ABSTRACT
We present an overview of recent experimental and computational advances in technology used to characterize the microbiome, with a focus on how these developments improve our understanding of inflammatory bowel disease (IBD). Specifically, we present studies that make use of flow cytometry and metabolomics assays to provide a functional characterization of microbial communities. We also describe computational methods for strain-level resolution, temporal series, mycobiome and virome data, co-occurrence networks, and compositional data analysis. In addition, we review novel techniques to therapeutically manipulate the microbiome in IBD. We discuss the benefits and drawbacks of these technologies to increase awareness of specific biases, and to facilitate a more rigorous interpretation of results and their potential clinical application. Finally, we present future lines of research to better characterize the relation between microbial communities and IBD pathogenesis and progression.
Introduction
Developments in microscopy by van Leeuwenhoek revolutionized our understanding of microbes. Similarly, technological advances, such as high-throughput sequencing, mass spectrometry, and computational tools for data analysis have tremendously contributed to the characterization of microbial communities in human health and disease. Specifically, it is now widely accepted that microbes play a fundamental role in the pathogenesis, disease progression, and response to treatment in inflammatory bowel disease (IBD). IBD is a devastating condition characterized by chronic inflammation of the digestive tract and for which there is currently no cure. While genetics is a determining factor in establishing disease risk,Citation1,Citation2 environmental factors, such as the microbiome, are thought to contribute as triggering elements that can modulate the natural history of the disease.Citation3–5 Understanding the interaction of host and microbes is therefore required for a more complete characterization of IBD and to potentially identify therapeutic targets.
Here, we review recent technological advances of relevance that are being utilized to understand how the host and the microbiome interact in the context of IBD (). We review experimental technologies that provide more accurate identification of microbes, describe methods to uncover microbial products responsible for pathogenic mechanisms, present novel techniques for data analysis, and discuss state-of-the-art approaches that aim at modifying the microbiome for therapeutic purposes. We further describe the limitations of these methods and discuss opportunities for future avenues of research.
Table 1. Tools for characterizing the IBD Microbiome.
Novel approaches to characterize the microbiome and its products
Combining flow cytometry with high-throughput sequencing
While the analysis of microbial components of the human microbiome can offer important insights into the etiology of IBD, changes in the abundance of specific bacteria do not necessarily reflect the immunological impact of the microbiome on the host. Fluorescence-activated cell sorting (FACS) coupled with 16S rRNA gene sequencing is a powerful flow cytometry-based technology that allows quantification and sorting of bacteria coated with immunoglobulin A antibodies (IgA-Seq).Citation6–10 Secretory IgA (sIgA) is the predominant antibody isotype at mucosal surfaces, and plays a direct role in intestinal immunity. Large amounts of sIgA are produced by plasma cells located in Peyer patches, and secreted into the intestinal lumen by transcytosis, where it coats invading pathogens with high affinity, blocking their binding to receptors on epithelial cells and facilitating their removal by immune exclusion.Citation73,Citation74 Simultaneously, sIgA maintains homeostasis by coating commensals in what is generally considered a relatively low-affinity and specificity interaction, containing commensals within the gut, regulating their composition, gene expression, and motility.Citation73,Citation75–78
By using labeled anti-human IgA antibodies, gut bacteria can be sorted into IgA-coated (IgA+) and non-coated (IgA-) fractions. Compositional profiles of the sorted bacterial fractions are then identified with 16S rRNA gene sequencing. IgA-Seq-based studies have identified aberrant sIgA-microbiota interactions in IBD. In particular, increased levels of sIgA coating and differential binding of pathobionts have been shown, compared to healthy controls,Citation7,Citation11,Citation79,Citation80 supporting the potential of this technology to identify immunologically reactive microbiota associated with disease. Some studies have further shown that many of the IgA responses to the microbiome seem to be strain-specific. Yang and colleagues showed that different strains of Bacteroides ovatus can drive either high or low mucosal IgA production in mice mono-colonized with different human fecal strains.Citation81 Similarly, Palm and collaborators isolated two strains of B. fragilis from IBD patients that showed either high or low IgA coating, and used them to colonize germ-free mice with induced colitis. Results showed that the IgA+ strain exacerbated inflammation and reduced colon length, compared to the IgA- strain.Citation7 Viladomiu and colleagues identified adherent- invasive E. coli (AIEC) strains in Crohn’s disease (CD) with and without associated spondyloarthritis (SpA).Citation79 Although no significant differences were observed in terms of relative abundance between both groups, IgA+ E. coli was significantly enriched in CD patients with SpA, and those strains were also enriched in virulence genes, compared to CD strains from non-SpA patients. These findings reinforce the idea that IgA-coating may not be dependent on bacterial abundances in the gut, but rather is directed to distinct immunologically reactive strains.Citation7,Citation9,Citation79–81
One of the main advantages of this methodology is its capacity to recover organisms that elicit IgA responses, which allow for further experimental validation such as testing the immunological impact of identified bacteria in germ-free mice or in vitro cell models.Citation7,Citation9,Citation79,Citation81,Citation82 These experiments have shown that specific, highly immune-reactive members of the human microbiota confer susceptibility to IBD upon transplantation into IBD-models of germ-free mice. In the study by Palm and colleagues, authors sorted IgA± from IBD patients, then isolated and rationally selected bacterial taxa based on their level of IgA coating. They then assembled highly coated and low-coated bacterial consortia and used them to colonize germ-free mice. Results showed a dramatic increase in intestinal inflammation and susceptibility to induced colitis when mice were exposed to the highly IgA+ consortium.Citation7 On the other hand, Viladomiu and colleagues used an in vitro model of Caco-2 intestinal epithelial cells to perform functional analysis of IgA-coated E. coli isolated from CD-SpA. This revealed the strain’s high adherence and invasion capability, as well as persistence following macrophage invasion, consistent with its characterization as adherent-invasive E.coli.Citation79 Authors also exposed germ-free mice to the highly invasive E. coli, which was able to induce Th17 mucosal immunity. They further showed that the same E. coli strain aggravated colitis or inflammatory arthritis after transplantation into interleukin-10 deficient or K/BxN mice, respectively. Finally, a recent study has also shown promising applications of IgA-Seq at identifying changes in IgA-binding profiles in response to IBD treatments. They found that IgA-coated taxa were predictive of progression to surgical resection.Citation80
