhibitor, BAY11-7082. Quantitative RT-PCR (e) and Western blot analysis (f) were performed. (g-h) HCT116 cells were transfected with two siRNAs targeting p65, and then co-cultured with F. nucleatum or PBS control. Quantitative RT-PCR (g) and Western blot analysis (h) were performed. (i-j) Adhesion assay (i) and migration assay (j) of HCT116 cells with indicated treatments. The migrated cells were observed at 18 hours. Scale bar, 100 μm. (k-l) P65-depleted HCT116 cells transfected with the indicated plasmids were applied for Western blot analysis (k) and adhesion assay (l). Scale bar, 100 μm. Data are shown as mean ± SD. * P < .05, ** P < .01, **** P < .0001, by Student’s t test.
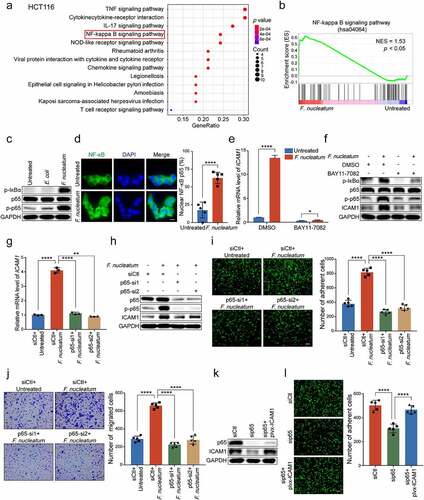
We were curious whether the activated NF-κB pathway was involved in the regulation of ICAM1. BAY11-7082, an inhibitor of NF-κB, was employed to treat CRC cells when infected with F. nucleatum. Both quantitative RT-PCR and Western blot analysis showed that the upregulation of ICAM1 induced by F. nucleatum infection was notably attenuated when HCT116 cells (-f)) and LoVo cells (Supplementary Figure 3c-d) were treated with NF-κB inhibitor. Moreover, the knockdown of p65 also inhibited F. nucleatum-induced ICAM1 upregulation in HCT116 cells (-h)) and LoVo cells (Supplementary Figure 3e-f). These results suggest that the upregulation of ICAM1 in CRC cells is due to the activation of NF-κB signaling by F. nucleatum.
As expected, the knockdown of p65 in HCT116 cells significantly decreased the promotion of F. nucleatum-mediated adhesion to endothelial cells, as well as the migration ability (-j)). Similar results were also observed in LoVo cells (Supplementary Figure 3g). Moreover, we reintroduced ICAM1 levels in p65-depleted CRC cells and found that ectopic expression of ICAM1 could rescue the inhibitory effects of p65 silencing on cell adhesion and migration (-l), Supplementary Figure 3h-i).
Taken together, these results indicate that F. nucleatum treatment upregulates ICAM1 through activating NF-κB signaling pathway.
ALPK1 mediates NF-κB-dependent responses to F. nucleatum infection.
We next investigated the upstream regulators which could transmit signals to NF-κB pathway and mediate the functions of F. nucleatum. It is reported that Toll-like receptor family is involved in the system of gut microbiota-host interactions, and the TLR4/MYD88 cascade is activated in response to F. nucleatum intervention.Citation29 However, we silenced TLR4 or MYD88 in F. nucleatum-treated HCT116 cells, and found that the knockdown of TLR4 or MYD88 could not reverse the promotion of adhesion ability induced by F. nucleatum (Supplementary Figure 4a). Intriguingly, previous studies have confirmed the distinct invasive potential of F. nucleatum into CRC cells undergoing a non-obligate intracellular life stage,Citation21 and ALPK1 may function as a new pattern recognition receptor in the cytoplasm for ADP-Hep present in Gram-negative bacterial.Citation23 Therefore, it is possible that F. nucleatum may activate NF-κB pathway through ALPK1 besides TLR4/MYD88 cascade in the cell membranes. To test this hypothesis, we treated HCT116 cells with F. nucleatum, E. coli or PBS control, and found that the protein level of ALPK1 was dramatically enhanced upon F. nucleatum treatment ()). To further determine whether ALPK1 is involved in the regulation of F. nucleatum-mediated NF-κB activation, we depleted ALPK1 using two siRNAs targeting different regions of ALPK1 in HCT116 cells (-c)). Western blot analysis showed that silencing of ALPK1 prevented NF-κB pathway from being activated by F. nucleatum infection ()), which was also verified by immunofluorescence assay on the nuclear localization of NF-κB p65 ()). Similar results were observed in LoVo cells (Supplementary Figure 4b-d). These results suggest that F. nucleatum activates NF-κB pathway through the upregulation of ALPK1.
Figure 5. ALPK1 mediates NF-κB-dependent responses to F. nucleatum infection. (a) Western blot analysis of ALPK1 in HCT116 cells treated with F. nucleatum, E. coli or PBS control. (b-c) Quantitative RT-PCR (b) and Western blot analysis (c) of ALPK1 were performed in HCT116 cells transfected with two siRNAs targeting ALPK1 or control siRNAs. (d) HCT116 cells transfected with the indicated siRNAs were co-cultured with F. nucleatum or PBS control. Western blot analysis of ALPK1, phosphor-IκBα, p65, phospho-p65 and ICAM1 were performed. (e) The distribution of NF-κB p65 in the indicated cells was measured by immunofluorescence assay. Scale bar, 20 μm. (f-g) HCT116 cells transfected with the indicated siRNAs were co-cultured with F. nucleatum or PBS control. Adhesion assay (f) and migration assay (g) were performed. The migrated cells were observed at 18 hours. Scale bar, 100 μm. (h-i) ALPK1-depleted HCT116 cells transfected with the indicated plasmids were applied for Western blot analysis (h) and adhesion assay (i). Scale bar, 100 μm. Data are shown as mean ± SD. *** P < .001, **** P < .0001, by Student’s t test.
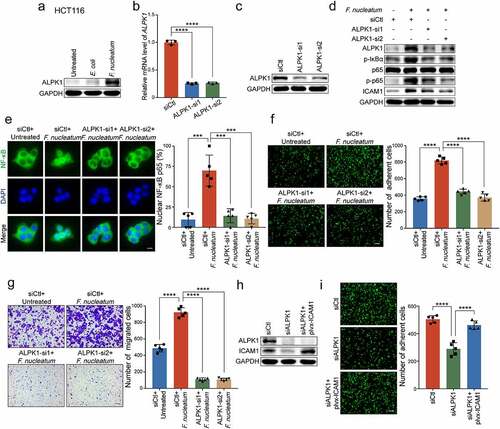
Since F. nucleatum is able to activate ALPK1/NF-κB pathway, we wondered whether this pathway could be involved in the regulation of ICAM1. Interestingly, we observed that F. nucleatum failed to upregulate the expression of ICAM1 when ALPK1 was silenced in HCT116 cells ()) and LoVo cells (Supplementary Figure 4c), highlighting the importance of ALPK1 in the upregulation of NF-κB-dependent ICAM1 by F. nucleatum. We then tried to determine whether ALPK1 mediated the functions of F. nucleatum in CRC progression. As expected, F. nucleatum-mediated CRC cells adhesion to endothelial cells was diminished when ALPK1 was knocked down in F. nucleatum-treated HCT116 cells ()). Meanwhile, silencing of ALPK1 significantly impaired the F. nucleatum’s oncogenic effect on cell migration (), Supplementary Figure 4e). Furthermore, we restored ICAM1 expression in ALPK1-depleted CRC cells and found that ectopic expression of ICAM1 significantly reversed the inhibitory effects of ALPK1 silencing on cell adhesion and migration (-i), Supplementary Figure 4f-g).
Together, these results suggest that F. nucleatum promotes CRC cell-endothelial cell adhesion and CRC cells migration through ALPK1/NF-κB/ICAM1 axis. To explore the specificity of F. nucleatum on ALPK1/NF-κB/ICAM1 axis, we further detected the protein levels of ALPK1, NF-κB signaling and ICAM1 in CRC cells with A. muciniphila or B. adolescentis treatment. Western blot analysis showed that A. muciniphila treatment, but not B. adolescentis, can slightly activate ALPK1/ NF-κB pathway (Supplementary Figure 4h-i). Interestingly, F. nucleatum treatment showed a much stronger stimulation than A. muciniphila on the ALPK1/NF-κB signaling, as well as the ICAM1 expression levels (Supplementary Figure 4h-i). Moreover, F. nucleatum strain 10953 could also notably activate ALPK1/NF-κB/ICAM1 signaling in HCT116 cells or LoVo cells comparable to the F. nucleatum strain 25586 (Supplementary Figure 4j-k).
Overabundance of F. nucleatum correlates with high expressions of ALPK1 and ICAM1 in CRC patients and indicates poor clinical outcome.
Recent studies have revealed that the abundance of F. nucleatum is increased in colorectal cancer tissues versus matched normal tissues.Citation14,Citation15 To establish a high-F. nucleatum environment in the colon, we administrated F. nucleatum or PBS into C57BL/6 mice, which received antibiotics by gavage for 3 days beforehand ()). After 15 days of treatment, mice were sacrificed and their colons were removed for the following experiments. Quantitative RT-PCR analysis revealed a significant increase of F. nucleatum in colon tissues with F. nucleatum treatment, indicating the model was successfully constructed ()). Notably, F. nucleatum significantly increased ICAM1 expression (). Moreover, we established an AOM/DSS-induced CRC model accompanying with gavage treatment of F. nucleatum, E. coli or PBS control as previously described (Supplementary Figure 5a-b).Citation27,Citation30 Quantitative RT-PCR analysis revealed a significant upregulation of ALPK1 and ICAM1 levels in orthotopic tumor tissues from mice receiving F. nucleatum treatment compared with E. coli or PBS control (Supplementary Figure 5c-d). These results confirm that the overabundance of F. nucleatum in mouse colon tissues leads to the upregulation of ALPK1 and ICAM1 in vivo.
Figure 6. Overabundance of F.nucleatum correlates with high expressions of ALPK1 and ICAM1 in CRC patients and indicates poor clinical outcome. (a) C57BL/6 mice pretreated with antibiotics were administrated with F. nucleatum or PBS control everyday by gavage and sacrificed after the treatment at 15 days. (b) Quantitative RT-PCR analysis of the relative abundance of F. nucleatum in colon tissues from mice with the indicated treatment (n = 6). Data are normalized to Universal Eubacteria 16S. (c) Quantitative RT-PCR analysis of ICAM1 mRNA expression in colon tissues from the indicated mice (n = 6). (d) Representative IHC images of ICAM1 protein expression in colon tissues from the indicated mice. (e-f) Quantitative RT-PCR analysis of ALPK1 (e) and ICAM1 (f) in Cohort 1 which was divided into two groups depends on the relative abundance of F. nucleatum. (g) The correlation between the relative mRNA levels of ALPK1 and ICAM1 in Cohort 1. (h) The relative mRNA levels of ICAM1 among CRC patients with different lymph nodes metastasis stages in Cohort 1. (i-j) IHC analysis of ALPK1 (i) and ICAM1 (j) in Tissue Array Cohort 2. Representative photographs of IHC staining in CRC tissues with or without liver metastasis were shown (left), and the relative expression was quantified by H-score (right). Scale bar, 50 μm. (k-l) Kaplan-Meier survival curve of ALPK1 (k) and ICAM1 (l) expression of CRC patients in Tissue Array Cohort 2. (m) The relative abundance of F. nucleatum in the feces of CRC patients with positive lymph nodes metastasis (N1+N2, n = 32) or without metastasis (N0, n = 40) in Cohort 3. Data are shown as mean ± SD. * P < .05, ** P < .01, *** P < .001, **** P < .0001, Mann-Whitney test (b), Student’s t test (c, e, f, h, i, j, m), Linear Regression (g), Log-rank test (k, l).