This promising technology, however, suffers from some limitations. Flow-cytometry is based on the optical properties of the sample and relies on the use of fluorescent antibodies and other cell markers that can result in nonspecific binding and differential-staining biases, which could affect IgA-Seq results and interpretation. To provide robust evidence for the specificity of the antibody to its target, antibody validation is recommended. Several methods are available for this purpose, including the use of positive and negative cells (i.e. testing the antibody against microbial communities from human samples vs microbial cultures where human IgA antibodies are not present, such as pure microbial cultures). In addition, when performing multicolor fluorescence assays (e.g. combining antibodies and viability dyes), fluorescent compensation should be applied. The principle of compensation is simple: for each fluorophore used in the experiment, a single-stained cell or bead sample must also be prepared. This process identifies spectral overlap between fluorophores and allows correction for signal “spillover” into other channels. In addition, a suspension of single cells is required for this technique to work accurately, as the detection of fluorescent signal from a cell forming part of an aggregate could result in the sorting of other non-positive particles, which could lead to misleading results. However, microbes often grow as aggregates or part of a biofilm, so filtering of larger particles and disaggregation through shaking or sonication are needed before sorting. Moreover, a high risk of microbial contamination within the cytometer is likely to occur if thorough cleaning and de-contamination protocols are not established. Importantly, software pipelines for microbiome analysis do not currently incorporate functionality specific for IgA-Seq data analysis, which can preclude the standardization of results. On the other hand, identification of disease-associated members of the microbiota can be hindered by the cross-reactivity of antibodies toward both pathobionts and commensals. The specificity of antibodies to microbial antigens and the underlying mechanisms by which the immune system distinguishes pathogens from commensals remains largely unknown.Citation8,Citation82,Citation83 Thus, comparison of IgA-coating patterns between disease and healthy controls seems imperative, which requires well-described IgA-coating profiles in healthy conditions before this methodology can be applied as a diagnostic tool. Besides IgA, other immunoglobulin isotypes are found in the gut lumen. In particular, previous studies have shown that IgG and IgM antibodies also bind differentially to the microbiota,Citation82,Citation84,Citation85 and increased levels in IgG-coating have been observed in CD and UC.Citation79,Citation82,Citation86 In a recent study, UC was associated with increased levels in both IgA- and IgG-bound bacteria, whereas only an increase in IgG was observed in CD, suggesting differential patterns of IgA- and IgG-binding to fecal bacteria in UC and CD.Citation11
In addition to bacteria, it has recently been shown that intestinal fungi can also be targeted by secretory IgA and systemic IgG and IgM.Citation14,Citation87,Citation88 Doron and colleagues sorted and sequenced IgA and IgG antibody-bound fungal cells from the stool of healthy individuals, which identified Candida albicans as the most immunoglobulin-reactive species in healthy conditions.Citation88 Subsequent studies identified that IgA preferentially targets fungal hyphal morphologies that are associated with invasion and virulence, rather than yeast morphologies, suggesting that IgA promotes a mutualistic relationship between the host and commensal fungi.Citation14,Citation87 Interestingly, Doron and collaborators showed a dysregulated antibody response against hyphal morphotypes of C. albicans, and an overall increase in fungal hyphal morphotypes in intestinal mucosal washings from CD patients. These results suggest that IgA mechanisms targeting fungal virulence factors could be disrupted in CD, and that Ig-SEQ also has potential applications for the analysis of mycobiota involvement in the pathogenesis of IBD.Citation14
Leveraging metabolomics to study microbial metabolites and molecules
Members of the microbiome interact with the host through metabolites, both self-synthesized de novo, or produced by modulating human- and diet-derived components.Citation25 Studying the metabolome, the combination of host and microbial-derived metabolic components, can offer important insights into molecular mechanisms observed in the gut microbiome of IBD patients. The most common approaches used to characterize the metabolome are separation of molecules by chromatography (mainly gas, GC, or liquid, LC), capillary electrophoresis (CE) with subsequent detection by mass spectrometry (MS), or nuclear magnetic resonance (NMR). All can be performed using untargeted or targeted methods, offering unique advantages and disadvantages.Citation26 In untargeted approaches, a complete profile of all the detectable metabolites present in the sample of interest can be obtained. Targeted methods, on the other hand, aim at identifying specific molecules from a predefined panel of targets. While targeted mass spectrometry offers better specificity and provides absolute quantification, this is at the expense of lower sensitivity (i.e. only metabolites in the panel can be measured), and requires a priori knowledge about what metabolites to target.Citation26 Untargeted metabolomics, on the other hand, can quantify a vast number of uncharacterized metabolites in a single run, which allows the identification of known and unknown molecules in the sample of interest, and is particularly useful for biomarker discovery.Citation15,Citation26 However, as only a subset of the metabolome is annotated, this method is generally applied to compare profiles of groups of samples. To resolve some of these issues, additional experimental and analytical approaches have been proposed, such as validation of metabolites against compound libraries, the use of ad hoc analytical pipelines to enhance metabolite clustering and annotation,Citation15 or the application of methods that couple targeted an untargeted metabolomics.Citation26 Although metabolomics theoretically identifies all small molecules present in a sample, in practice this is limited by how molecules are extracted, ionized, and detected. In addition, the different types of metabolomic tools provide different levels of resolution and thus the choice of analytical methods can produce different results. Therefore, the combination of complementary instrumental platforms and analytical methods is required for a comprehensive profiling of the metabolome.Citation25
IBD is associated with an aberrant microbiome that has a reduced capacity to produce metabolites that are important for intestinal homeostasis, while producing pathogenic molecules that trigger pro-inflammatory processes in the host. Integrating metabolomic and metagenomic profiling has allowed the identification of metabolite biomarkers that differentiate IBD patients and healthy controls.Citation15,Citation89 Such differences and the knowledge that specific microbial-derived metabolites promote intestinal barrier integrity and regulate inflammatory processes have inspired numerous translational studies seeking to apply microbiome-derived metabolites in novel therapeutics for IBD.Citation90
Short-chain fatty acids
Short-chain fatty acids (SCFA), including butyrate, acetate, and propionate, are produced by gut bacteria through the digestion of dietary fibers. These metabolites participate in numerous cellular and immunological processes, such as stimulating the production of mucins, reducing intestinal permeability, and promoting anti-inflammatory pathways.Citation27,Citation33 Both gas chromatography – in combination with various detectors – and liquid chromatography are commonly used to analyze SCFA in fecal samples.Citation28,Citation29 However, these methods have low sensitivity for the detection of SCFA in complex samples, due to the small mass of the compounds and spectral overlap with interfering peaks from solvents and additives. In addition, SCFA’s hydrophilicity results in poor chromatographic separation. To overcome these problems, SCFA can be derivatized, a process that alters the structure of the analyte to facilitate its isolation, separation, or detection.Citation27–29 Nevertheless, most chemical derivatization procedures for GC/MS and LC/MS are not suitable for the analysis of SCFA in biological samples due to their volatility and thermal instability. Consequently, several protocols have been specifically adapted for metabolomic analysis of SCFA, including chemical derivatization methods,Citation28,Citation30 as well as isotopically labeled derivatization methods, which introduce isotope tags for accurate compound quantification while improving the chromatographic retention of metabolites.Citation27,Citation31,Citation32
It has been observed that levels of butyrate and their bacterial producers are lower in patients with active IBD compared to controlsCitation16–18,Citation89,Citation91. This suggests that restoring gut microbial homeostasis can increase luminal levels of butyrate and exert an anti-inflammatory function. However, the complex ecology of the gut microbiome makes it difficult to modulate the butyrate-producing capacity of these communities, and direct supplementation of SCFAs has been explored as a more practical alternative. Butyrate administration has shown some promise in the treatment of IBD, including reduction of inflammation, symptoms and development of the disease.Citation34–36 In vivo and in vitro studies have shown that butyrate’s anti-inflammatory properties are associated with the inhibition of nuclear factor kappa B, which reduces the expression of pro-inflammatory genes.Citation36,Citation92,Citation93 In addition, several studies have shown that the combination of butyrate with biologics can significantly improve their therapeutic effect. In one study, mesalazine (5-aminosalicylic acid, 5-ASA) and butyrate significantly ameliorated disease based on endoscopic and histologic scores, and maintained clinical remission compared with treatment with isolated 5-ASA alone.Citation34,Citation35,Citation94 The levels of butyrate and substrates involved in butyrate synthesis have also been associated with anti-TNFα therapy efficacy in IBD patients.Citation91 Multiple clinical studies are currently investigating the efficacy of SCFA in the treatment of IBD, including the administration of encapsulated butyrate to reduce intestinal inflammation in IBD and irritable bowel syndrome patients (www.clinicaltrials.gov: NCT04879914), butyrate in combination with other therapeutics (e.g. hydroxocobalamin to treat UC, trial NCT04259060), or dietary interventions to modulate the microbiome and increase the amount of colonic SCFA (NCT04520594, NCT04522271, and NCT04065048).
Dietary amino acids
IBD is also characterized by a reduction in specific dietary amino acids that are affected by microbial activity.Citation20,Citation21 An important example is tryptophan (Trp), which serves as substrate to gut microbial enzymes that convert dietary Trp into tryptamine and other indole derivatives. Trp derivatives can function as endogenous ligands for the aryl hydrocarbon receptor (AhR), that is essential to maintain gut homeostasis and immune responses.Citation21,Citation22 Therefore, a reduction of Trp and other Trp-derived AhR ligands can have important effects on the anti-inflammatory and protective functions associated to AhR activity. In a murine model of dextran sulfate sodium (DSS)-induced colitis, administration of a Trp-deficient diet or depletion of AhR encoding genes exacerbated colitis and led to weight loss and reduction of the production of antimicrobial peptides and other important chemokines.Citation95,Citation96 Administration of Trp in the diet attenuates inflammation and protects against colitis symptoms. Similar results have been reported in other studies with DSS-induced colitis animal models treated with Trp or its derivatives.Citation96–99 Among the Trp-derivative metabolites that can ameliorate induced colitis in animals are the microbially produced indole (IND) and indole-3-propionate (IPA), which have been shown to regulate intestinal epithelial function via activation of IL-10 expression.Citation97,Citation100 Other derivatives, such as indole-3-pyruvate (IPyA), indole-3-aldehyde (I3A), and indole-3-ethanol (I3E) are essential to modulate gut barrier integrity via tight junctions and adherent junctions.Citation21,Citation99 Scott and colleagues demonstrated that IPyA, I3A, and I3E protected against intestinal permeability and colitis in mice, and that removal of Trp-metabolizing bacteria by antibiotic treatment depleted their production.Citation99 Therefore, bacterial tryptophan metabolites have an important potential use as treatments for IBD symptoms, although evidence from clinical studies in humans is not yet available.