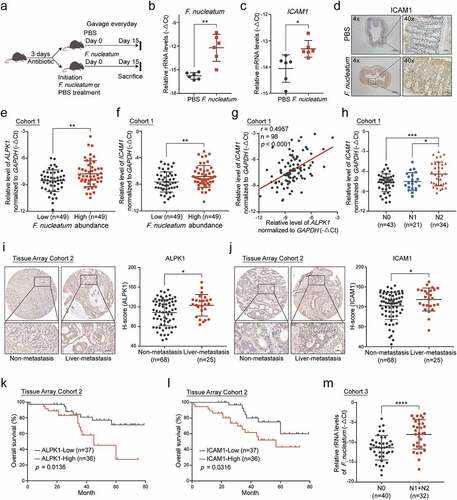
To further investigate the clinical significance of F. nucleatum/ALPK1/ICAM1, we quantified F. nucleatum and the expression of ALPK1 and ICAM1 with quantitative RT-PCR analysis in 98 paired fresh CRC and adjacent non-tumor tissues (Cohort 1). The relative amount of F. nucleatum was higher in CRC tissues compared with the non-tumor controls (Supplementary Figure 5e). We ranked these patients (n = 98) according to the levels of F. nucleatum in CRC tissues, and defined the top 1/2 patients as the F. nucleatum-high group (n = 49) and the bottom 1/2 patients as F. nucleatum-low group (n = 49) as previously described.Citation11 Notably, we found the expression of ALPK1 and ICAM1 was significantly higher in the F. nucleatum-high group (-f)). Similarly, there was a positive correlation between the relative abundance of F. nucleatum and ALPK1 levels in CRC tumor tissues (Supplementary Figure 5f), as well as the correlation between F. nucleatum and ICAM1 levels (Supplementary Figure 5g). Furthermore, the mRNA level of ICAM1 was positively associated with ALPK1 expression both in Cohort 1 and The Cancer Genome Atlas (TCGA) database (), Supplementary Figure 5h). More importantly, tumor tissues from CRC patients with lymph nodes metastasis showed an upregulation of ICAM1 compared with tissues from non-metastasis patients, suggesting that ICAM1 was positively associated with advanced lymph nodes metastasis in CRC patients ()). To further analyze the clinical significance of ALPK1 and ICAM1 in CRC patients with distant metastasis, we confirmed the expression of ALPK1 and ICAM1 in human CRC tissue array by IHC analysis (Tissue Array Cohort 2). Importantly, the high expression of ALPK1 or ICAM1 was significantly correlated with positive hepatic metastases (-j)), and the protein level of ICAM1 was positively associated with ALPK1 expression (Supplementary Figure 5i). In addition, we followed up patients in Cohort 2 and analyzed the relevance of ALPK1 and ICAM1 expression to patient outcomes. Kaplan-Meier analysis showed that high expression of ALPK1 or ICAM1 was significantly associated with a shorter overall survival time of CRC patients (-l)).
To further determine the role of F. nucleatum in CRC metastasis, we quantified the relative abundance of F. nucleatum in 32 fecal samples from CRC patients with positive lymph nodes metastasis (N1+N2) and 40 fecal samples without lymph nodes metastasis (N0) (Cohort 3). As expected, the level of F. nucleatum was specifically much higher in CRC patients with positive lymph nodes metastasis ()). Meanwhile, there was an enrichment of F. nucleatum in the feces of CRC patients compared with the normal controls (Supplementary Figure 5j).
Taken together, our results indicate that the overabundance of F. nucleatum is associated with high expressions of ALPK1 and ICAM1 in CRC tissues. Furthermore, patients with high levels of F. nucleatum, ALPK1 or ICAM1 have a higher risk for poor clinical outcomes, which has important implications for their clinical management.
Discussion
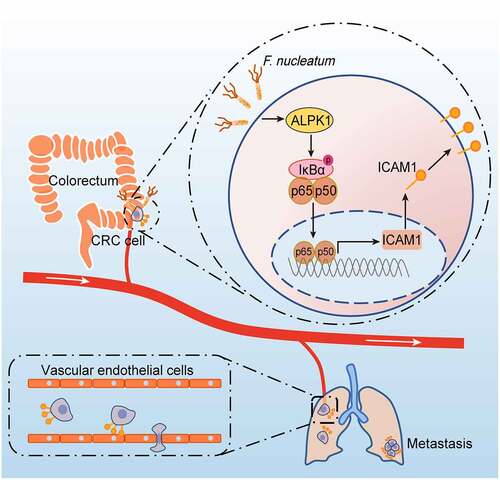
In this study, we for the first time reveal that F. nucleatum enhances the intercellular adhesion ability of CRC cells, promotes CRC cells adhesion to endothelial cells, facilitates extravasation and metastasis, providing a new microbiota-host interaction model for controlling CRC metastasis (). Mechanistically, F. nucleatum treatment activates NF-κB signaling of CRC cells through a newly identified pattern recognition receptor ALPK1, thereby promotes the expression of ICAM1, which is critical for F. nucleatum-induced CRC cell-endothelial cell adhesion and metastasis.
Figure 7. Schematic diagram of F. nucleatum-induced ALPK1/NF-κB/ICAM1 axis regulating the metastasis of CRC. The high abundance of F. nucleatum in gut activates NF-κB signaling of CRC cells through a newly identified pattern recognition receptor ALPK1, thereby promotes the expression of ICAM1, which is critical for F. nucleatum-induced CRC cell-endothelial cell adhesion, extravasation and metastasis.
Metagenomic and transcriptomic analyses have revealed that gut microbiota, such as Fusobacterium nucleatum,Citation17 Akkermansia muciniphilaCitation31 and Streptococcus thermophilusCitation32 were involved in CRC development. Recently, the presence of F. nucleatum was detected in metastases and it may promote colorectal cancer metastasis. Several research groups have reported that F. nucleatum promotes colorectal cancer metastasis through the activation of KRT7-AS,Citation26 the upregulation of CARD3 to activate autophagy,Citation33 the induction of IL-8 and CXCL1Citation21 or exosomes derived from F. nucleatum-infected colorectal cancer cells.Citation34 However, tumor metastasis is an extremely complex process, including cancer cells detaching from the primary tumor, entering the blood circulation, adhering to endothelial cells and extravasation, reaching the distal part to form visible metastases.Citation5 The direct interaction between tumor cells and endothelial cells is a crucial step for trans-endothelial invasion and colonization. Modulation of the expression of adhesion-related molecules may be an effective strategy for improving the prognosis of CRC. Our present study for the first time revealed that F. nucleatum enhanced the adhesion of CRC cells to endothelial cells in vitro and in vivo, which is a F. nucleatum-induced novel crosstalk between F. nucleatum and CRC cell-endothelial cell adhesion in promoting CRC metastasis.
In this study, we analyzed the mechanisms by which F. nucleatum induced the enhancement of CRC cell-endothelial cell adhesion. We identified ICAM1 overlapping between cell adhesion-related genes and F. nucleatum-upregulated genes in our RNA-sequencing results, as a direct downstream target of F. nucleatum. ICAM1, a cell surface adhesion molecule, could promote pancreatic ductal adenocarcinoma,Citation35 hepatocellular carcinomaCitation36 and colorectal cancerCitation37 development. We found that the downregulation of ICAM1 decreased the promotion of F. nucleatum-induced CRC cell-endothelial cell adhesion and metastasis in vitro and in vivo.