LC-MS/MS has been widely used to quantify Trp and its metabolites.Citation38,Citation101 However, this method often focuses only on major Trp metabolites, such as kynurenine and tryptophan. Nevertheless, the majority of Trp is metabolized through the kynurenine pathway into a variety of less-abundant compounds, and changes in concentrations of these less-abundant metabolites have been observed in IBD.Citation20,Citation38 Whiley and colleagues developed a specific method to quantify Trp and its downstream metabolites, called targeted ultrahigh-performance liquid chromatography tandem mass spectrometry with electrospray ionization (UHPLC-ESI-MS/MS), and validated it in a cohort of UC patients.Citation37 Besides Trp, the metabolism of other amino acids, such as arginine and glutamine, are also altered in IBD and could represent additional targets to treat the disease.Citation21 However, findings from studies in animal models have not been able to be replicated in human studies in pediatric CD, adult CD and UC patients.Citation102,Citation103
Secondary bile acids
Secondary bile acids (SBA) are also bacterial-derived gut metabolites, which play key roles as signaling molecules.Citation104 While primary bile acids (PBA) are produced from cholesterol in the liver, microorganisms are responsible for transforming them into secondary and tertiary bile acids in the gut via deconjugation of glycine or taurine by bile salt hydrolases, and dehydroxylation, oxidation, and epimerization of the cholesterol core.Citation105 Alterations in the chemistry of SBA have been observed in IBD patients.Citation15,Citation23,Citation24 For example, decreased bile acid deconjugation has been associated with UC and CD, and the alterations in bile acid metabolism have been directly linked with the microbial dysbiosis occurring in IBD.Citation106 Sinha and collaborators recently combined metabolomic, microbiome, metagenomic, and transcriptomic profiling of stool from ileal pouches in colectomy-treated UC patients. Results showed that, compared to controls, patients had reduced levels of the two most abundant SBA in the gut, lithocholic acid (LCA) and deoxycholic acid (DCA), as well as genes involved in SBA transformation from PBA, and reduced abundance of Ruminococcaceae, a family of bacteria known to produce SBA.Citation24 Although approximately 26% of bacterial strains identified in the human gut contain bile salt hydrolases that can deconjugate PBA, only a few (from the Lachnospiraceae and Ruminococcaceae families) are currently known to perform subsequent dehydroxylation to generate DCA and LCA.Citation105,Citation107
Therapeutics targeting receptors involved in bile acid metabolism can affect microbiome composition.Citation108,Citation109 In parallel, the expression of ileal bile acid transporters, particularly the main transporter, apical sodium-dependent bile acid transporter (ASBT), have been shown to be decreased in animal models of intestinal inflammation, as well as in IBD patients.Citation23,Citation110–112 Therefore, the restoration of bile acid metabolism is gaining attention for its therapeutic potential. Clinical studies have demonstrated that patients with bile acid malabsorption (BAM) associated diarrhea respond well to some bile acid sequestering agents, including cholestyramine, colestipol, or colesevelam.Citation110,Citation113,Citation114 Colesevelam therapy has also been shown to be effective at reducing the number of liquid stools/day, improving stool consistency in CD patients, and treating post-operative BAM in CD compared to other therapeutics.Citation114,Citation115 Because bile acid sequestrants are not absorbed in the intestine, systemic effects are generally low. However, they can bind to other vitamins and drugs and interfere with their absorption, which could result in vitamin deficiencies and inefficacy of other treatments.Citation115
Promising results have also been observed with direct administration of SBA in experimental models of colitis. However, an important limitation of the oral administration of SBA is their rapid uptake into the enterohepatic circulation in the small intestine and delivery to the liver, which can result in low concentrations reaching the colon.Citation108,Citation116 Therefore, alternative delivery methods may be required to achieve therapeutic concentration in the colon. Recently, rectal administration of DCA and LCA mitigated inflammation in three different murine models of acute and chronic colitis, and reduced the expression of key pro-inflammatory cytokines and chemokines.Citation24 DCA and LCA were shown to exert their effects via activation of the widely expressed G protein-coupled receptor TGR5. Together with TGR5, the farnesoid X receptor (FXR) were both activated by bile salts and involved in immune modulation and intestinal barrier functions. Several FXR-agonists are being explored as novel therapeutics to treat intestinal inflammatory conditions. Among them, ursodeoxycholic acid (UDCA) is an SBA with cytoprotective and immunomodulatory properties that is currently used to treat certain types of cholestasis and primary biliary cirrhosis.Citation116 UDCA therapies have also shown anti-inflammatory effects in animal models of colitis,Citation117 which supports the idea that UDCA may also be useful in preventing or treating IBD in humans. This hypothesis is currently being tested in a phase-II clinical trial that investigates the ability of UDCA to reduce inflammation and improve quality of life in UC pouch patients (NCT03724175). UDCA can also modify the microbiome: a clinical trial of long-term (3 years) treatment with UDCA to prevent colorectal adenomatous polyps resulted in an overrepresentation of F. prausnitzii and underrepresentation of R. gnavus, two species that show abnormally low and high levels in IBD, respectively.Citation109 Given the clear association between microbial imbalances and bile acid metabolism, approaches that seek to restore key bacterial species (e.g. SBA-producers) and bile acid metabolism hold great clinical promise in the treatment of IBD.
Bacterial cell components
Bacterial cell components may also be used as additional targets to treat intestinal inflammatory conditions. Capsular polysaccharide A (PSA), found on the cell surface of the gut commensal Bacteroides fragilis, has been extensively studied for its immune modulatory properties and its key role in reducing inflammation.Citation118 Colonization of mice with PSA-producer strains of B. fragilis or with purified PSA can protect animals against experimentally induced colitis and cure the disease. The mechanism of action of PSA involves the induction of IL-10-secreting Tregs mediated by TLR2 signaling.Citation119,Citation120 In a recent study, the zwitterionic polysaccharide TP2, a capsular polysaccharide of B. fragilis, was extracted and administered to rats with induced enteritis.Citation121 Treatment with TP2 significantly alleviated inflammation and reduced adhesion score and ulcer incidence. In addition, TP2 was not degraded during passage through the intestinal tract in models in vivo and in vitro simulating gastric, intestinal, and colonic conditions. TP2 integrity was maintained during absorption, distribution, metabolism, and excretion. However, safety and pharmacokinetic studies of PSA and other capsular bacterial polysaccharides for its use in humans are not yet available.
Although the characterization of microbial-derived metabolites and molecules in IBD is a promising research approach, their application encompasses some drawbacks. Metabolite biomarkers of IBD are commonly identified from fecal samples, but as they undergo different processes of absorption and metabolic changes before being excreted, they may not be the same molecules involved in relevant host interactions. Further, the use of single therapeutic metabolites has important pharmacological limitations related to dose, administration, and metabolic stability.Citation90 The use of crude metabolites can often lead to high variability in the amounts that are able to reach the target site. If administered systematically, there is a risk that the metabolite does not reach its target, as the serum half-life of certain molecules can be very short. Oral administration is an attractive alternative, although as soluble metabolites pass through the gastro-intestinal tract they can be partially broken down by digestive enzymes, which makes it difficult to estimate how much product reaches the gut and complicates reproducibility. Additional studies are thus required that accurately estimate the efficacy of microbial-derived molecules and their optimal method of administration for the treatment of IBD.