Furthermore, we attempted to reveal the mechanisms for the regulation of ICAM1 by F. nucleatum. Our KEGG analysis based on RNA-sequencing results demonstrated that NF-κB pathway was activated by F. nucleatum treatment. Consistently, several other studies also confirmed the activation of NF-κB pathway in F. nucleatum-infected cells.Citation17,Citation26,Citation38,Citation39 In addition, the recognition of TLR4 on the surfaces of CRC cells was suggested to be involved in the sustained activation of NF-κB signaling pathway.Citation17 However, F. nucleatum was reported to adhere to and invade epithelial cells via its virulence factors such as FadA,Citation39 Fap2Citation40 and FomA.Citation41 Moreover, a recent study confirmed the invasive potential of F. nucleatum into HCT116 cells by fluorescence microscopy and flow cytometry,Citation21 suggesting that there may exist other ways to activate NF-κB pathway. Intriguingly, a recent study identified ALPK1 as a key mediator of microbiota-induced NF-κB activation, which is a member of the atypical kinase family alpha kinases that recognize phosphorylation sites within α helices.Citation42,Citation43 It is generally acknowledged that ALPK1 accepted the signal of ADP-Hep present in Gram-negative bacterium.Citation23 Recognition of ADP-Hep by ALPK1 represents a generic form of innate sensing, which mediates immune responses to diverse bacterial pathogens. Here, we found that F. nucleatum treatment led to an induction of ALPK1 and resulted in the sustained activation of NF-κB signaling, and silencing of ALPK1 inhibited NF-κB activation and ICAM1 upregulation by F. nucleatum infection. In addition, our results suggested that F. nucleatum exhibited a dramatic activation on ALPK1/NF-κB/ICAM1 pathway compared with other Gram-negative bacteria. ALPK1/NF-κB/ICAM1 is relatively a specific pathway for F. nucleatum infection. However, we did not dissect the detail mechanisms by which F. nucleatum activated ALPK1 pathway. Several research groups reported a similar manner that Helicobacter pylori activated ALPK1/NF-κB axis via the type-IV secretion system (T4SS).Citation42,Citation44–46 It will be of great interest to elucidate how F. nucleatum stimulates the expression of ALPK1 in CRC cells and further influences NF-κB pathway.
In addition, we found that the abundance of F. nucleatum and ICAM1/ALPK1 were highly enriched in CRC patients with metastasis. Moreover, the relative abundance of F. nucleatum was positively correlated with ICAM1/ALPK1 levels in CRC tissues, and the high expression of ICAM1/ALPK1 was significantly associated with a shorter overall survival time of CRC patients. Recent studies have reported that F. nucleatum promotes colorectal carcinogenesis by modulating Wnt/β-catenin pathway,Citation34,Citation39,Citation47 NF-κB pathway,Citation17,Citation26,Citation39 autophagy,Citation18,Citation33 glucose metabolism,Citation48 etc. ALPK1/NF-κB/ICAM1 axis could, at least in part, explain how F. nucleatum treatment promotes CRC metastasis. However, it is possible that other mechanisms may be involved in the F. nucleatum-induced interactions between CRC cells and endothelial cells, and subsequent CRC metastasis. As the amount of F. nucleatum is associated with the risk of CRC metastasis, the measurement of F. nucleatum abundance may be an effective approach to predict patient outcome. Furthermore, our findings support the hypothesis that targeting F. nucleatum, host epithelial ICAM1 expression and/or ALPK1 may provide a means to block F. nucleatum-induced CRC metastasis and create diagnostic opportunities.
In summary, our current findings provide important insights into the molecular mechanisms underlying the regulation of an intercellular molecule, ICAM1, by F. nucleatum in CRC metastasis. F. nucleatum induces NF-κB pathway through ALPK1 activation, resulting in the upregulation of ICAM1 and enhanced CRC cell-endothelial cell adhesion and metastasis. Since the compelling evidences in vitro and in vivo highlight the emergence of F. nucleatum, ICAM1 or ALPK1 as pro-metastatic roles in CRC patients, a potential therapeutic strategy targeting F. nucleatum, ICAM1 and/or ALPK1 could be well-utilized.
Methods
Cell culture
Human colorectal cancer cell lines (LoVo, HCT116) and Human Umbilical Vein Endothelial Cells (HUVECs) were purchased from American Type Culture Collection (ATCC). HCT116 cultured in Maccoy 5A (Genom, China), LoVo cultured in F-12 K (Genom, China) and HUVECs cultured in high-glucose DMEM (GIBCO, China) were supplemented with 10% FBS (Sijiqing, China) at 37°C in a humidified 5% CO2 atmosphere.
Bacterial strains and culture conditions
F. nucleatum, A. muciniphila and B. adolescentis were purchased from American Type Culture Collection (ATCC, F. nucleatum strain 25586 and 10953, A. muciniphila BAA-835 and B. adolescentis strain 15703). F. nucleatum was grown in Columbia blood agar (Comagal, China) in an anaerobic glove box at 37°C. A. muciniphila in Brain Heart Infusion (BD Difco, USA) supplemented with 0.05% mucin Type II (Sigma-Aldrich, USA), B. adolescentis in Reinforced Clostridium Medium (BD Difco, USA) were cultured under an atmosphere of 10% H2, 10% CO2, and 80% N2 in an AW500SG anaerobic workstation (ELECTROTEK) at 37°C. The E. coli strain DH5α (Takara, Japan) was cultured in Luria-Bertani medium overnight at 37°C.
Human specimens
A total of 98 paired fresh tumor and adjacent non-tumor tissues were obtained from patients with primary CRC who did not receive preoperative anti-tumor treatment from Sir Run Run Shaw Hospital of Zhejiang University (Hangzhou, China). Fecal samples were collected from 72 CRC patients and 66 healthy people. All samples were frozen in liquid nitrogen until use. Another clinical tissue array cohort with 93 paired tumor and adjacent non-tumor tissues with or without liver metastasis were obtained from The Second Affiliated Hospital of Zhejiang University (Hangzhou, China).
Patient-derived xenograft (PDX)
CRC surgical specimen was physically separated into small pieces (2–3 mm) and subcutaneously transplanted into nude mice. The xenografts were resected from mice and cut into similarly sized PDX tissues. Then the obtained PDX tumor tissues were randomly divided into two groups, and treated with PBS or F. nucleatum separately at a multiplicity of infection (MOI) of 100:1. Total RNAs were extracted from the PDX tissues by Trizol reagent (Invitrogen, USA) following the manufacturer’s instructions for subsequent experiments.
Adhesion assay
Adhesion assay was performed as described previously.Citation49 Briefly, HUVECs were seeded on 96-well plates and grown overnight to reach full confluence. 5 × 104 GFP-labeled HCT116 cells pretreated with F. nucleatum were washed with PBS for three times, and re-suspended in serum-free culture medium to add onto the monolayer HUVECs and co-cultured at 37°C for 15 minutes. After washing by PBS softly for three times to remove the non-adherent cells, the adherent tumor cells on the monolayers were observed and imaged with a fluorescence microscope. Each experiment was analyzed in triplicate.
Migration assay
CRC cells with the indicated treatment were incubated with F. nucleatum (MOI of 100:1) for 6 hours, and migration assay was performed as previously described.Citation50 The migrated cells were observed and photographed under inverted microscope at 18 or 24 hours. Each experiment was analyzed in triplicate.
Animal assays
SCID mice (6–8 weeks old), BALB/c nude mice (6–8 weeks old) and C57BL/6 mice (5–6 weeks old, 6–8 weeks old) were used for indicated studies, cultured in specific-pathogen-free (SPF) facilities.
For cancer cell extravasation model, GFP-labeled HCT116 cells pretreated with PBS or F. nucleatum (MOI of 100:1) were intravenously injected into SCID mice (2 × 106 cells/per mouse). 24 hours after inoculation, the lungs were perfused with PBS by using gravity pressure through the heart. When the effluent became clear, the lungs were resected and the colonized HCT116 cells were measured by immunofluorescence assay, in which lung sections were stained for CD31 (red) as previously reported.Citation35,Citation51
For pulmonary metastasis model, HCT116 control cells or ICAM1-knockdown HCT116 cells were incubated with PBS or F. nucleatum (MOI of 100:1) for 24 hours and tail-vein injected into BALB/c nude mice (2 × 106 cells/per mouse). The lungs were surgically excised after 8 weeks, and the gross images and H&E staining of lung tissue sections were used to evaluate tumor metastasis.
For F. nucleatum-treatment C57BL/6 mice model, PBS-resuspended 109 colony-forming units (CFU) F. nucleatum or PBS were administrated into C57BL/6 mice (5–6 weeks old) every day, which were fed with 2 mg/ml streptomycin in the drinking water for 3 days beforehand to ensure the consistency of regular microbiota and facilitate F. nucleatum colonization as previously described.Citation17 After 15 days, mice were sacrificed and the colons of mice were surgically excised for further analysis.
For AOM/DSS-induced CRC model, C57BL/6 mice (6–8 weeks old) were given one single intraperitoneal injection of carcinogen azoxymethane (AOM, Sigma) at 10 mg/kg body weight following by 5 successive days of 2.5% dextran sodium sulfate (DSS) in the drinking water, and then were given regular drinking water for 2 weeks. This cycle was then repeated for twice. Mice in the F. nucleatum and E. coli groups were administrated with 109 CFU F. nucleatum or E. coli suspended in 300 μL sterile anaerobic PBS every 2 days, and the control group was administered with PBS. After 4 months, mice were sacrificed and the colons were surgically excised for further analysis.
RNA sequencing
For RNA sequencing, HCT116 cells were co-cultured with PBS or F. nucleatum (MOI of 100:1) for 6 hours. Total RNA was isolated from indicated cells by Trizol reagent (Invitrogen, USA), and mRNA was purified using poly-T oligo-attached magnetic beads. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations. 150 bp Paired-end libraries were sequenced by Illumina PE150 platform. Paired-end clean reads were aligned to the human genome version hg19 using Hisat2 v2.0.5. Differential expression analysis of two groups was performed using the DESeq2. P value < .05 and | log2 (fold change) | > 1 were considered as significant.
RNA extraction and quantitative RT-PCR
Total RNAs were extracted from CRC cell lines or tissue samples using Trizol reagent (Invitrogen, USA), and cDNAs were reversed by Evo M-MLV RT Kit (Accurate Biology, China) according to the manufacturer’s instructions. Quantitative RT-PCR analysis was performed in triplicate in ROCHE LightCycler480 System (Rotor gene 6000 Software, Sydney, Australia). Each reaction was tested in triplicate in 10 μl reaction system containing SYBR Premix Ex Taq (Takara, Japan), primers and template cDNA. Relative abundance was calculated by -ΔCt method as previously described.Citation27,Citation52 GAPDH or ACTIN were served as an internal reference gene. The following primer sets were used:
Human ICAM1: Forward: 5ʹ-CACCCTAGAGCCAAGGTGAC-3ʹ,
Reverse: 5ʹ-GGGCCATACAGGACACGAAG-3ʹ;
Human ALPK1: Forward: 5ʹ-TGACCACCATTTGCTGTCC-3ʹ,
Reverse: 5ʹ-ACGTGCCACGGATATTCAC-3ʹ
Human GAPDH: Forward: 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ,
Reverse: 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ
Mouse ALPK1: Forward: 5ʹ-CGCTATTGTCTTCTTGATGGACC-3ʹ
Reverse: 5ʹ-TTGACGGAGACTCGGGCTT-3ʹ;
Mouse ICAM1: Forward: 5ʹ-CACCCCGCAGGTCCAATTC-3ʹ,
Reverse: 5ʹ-TCCAGCCGAGGACCATACAG-3ʹ;
Mouse ACTIN: Forward: 5ʹ-AGCCATGTACGTAGCCATCC-3ʹ
Reverse: 5ʹ-CTCTCAGCTGTGGTGGTGAA-3ʹ.
DNA extraction and F. nucleatum quantification
Genomic DNAs were extracted from clinical or mice tissues by QIAamp DNA Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. Genomic DNAs were extracted from clinical stool samples with TIANamp Stool DNA Kit (Tiangen, China). Genomic DNAs from each specimen were subjected to quantitative RT-PCR analysis to determine the relative abundance of F. nucleatum as described above. Universal Eubacteria 16S was used as an internal reference gene for stool samples. The PGT gene was used as an internal control for tissue samples as previously described.Citation27 The following primer sets were used:
F. nucleatum: Forward: 5ʹ-CGGGTGAGTAACGCGTAAAG-3ʹ,
Reverse: 5ʹ-GCATTCGTTTCCAAATGTTGTCC-3ʹ;
PGT: Forward: 5ʹ-ATCCCCAAAGCACCTGGTTT-3ʹ,
Reverse: 5ʹ-AGAGGCCAAGATAGTCCTGGTAA-3ʹ;
Universal Eubacteria 16S: Forward: 5ʹ-CGGCAACGAGCGCAACCC-3ʹ,
Reverse: 5ʹ-CCATTGTAGCACGTGTGTAGCC-3ʹ.