Analyzing microbiome data in IBD
Identifying IBD-associated bacterial strains
It has been hypothesized that, similar to Koch’s postulates, the identification of strain-level variation in IBD patients has the potential to reveal novel insights into its pathogenicity and pave the way to new treatments.Citation122,Citation123 This has proven true in the case of E. coli,Citation124–128 where certain strains have indeed been associated with IBD. The advent of high-throughput sequencing has provided a novel approach to identify IBD-associated strains through the analysis of shotgun metagenomic data.
More recent work has implicated additional strains in IBD pathogenesis. In a study of IBD twins,Citation46 eight species were found to have higher relative abundance in IBD-twins relative to healthy controls, as well as between healthy co-twins and healthy controls, including Gordonibacter pamelaeae, Escherichia unclassified, and Eggerthella unclassified.Citation46 Multiple studies have shown that Ruminococcus gnavus appears more frequently in IBD patients,Citation15,Citation47 particularly in CD patients. In a study of IBD patients receiving anti-integrin treatment, Roseburia inulinivorans and Burkholderiales species were found in significantly higher abundance in CD patients achieving week 14 remission compared to patients who did not enter remission,Citation49 suggesting that bacterial species and strains could also play an important role in disease progression.
An important caveat of these studies is that multiple software pipelines exist for identifying bacterial taxa at high resolution from metagenomic dataCitation129 and the interpretation of these results depends significantly on the specific tool used.Citation42,Citation130,Citation131 While some tools construct k-mers and assemble them using de Bruijn graphs,Citation132 others assign reads to specific genomes using a set of marker genes.Citation39–41 While computationally less expensive, this marker-based approach depends strongly on the reference database used. Methods such as MetaPhlAn have been often utilized in IBD studies,Citation15,Citation46–49 and although recently expanded with the inclusion of additional genomes,Citation39 it remains to be determined to what extent these results are limited by existing genome collections. Recently developed methods based on strain collectionsCitation42 have increased potential to identify strains with high confidence and resolution, although they also suffer from limitations related to the size of the collection being used.
Overall, the use and development of software tools to identify bacterial strains from shotgun metagenomic data shows great promise to characterize potentially pathogenic microbes in IBD in a high-throughput manner and without the limitations of approaches that require microbial isolation. However, the variety of analysis pipelines and the lack of a gold standard to compare them suggests the variability of strains associated with IBD could not only be due to cohort-specific factors, but also partially driven by the analysis itself. Thus, methods that compare and validate multiple-software tools are needed.
The temporal variability of the IBD microbiome
IBD is a chronic disease that undergoes different stages during its natural history, with periods of quiescence punctuated by acute inflammatory episodes (“flares”). Patients often require partial or total colectomy, which can happen several years after the disease is initially diagnosed. The microbiome of IBD patients differs with that of healthy subjects,Citation133,Citation134 varies with IBD type,Citation18 and treatment.Citation12,Citation135–139 Moreover, the history of the patient’s treatment course matters in their response to future treatments.Citation135 This rising appreciation of the temporal variability of the disease has led to a call for longitudinal studies.Citation138,Citation140,Citation141
Some studies have partially characterized temporal changes in cohorts of IBD patients,Citation142,Citation143 in mouse models,Citation60,Citation144–147 and during flares.Citation148 However, many of these have been exploratory studies, with either small patient cohortsCitation139,Citation143,Citation149 or large cohorts but with limited sampling.Citation150 Temporal sampling differs significantly between studies ranging from weekly,Citation151 or monthly,Citation18 to annual or longer intervals between samples,Citation133,Citation136,Citation150 which further complicates analysis and interpretation. Some results suggest that daily sampling is not necessary to fully characterize the temporal variability of microbial communities, and that samples taken every few weeks may be enough to capture disease-relevant information.Citation152 It is, however, unclear how frequent sampling the needs to be to capture features of clinical importance, such as the prediction of flares, the need to escalate therapy, or the future likelihood of requiring surgery.
Importantly, most longitudinal studies of microbiome variation in IBD bin temporal changes into categories so that comparisons can be made at the group level. The intent here is to determine which taxa are higher in healthy subjects compared to IBD patients. This approach has facilitated the characterization of temporal variation in IBD.Citation19,Citation61,Citation62,Citation153,Citation154 It has identified signals associated with response to anti-TNF therapy,Citation19 as well as the likelihood of successful clinical and histological remission of UC after FMT.Citation62 However, this approach also limits our ability to uncover subject-specific characteristics of the disease or the likelihood a specific patient will respond to treatment, one of the premises of personalized medicine. Therefore, new methodologies that allow the analysis of longitudinal microbiome data at the subject level are an important unmet need that will be required for better patient stratification and treatment efficacy.
Fungi and viruses: overlooked contributors in IBD studies
Most studies exploring the gut microbiome have focused their attention in the bacterial component, largely overlooking the fungal (mycobiome) and viral (virome) elements. Viruses and fungi also play important roles in the intestinal ecosystem, and they are now receiving increased attention in the context of human health and disease.Citation63–67
Albeit essential to human health, the mycobiome comprises a lower proportion of the overall microbiome compared to bacteria and viruses,Citation66 which partially explains why most IBD mycobiome sequencing studies to date are based on amplicon sequencing.Citation53–56 Two ribosomal regions have been commonly used in mycobiome analyses: the 18S rRNA gene and the internal transcribed spacer (ITS) regions, internal to the 18S, 5.8S, and 28S rRNA sequences.Citation66,Citation155 The ITS regions have been identified as the most effective to characterize fungi in complex communities, and are currently used as the primary approach for fungal analyses.Citation66,Citation156 ITS regions are present in high copy number and are conserved across fungi, but contain two highly variable fragments, ITS1 and ITS2, that, combined with the low variable 5.8S fragment, improves resolution at low taxonomic levels. The use of ITS regions, however, does not come without complications, including amplificationCitation67 and sequencing biases.Citation157,Citation158 While shotgun sequencing offers multiple advantages over amplicon-based approaches (improved taxonomic resolution, no amplification biases, functional information), the number of fungal metagenomic studies in IBD are few,Citation66,Citation137 partially due to the inherent difficulty of working with the low amounts of fungal (compared to bacterial or viral) DNA present in stool.
The number of research studies interrogating the IBD virome are also scarce, but there is growing evidence that suggests that IBD viral communities also exhibit differences in composition, with increased viral diversity and abundance.Citation64,Citation159–165 The viral component of the microbiota is mainly composed of prokaryotic-infecting viruses (bacteriophages or phages),Citation64,Citation65,Citation166 which have the ability to shape bacterial communities and can have important repercussions in IBD.Citation65,Citation159,Citation167,Citation168
The vast diversity of viruses and their variability in morphology, genetic material (DNA or RNA), configuration (single- or double-stranded), and genes sense of orientation represent important limitations for the study of viral communities.Citation169,Citation170 Phages can also vary depending on their interactions with the bacterial host, being able to infect as lytic or temperate phages.Citation170 As viruses do not have a single, universal marker gene, studying the virome relies heavily in non-targeted, shotgun sequencing approaches.Citation170,Citation171 Although the amounts of viral particles in the gut is estimated to be at least equal to the number of bacterial cells, the size of viral genomes is smaller and viral nucleic acids only account for a small proportion of the gut microbiome.Citation166,Citation170,Citation172 Thus, untargeted shotgun metagenomic sequencing methods often fail to yield sufficient viral reads. To overcome this problem, viral particles are usually concentrated and purified before performing sequencing.Citation160,Citation164,Citation173 In some cases, however, absolute yields of viral nucleic acid preparations are low and efforts to specifically favor viral yields still fail to deliver sufficient information. For example, Lloyd-Price and colleagues performed multi-omic analyses on various samples from IBD patients, obtaining an in-depth profile of the bacterial communities, and at the same time, purified viral RNA from stool, which was amplified and subsequently sequenced. Despite the efforts to investigate the virome, only a small number of viruses could be identified, and only a single phage could be identified as being differentially enriched in IBD and non-IBD dysbiosis.Citation89 Alternatively, methods such as Multiple Displacement Amplification (MDA) can increase viral and fungal genomic concentrations by performing whole-genome amplification without sequence-specific primers prior to shotgun sequencing.Citation65,Citation174,Citation175 However, MDA is a nonspecific reaction that can amplify any DNA present in the sample, including contaminant DNA from reagents or the environment.
An additional challenge in fungal and viral analysis is that residual genomic material from human and bacterial cells are also extracted and need to be filtered out prior to analysis. Removal of host-contaminant sequences can be achieved by mapping the reads against reference genomes with computational tools like Bowtie,Citation176 DeconSeq,Citation177 BBMap (https://sourceforge.net/projects/bbmap/), and BMTagger (ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/bmtagger/). Tools such as ViromeQC can be used to quantify bacterial, fungal, and archaeal contaminations in virome sequencing data.Citation178 Specific tools for viral identification in metagenomic data are also available, including PhiSpy,Citation57 VirSorter,Citation58 and PHASTER.Citation59 Similarly, tools to identify fungal sequences in metagenomic datasets include FindFungi,Citation50 HumanMycobiomeScan,Citation51 and CCMetagen,Citation52 which can identify reads originating from eukaryotic and prokaryotic genomes.
Finally, comprehensive pipelines for decoding, quality filtering, sequence clustering, and taxonomic assignment of high-throughput sequencing data are also skewed towards bacterial analyses. QIIME, RDP, or Mothur can be adapted to incorporate fungal analysis. Other software pipelines exclusive for fungal analysis include BROCC (http://sourceforge.net/projects/brocc/), CloVR-ITSCitation179 or PIPITS.Citation180 On top of ad hoc software tools and algorithms for viral contig annotation and viral databases for sequencing data processing, recent stand-alone pipelines specific to viral metagenomic analysis have been developed, such as ViroMatch,Citation181 Sunbeam,Citation182 and VirusSeeker.Citation183
Microbial networks and compositional data analysis
The microbiome is a complex ecosystem, composed of many interacting entities that form a dynamic community.Citation184 Networks built to represent such high-dimensional systems are powerful tools because they can capture putative relationships between members of the community that might be predictive of outcomes not associated with individual components of the network. The construction and analysis of microbiome networks, however, requires approaches specifically designed for them, given the sparse and compositional nature of the data.Citation185 Microbiome data is sparse since most samples only contain a small subset of all possible taxa. It is also compositional, as data is often expressed as relative – as opposed to absolute – abundances.Citation186 Because compositional data must add up to unity, an increase in the relative abundance of one taxon induces a decrease in relative abundance of other taxa, even when the absolute number of those taxa have not changed.