Immunohistochemistry (IHC) and Western blot analysis
IHC staining was performed as previously described.Citation53 Tissue microarrays were scanned with a digital slide scanner (Pannoramic MIDI, 3D HISTECH) after staining and processed with Pannoramic viewer software. Intensity of staining in cells was automatic calculated by Quant center software. Histochemistry score (H-score) was acquired according to the formula: H-score = (percentage of weak intensity area ×1) + (percentage of moderate intensity area ×2) + (percentage of strong intensity area ×3). Western blot analysis was conducted as previously described.Citation54 The following primary antibodies were used: ICAM1 (10831-1-AP, Proteintech), phospho-IκBα (#2859, CST), NF-κB p65 (#8242, CST), phospho-NF-κB p65 (#3033, CST), ALPK1 (GTX01077, GeneTex) and GAPDH (10494-1-AP, Proteintech).
Immunofluorescence and confocal microscopy
Immunofluorescence staining was performed as previously described.Citation55 Images were collected with a confocal microscope (LSM 880, Zeiss) and processed with ZEN image software. The following primary antibodies were used: CD31 (11265-1-AP, Proteintech), NF-κB p65 (#8242, CST).
Cell transfection
The siRNAs were transfected into cells using Lipofectamine RNAiMAX (Thermo Fisher Scientific, USA) in opti-MEM (Genom, China) according to the manufacturer’s instructions. The following sequences of siRNAs were used:
siCtl: 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ;
siICAM1-1: 5ʹ-CCUAUGGCAACGACUCCUUTT-3ʹ;
siICAM1-2: 5ʹ-CCUCAGCACGUACCUCUAUTT-3ʹ;
sip65-1: 5ʹ-CAGAUACAGACGAUCGUCATT-3ʹ;
sip65-2: 5ʹ-CCCUAUCCCUUUACGUCAUTT-3ʹ;
siALPK1-1: 5ʹ-GCAGGUUGGAUAAACUUAUTT-3ʹ;
siALPK1-2: 5’-CGCAGGCCCUACAUUUAAATT-3ʹ;
siTLR4: 5ʹ-GGGCUUAGAACAACUAGAATT-3ʹ;
siMYD88: 5ʹ-GGCAACUGGAACAGACAAATT-3ʹ.
Plasmid construction and transfection
To generate shRNA constructs of ICAM1, oligos targeting mRNA of ICAM1 were synthesized and cloned into lentiviral pLKO.1-puro vector (Addgene plasmid #8453). The full-length OFR of human ICAM1 (NM_000201.3) were prepared by cloning the cDNA from HEK293 cells into the pLVX-puro vector (Clontech #PT4002-5). Transient plasmid transfection was carried out using FuGENE HD transfection reagent (Promega, USA) according to the manufacturer’s instructions. The following primer sequences for vectors construction were used:
pLKO.1-sh ICAM1:
Forward: 5ʹ-CCGGGCTGACGTGTGCAGTAATACTTCTCGAGAAGTATTACTGCACACGTCAGCTTTTTTG-3ʹ,
Reverse: 5ʹ-AATTCAAAAAAGCTGACGTGTGCAGTAATACTTCTCGAGAAGTATTACTGCACACGTCAGC-3ʹ;
plvx- ICAM1-FL:
Forward: 5ʹ-ATGGCTCCCAGCAGCCCCCGG-3ʹ,
Reverse: 5ʹ-TCAGGGAGGCGTGGCTTGTGT GTTCG-3ʹ.
Lentivirus production, precipitation and infection
To generate cells stably expressing short-hairpin RNAs (shRNAs), lentivirus production, precipitation and infection were performed as previously described.Citation56
Statistical analysis
Statistical analysis was performed using the GraphPad Prism 7.0 software. Data were analyzed with paired or unpaired Student’s t test, Mann-Whitney test, linear regression or Log-rank test as showed in figure legends. P value less than 0.05 was considered statistically significant.
Abbreviations
CRC: colorectal cancer; F. nucleatum: Fusobacterium nucleatum; E. coli: Escherichia coli; A. muciniphila: Akkermansia muciniphila; B. adolescentis: Bifidobacterium adolescentis; PBS: phosphate buffer saline; HUVECs: Human Umbilical Vein Endothelial Cells; PDX: patient-derived xenograft; MOI: multiplicity of infection; SPF: specific-pathogen-free; CFU: colony-forming units; NES: Normalized Enrichment Score; TCGA: The Cancer Genome Atlas; ATCC: American Type Culture Collection.
Data availability
The accession number for the data for RNA-sequencing reported in this study is NCBI GEO: GSE171611.
Study approval
All animal experiments used in this study were approved by the Institutional Animal Care and Use Committee of Zhejiang University (SRRSH202102222). The collection of human samples was approved by the ethics committee of Sir Run Run Shaw Hospital and The Second Affiliated Hospital of Zhejiang University with informed written consent obtained from all participants (20180226-80, 2020-446).
Author Contributions
Y.Z, L.Z and W.Z designed the experiments and interpreted the results. Y.Z, L.Z, S.Z, MJ.L, CC.X and DJC.J performed the experiments. YD.Q analyzed the RNA-sequencing data. TY.H, L.W and AQ.L provided clinical CRC tissues. Y.Z and BY.W analyzed the clinical data. SJ.C and JM.S improved the project design and interpreted the results. Y.Z, W.Z and L.Z wrote the manuscript, which was further refined by all authors. W.Z supervised the overall study.
Supplemental Material
Download Zip (51.8 MB)Acknowledgments
This work was derived from the conference abstract in Journal of Digestive Diseases (Volume 22, Issue S1 PO-035).Citation57
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary Material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, and Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–20. doi:10.3322/caac.21492.
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–164. doi:10.3322/caac.21601.
- Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi:10.1038/nrc2618.
- Reymond N, d’Agua BB, Ridley AJ, and d’Água BB. Crossing the endothelial barrier during metastasis. Nat Rev Cancer. 2013;13:858–870. doi:10.1038/nrc3628.
- Kobayashi H, Boelte KC, Lin PC. Endothelial cell adhesion molecules and cancer progression. Curr Med Chem. 2007;14:377–386. doi:10.2174/092986707779941032.
- Makrilia N, Kollias A, Manolopoulos L, Syrigos K. Cell adhesion molecules: role and clinical significance in cancer. Cancer Invest. 2009;27:1023–1037. doi:10.3109/07357900902769749.