Compositionality is partially a consequence of the rarefaction process performed to normalize data across samples and groups.Citation187 While there is no consensus on the extent to which rarefaction techniques might reduce statistical power,Citation188,Citation189 compositional data introduces important biases in the analysis of correlations or when identifying differentially abundant features, and can result in incorrect conclusions if not properly accounted for.Citation190,Citation191 Several recent methods have been developed to analyze microbiome data in a compositional context. The use of a proportionality statistic based on log-ratios has been shown to estimate when variables are proportional in an accurate manner,Citation190 and reference frames have also been proposed for differential abundance analysis.Citation192 The sparse nature of microbiome data can also lead to a large number of false correlations driven by influential observations. We have recently proposed an approach based on jackknifing that significantly reduces the number of such false positives.Citation193
Among the most commonly used methods for network analysis in microbiome data are SparCC,Citation68 SPIEC-EASICitation69 and MENAP.Citation70 SparCC uses log-transformed data to infer co-occurrence relationships between taxa. SPIEC-EASI employs a sparse neighborhood and inverse covariance selection framework to determine the underlying ecological network. MENAP uses a Random Matrix Theory-based approach to determine the adjacency matrix used to construct the network. Other tools such as ReBoot/CoNet,Citation194 REBACCA,Citation195 CCLasso,Citation196 Mint,Citation197 gCoda,Citation198 have emerged more recently, and additional work has been proposed to construct microbe–metabolite interaction networks.Citation199 When longitudinal data is available, ecological network characterization using the Lotka-Volterra framework has provided novel insights into Clostridium difficile colonization,Citation200 microbial and immune cell interactions,Citation201 and inter-kingdom interactions in the infant microbiome.Citation202 This approach has been implemented in MDSINE, a suite of algorithms for microbiome time-series data analysis and prediction.Citation71 Overall, the application of network analysis methods to IBD data is still in early stages. Recent work combining SparCC and SPIEC-EASI on IBD patients and healthy controls showed that a signature of IBD can be reflected in microbial co-abundance analysis, particularly implicating E. coli and O. formigenes as IBD-associated taxa.Citation72
While the use of these methods can partially reduce some of the complexities associated with compositionality, they are not without issues. Log-ratio approaches cannot work with zeroes, which must be replaced by pseudo-counts, introducing additional biases.Citation203 Further, the lack of information on microbial load when using relative abundances is critical in most microbiology studiesCitation204 and limits the application of tools like ecological network modeling. The use of methods to quantify absolute counts is an alternative to relative abundance-based methods that should be considered, as they have been shown to be of high relevance in microbiome studies of IBD.Citation205
Therapeutic engineering of the microbiome
Microbial transplants: a promising therapeutic approach
Among the different approaches to treat IBD, microbiome-based therapeutics have gained tremendous attention as a novel approach to efficiently induce remission (). Although pre- and probiotics have been utilized in the past with mixed results,Citation4 fecal microbiota transplants (FMTs) are arguably a more commonly tested method in current research and clinical studies. The extremely high efficacy of FMTs in recurrent Clostridioides difficile infection (CDI) refractory to antibioticsCitation206–209 has paved the way for their use in the treatment of IBD. Moayyedi and colleagues demonstrated how FMTs can induce remission at 7 weeks post-FMT in ulcerative colitis (UC).Citation210 Intriguingly, 7 of the 9 patients who responded to treatment had received fecal material from a single donor, suggesting that the microbiome of this donor was different from that of other donors and particularly efficient at inducing remission. A later study of an intense FMT regime demonstrated remission in 27% of UC patients, compared to only 8% with placebo.Citation211 No donor-specific effects were noted, and several bacterial and metabolite biomarkers could be identified to be associated with remission.Citation62
Figure 1. Opportunities for microbiome therapeutics in IBD. a. Fecal Microbiota Transplant (FMT) transplants whole microbial communities from healthy donors into IBD patients and has shown promising results particularly in the treatment of UC. b. Bacterial engineering can be used to enhance the beneficial properties of bacterial strains, including targeted delivery of therapeutic molecules, which offers important efficacy and safety advantages compared to the use of FMTs or molecules delivered systemically. c. Phage therapy or “decolonization”, currently being explored for CD and UC, selectively targets and removes specific bacteria associated with disease, without disrupting other members of the gut microbiota. d. Biologic therapies, which can induce clinical and histological healing, are the current standard of care in IBD. Lack of response, development of resistance and side effects remain challenges for some patients. e. The aberrant microbiome of IBD patients has a reduced capacity to produce metabolites that modulate intestinal homeostasis. Direct administration or modulation of the microbiome to enhance the production of such metabolites is being explored as potential therapeutics for IBD. f. Microbial consortia designed to induce specific immune responses are also being investigated as therapeutics in animal models and pioneered in human studies. Image created with BioRender.com.
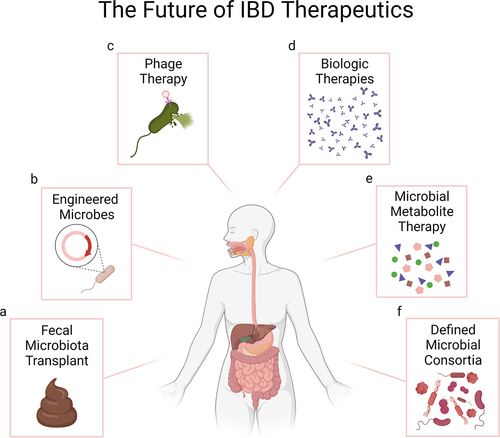
While results of FMTs in IBD are highly encouraging, there are issues that moderate the general applicability of this approach. Safety is commonly discussed as an important limitation of FMTs, given the risk of transferring pathogenic agents to the recipient.Citation212 The global COVID-19 pandemic has further stressed the need to rigorously test fecal material prior to transplantation. The use of a procedure that is still experimental requires a careful assessment of risks and benefits for different patient cohorts. A more rigorous exclusion criteria could potentially prevent or limit the severity of adverse events in future studies. Indeed, a nation-wide study of FMT in CDI has recently shown the procedure to have a good safety profile.Citation213 However, the invasive nature of the procedure and its associated economic cost are disadvantages compared to other therapies. Finally, the heterogeneity of the product used in FMTs presents two important challenges: because the fecal material varies donor to donor, response to intervention (or lack of it) depends both on patient and donor, which complicates interpretation of results. It is also unclear how fecal material can be regulated, if at all, in terms of intellectual property and commercialization, which partially hinders the involvement of the private sector in developing FMT therapeutics.
To address some of these problems, fecal material in capsule form has been pioneered in the study of CDI.Citation214 As they are taken orally, fecal pills are less invasive and more cost-efficient, at the expense of a diminished (although still high) efficacy, at least in CDI.Citation215 Their efficacy in IBD, however, remains to be tested. An alternative that is gaining traction in microbial therapeutics is the use of bacterial communities, which contain defined sets of isolates with desirable properties to treat a given condition. This approach circumvents several of the main issues of solutions based on fecal material, and they provide a higher level of safety and homogeneity, although at the potential expense of having lower efficacy. A recent study of an oral formulation of defined Firmicutes spores found that, in conjunction with vancomycin, clinical response was higher than in no vancomycin plus spores.Citation216 It should be noted that this study did not consider histological remission, a stricter endpoint that has been reached with full FMTs in UC,Citation210,Citation217 and that clinical remission was most notable in those patients who also received vancomycin, suggesting that efficacy of the defined community alone might be lower than that observed with FMTs.