- Wai Wong C, Dye DE, Coombe DR. The role of immunoglobulin superfamily cell adhesion molecules in cancer metastasis. Int J Cell Biol. 2012;2012:340296. doi:10.1155/2012/340296.
- Maeda K, Kang SM, Sawada T, Nishiguchi Y, Yashiro M, Ogawa Y, Ohira M, Ishikawa T, Hirakawa-YS Chung K. Expression of intercellular adhesion molecule-1 and prognosis in colorectal cancer. Oncol Rep. 2002;9:511–514.
- Tung SY, Chang SF, Chou MH, Huang WS, Hsieh YY, Shen CH, Kuo H-C, Chen C-N. CXC chemokine ligand 12/stromal cell-derived factor-1 regulates cell adhesion in human colon cancer cells by induction of intercellular adhesion molecule-1. J Biomed Sci. 2012;19:91. doi:10.1186/1423-0127-19-91.
- Paschos KA, Canovas D, Bird NC. The role of cell adhesion molecules in the progression of colorectal cancer and the development of liver metastasis. Cell Signal. 2009;21:665–674. doi:10.1016/j.cellsig.2009.01.006.
- Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi:10.1136/gutjnl-2015-310101.
- Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, Brim H, Ashktorab H, Ng SC, Ng SSM, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. 2017;23:2061–2070. doi:10.1158/1078-0432.CCR-16-1599.
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi:10.1038/nrg3182.
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi:10.1101/gr.126516.111.
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi:10.1101/gr.126573.111.
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy T, Chung D, Lochhead P, Hold G, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi:10.1016/j.chom.2013.07.007.
- Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of microRNA-21. Gastroenterology. 2017;152:851–66 e24. doi:10.1053/j.gastro.2016.11.018.
- Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, and Nagarsheth N, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170:548–63 e16. doi:10.1016/j.cell.2017.07.008.
- Taniguchi K, Karin M. NF-kappaB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18:309–324. doi:10.1038/nri.2017.142.
- Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi:10.1126/science.aal5240.
- Casasanta MA, Yoo CC, Udayasuryan B, Sanders BE, Umana A, Zhang Y. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal. 2020;13. doi:10.1126/scisignal.aba9157.
- Tang W, Guo Z, Cao Z, Wang M, Li P, Meng X, Zhao X, Xie Z, Wang W, and Zhou A, et al. d-Sedoheptulose-7-phosphate is a common precursor for the heptoses of septacidin and hygromycin B. Proc Natl Acad Sci U S A. 2018;115:2818–2823. doi:10.1073/pnas.1711665115.
- Zhou P, She Y, Dong N, Li P, He H, Borio A, Wu Q, Lu S, Ding X, Cao Y, et al. Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose. Nature. 2018;561:122–126. doi:10.1038/s41586-018-0433-3.
- Gaudet RG, Sintsova A, Buckwalter CM, Leung N, Cochrane A, Li J, Cox AD, Moffat J, Gray-Owen SD. INNATE IMMUNITY. Cytosolic detection of the bacterial metabolite HBP activates TIFA-dependent innate immunity. Science. 2015;348:1251–1255. doi:10.1126/science.aaa4921.
- Milivojevic M, Dangeard AS, Kasper CA, Tschon T, Emmenlauer M, Pique C, Schnupf P, Guignot J, Arrieumerlou C. ALPK1 controls TIFA/TRAF6-dependent innate immunity against heptose-1,7-bisphosphate of gram-negative bacteria. PLoS Pathog. 2017;13:e1006224. doi:10.1371/journal.ppat.1006224.
- Chen S, Su T, Zhang Y, Lee A, He J, Ge Q, Wang L, Si J, Zhuo W, Wang L, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis by modulating KRT7-AS/KRT7. Gut Microbes. 2020;11:511–525. doi:10.1080/19490976.2019.1695494.
- Fan L, Xu C, Ge Q, Lin Y, Wong CC, Qi Y, Ye B, Lian Q, Zhuo W, Si J, et al. A. Muciniphila suppresses colorectal tumorigenesis by inducing TLR2/NLRP3-Mediated M1-Like TAMs. Cancer Immunol Res. 2021;9:1111–1124. doi:10.1158/2326-6066.CIR-20-1019.
- Fan L, Qi Y, Qu S, Chen X, Li A, Hendi M, Xu C, Wang L, Hou T, Si J, et al. B. adolescentis ameliorates chronic colitis by regulating Treg/Th2 response and gut microbiota remodeling. Gut Microbes. 2021;13:1–17. doi:10.1080/19490976.2020.1826746.
- Abreu MT, Peek RM Jr. Gastrointestinal malignancy and the microbiome. Gastroenterology. 2014;146:1534–46 e3. doi:10.1053/j.gastro.2014.01.001.
- Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat Protoc. 2007;2:1998–2004. doi:10.1038/nprot.2007.279.
- Wang L, Tang L, Feng Y, Zhao S, Han M, Zhang C, Yuan G, Zhu J, Cao S, Wu Q, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8 + T cells in mice. Gut. 2020;69:1988–1997. doi:10.1136/gutjnl-2019-320105.
- Li Q, Hu W, Liu WX, Zhao LY, Huang D, Liu XD, Chan H, Zhang Y, Zeng J-D, Coker OO, et al. Streptococcus thermophilus inhibits colorectal tumorigenesis through secreting beta-Galactosidase. Gastroenterology. 2021;160:1179–93 e14. doi:10.1053/j.gastro.2020.09.003.
- Chen Y, Chen Y, Zhang J, Cao P, Su W, Deng Y, Zhan N, Fu X, Huang Y, Dong W, et al. Fusobacterium nucleatum promotes metastasis in colorectal cancer by activating autophagy signaling via the upregulation of CARD3 expression. Theranostics. 2020;10:323–339. doi:10.7150/thno.38870.
- Guo S, Chen J, Chen F, Zeng Q, Liu WL, Zhang G. Exosomes derived from Fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut. 2020. doi:10.1136/gutjnl-2020-321187.
- Huang C, Li N, Li Z, Chang A, Chen Y, Zhao T, Li Y, Wang X, Zhang W, Wang Z, et al. Tumour-derived Interleukin 35 promotes pancreatic ductal adenocarcinoma cell extravasation and metastasis by inducing ICAM1 expression. Nat Commun. 2017;8:14035. doi:10.1038/ncomms14035.
- Liu S, Li N, Yu X, Xiao X, Cheng K, Hu J, Wang J, Zhang D, Cheng S, Liu S, et al. Expression of intercellular adhesion molecule 1 by hepatocellular carcinoma stem cells and circulating tumor cells. Gastroenterology. 2013;144:1031–41 e10. doi:10.1053/j.gastro.2013.01.046.
- Mao L, Yang J, Yue J, Chen Y, Zhou H, Fan D, Zhang Q, Buraschi S, Iozzo RV, Bi X, et al. Decorin deficiency promotes epithelial-mesenchymal transition and colon cancer metastasis. Matrix Biol. 2021;95:1–14. doi:10.1016/j.matbio.2020.10.001.
- Garcia-So J, Zhang X, Yang X, Rubinstein MR, Mao Y, Kitajewski J. Omega-3 fatty acids suppress Fusobacterium nucleatum-induced placental inflammation originating from maternal endothelial cells. JCI Insight. 2019;4. doi:10.1172/jci.insight.125436.
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi:10.1016/j.chom.2013.07.012.
- Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan K, et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi:10.1016/j.chom.2016.07.006.
- Nakagaki H, Sekine S, Terao Y, Toe M, Tanaka M, Ito HO, Kawabata S, Shizukuishi S, Fujihashi K, Kataoka K, et al. Fusobacterium nucleatum envelope protein FomA is immunogenic and binds to the salivary statherin-derived peptide. Infect Immun. 2010;78:1185–1192. doi:10.1128/IAI.01224-09.
- Zimmermann S, Pfannkuch L, Al-Zeer MA, Bartfeld S, Koch M, Liu J, Rechner C, Soerensen M, Sokolova O, Zamyatina A, et al. ALPK1- and TIFA-dependent innate immune response triggered by the Helicobacter pylori Type IV secretion system. Cell Rep. 2017;20:2384–2395. doi:10.1016/j.celrep.2017.08.039.
- Ryazanov AG, Pavur KS, Dorovkov MV. Alpha-kinases: a new class of protein kinases with a novel catalytic domain. Curr Biol. 1999;9:R43–5. doi:10.1016/s0960-9822(99)80006-2.
- Gall A, Gaudet RG, Gray-Owen SD, Salama NR. TIFA signaling in gastric epithelial cells initiates the cag type 4 secretion system-dependent innate immune response to Helicobacter pylori infection. mBio. 2017:8. doi:10.1128/mBio.01168-17.
- Stein SC, Faber E, Bats SH, Murillo T, Speidel Y, Coombs N, Josenhans C. Helicobacter pylori modulates host cell responses by CagT4SS-dependent translocation of an intermediate metabolite of LPS inner core heptose biosynthesis. PLoS Pathog. 2017;13:e1006514. doi:10.1371/journal.ppat.1006514.
- Pfannkuch L, Hurwitz R, Traulsen J, Sigulla J, Poeschke M, Matzner L, Kosma P, Schmid M, Meyer TF. ADP heptose, a novel pathogen-associated molecular pattern identified in Helicobacter pylori. FASEB J. 2019;33:9087–9099. doi:10.1096/fj.201802555R.
- Rubinstein MR, Baik JE, Lagana SM, Han RP, Raab WJ, Sahoo D, Dalerba P, Wang TC, Han YW. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator Annexin A1. EMBO Rep. 2019;20(20). 10.15252/embr.201847638.
- Hong J, Guo F, Lu SY, Shen C, Ma D, Zhang X, Xie Y, Yan T, Yu T, Sun T, et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. 2021;70:2123–2137. doi:10.1136/gutjnl-2020-322780.
- Kukreja P, Abdel-Mageed AB, Mondal D, Liu K, Agrawal KC. Up-regulation of CXCR4 expression in PC-3 cells by stromal-derived factor-1alpha (CXCL12) increases endothelial adhesion and transendothelial migration: role of MEK/ERK signaling pathway-dependent NF-kappaB activation. Cancer Res. 2005;65:9891–9898. doi:10.1158/0008-5472.CAN-05-1293.
- Zhang M, Miao F, Huang R, Liu W, Zhao Y, Jiao T. RHBDD1 promotes colorectal cancer metastasis through the Wnt signaling pathway and its downstream target ZEB1. J Exp Clin Cancer Res. 2018;37:22. doi:10.1186/s13046-018-0687-5.
- Saito K, Ozawa Y, Hibino K, Ohta Y, Kaibuchi K. FilGAP, a Rho/Rho-associated protein kinase-regulated GTPase-activating protein for Rac, controls tumor cell migration. Mol Biol Cell. 2012;23:4739–4750. doi:10.1091/mbc.E12-04-0310.
- He J, Ge Q, Lin Z, Shen W, Lin R, Wu J, Wang B, Lu Y, Chen L, Liu X, et al. MiR-129-5p induces cell cycle arrest through modulating HOXC10/Cyclin D1 to inhibit gastric cancer progression. FASEB J. 2020;34:8544–8557. doi:10.1096/fj.201903217R.
- Hawinkels LJ, Paauwe M, Verspaget HW, Wiercinska E, van der Zon JM, van der Ploeg K, Koelink PJ, Lindeman JHN, Mesker W, ten Dijke P, et al. Interaction with colon cancer cells hyperactivates TGF-beta signaling in cancer-associated fibroblasts. Oncogene. 2014;33:97–107. doi:10.1038/onc.2012.536.
- Xiong H, Hong J, Du W, Lin YW, Ren LL, Wang YC, Su W-Y, Wang J-L, Cui Y, Wang Z-H, et al. Roles of STAT3 and ZEB1 proteins in E-cadherin down-regulation and human colorectal cancer epithelial-mesenchymal transition. J Biol Chem. 2012;287:5819–5832. doi:10.1074/jbc.M111.295964.
- Jeong MH, Park SY, Lee SH, Seo J, Yoo JY, Park SH, Kim MJ, Lee S, Jang S, Choi H-K, et al. EPB41L5 mediates TGFbeta-induced metastasis of gastric cancer. Clin Cancer Res. 2019;25:3617–3629. doi:10.1158/1078-0432.CCR-18-2959.
- Li Y, Sun Q, Jiang M, Li S, Zhang J, Xu Z, Guo D, Gu T, Wang B, Xiao L, et al. KLF9 suppresses gastric cancer cell invasion and metastasis through transcriptional inhibition of MMP28. FASEB J. 2019;33:7915–7928. doi:10.1096/fj.201802531R.
- Zhang Y, Chen S. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-κB/ICAM1 axis. J Dig Dis. The 21st Congress of Gastroenterology China, Nanchang, China. 2021;22(S1 Suppl):S61. doi:10.1111/1751-2980.13054.