The curation of defined microbial consortia built to prevent or treat disease has several advantages over FMTs, including replicability, safety, and scalability. Being able to select subsets of bacteria that exhibit desirable properties would be a fundamental step forward in the development of efficient microbial therapeutics.Citation218 As with FMTs, the rational design of microbial consortia also has its roots in CDI studies.Citation219 Recent work has demonstrated that consortia with less than 20 strains can prevent or treat colitis in murine models after two weeks of treatment.Citation220 The mice further showed reduced tissue inflammation (histology, inflammatory cytokines) and increased levels of metabolites associated with a healthy mucosa, demonstrating the feasibility of this approach in a model organism. While these findings need to be replicated and likely will require refinements to be effective in humans, results suggest that remission, both clinical and histological, may be possible with defined microbial communities.
Next-generation probiotics: genetic engineering of bacterial strains
Due to the difficulties and limited success of using single metabolites and pre- and probiotics as therapeutics for IBD, synthetic biology approaches have begun to be applied to produce engineered strains that can deliver selected therapeutic molecules, thus enhancing their probiotic capacity.Citation221,Citation222 One of the advantages of using orally administered engineered bacteria to deliver therapeutics is their ability to survive the passage through the intestinal tract while expressing the molecules of interest. This can circumvent the need for large amounts of crude metabolites that are mostly broken down by digestive enzymes. In addition, oral administration of bacteria ensures that therapeutic molecules are synthesized and delivered at the mucosal surfaces, which increases their efficacy compared to classic systemic therapies. At the same time, intestinal delivery of bacterial-derived molecules improves safety and prevents secondary effects related to systemic exposure to drugs, such as generalized immune-suppression.
Classic tools to genetically engineer bacteria use recombinant plasmids as cloning vectors to deliver genes of interest. Cloning vectors allow the production of large amounts of protein, but embody certain limitations including low efficiency of transformation and limitations of insert size. Recent discoveries and development of new tools, such as the CRISPR-Cas systems, have revolutionized the genetic engineering scene, offering an enormous potential to engineer genomes with greater efficacy than previously achieved.Citation223 CRISPR (clustered regularly interspaced short palindromic repeats) and its associated Cas proteins are tools derived from the prokaryotic immune system that have been co-opted as genetic editing tools.Citation223,Citation224 This methodology has been applied to bacteria and yeasts to modify their functional repertoire, exploited for industrial applications or used for the direct removal of specific genes or pathogens.Citation222–225 Among the different CRISPR systems, CRISPR-Cas9 and CRISPR-Cas12a (also known as Cpf1) are two major nucleases that have been used in bacterial genetic editing experiments. By inserting a guide RNA sequence targeting a specific region of the bacterial DNA, Cas nucleases introduce a break in the pathogen’s genome which allows the removal of specific genes or causes bacterial death.Citation223,Citation226,Citation227 If a template DNA is provided, the genomic break introduced by the Cas nucleases can be repaired by homologous recombination, inserting the new DNA fragment into the bacterial genome.
Some of the current standardized treatments for IBD consist of systemic administration of anti-inflammatory drugs and antagonists of pro-inflammatory molecules. However, some patients do not respond to such treatments, or stop responding to treatment after some time.Citation228,Citation229 As an alternative, recombinant bacterial strains have been successfully engineered to synthesize active biotherapeutics including cytokines and immunoglobulins that can be delivered at mucosal sites. Lactococcus lactis, a nonpathogenic, lactic-acid bacterium commonly used in the production of fermented foods, is the gold standard for probiotic gene-editing strategies, due to its well-studied genome and its safety properties.Citation221 One of the first recombinant L. lactis, from the pioneer work by Steidler and colleagues, was designed to treat IBD by secreting IL-10 in-situ in the colon in two murine models of disease.Citation230 In one of the models, chronic colitis was induced by administration of DSS and the efficacy of L. lactis-secreting IL-10 (LL-mIL10) to treat symptoms was tested. In the second model, the ability of LL-mIL10 to prevent disease was tested in IL-10-/- mice, a model that spontaneously develops colitis. Results showed that LL-mIL10 was able to both reduce pathological symptoms and inflammation and prevent onset of colitis. In addition, the protective effect of bacterial-synthesized IL-10 performed better than parenteral administration of recombinant IL-10 and other systemic drugs as lower amounts were required to reduce inflammation. Based on these results, a small clinical trial with 10 CD patients was conducted with the purpose of testing the safety of L. lactis secreting human IL-10.Citation231 Because of the safety concerns of using genetically engineered organisms in humans, the human IL-10 encoding cassette was used to replace a thymidylate synthase gene, essential for the growth of L. lactis, with the purpose of blocking its growth and preventing colonization in the host.Citation230,Citation231 An improvement in disease severity in 8 out of 10 participants encouraged a larger phase-II trial (www.clinicaltrials.gov: NCT00729872). However, no statistically significant results were observed compared to placebo treatment, which may have been related to insufficient bacterial viability or localized delivery at the site of inflammation. In another study, a L. lactis strain was engineered to secrete a low calcium response V (LcrV) protein, naturally present in enteropathogenic Yersinia pseudotuberculosis that evades the host immune system by stimulating IL-10 production and preventing inflammatory cascades. Results demonstrated the ability of LL-LrcV to treat and prevent colitis in two murine models of acute disease.Citation232 Moreover, L. lactis strains have been engineered to secrete neutralizing anti-TNF-α nanobodies (small and highly stable single-domain antibodies) that can block TNF-α pro-inflammatory effects and ameliorate DSS-induced colitis in mice.Citation233 Although L. lactis has been most frequently used in IBD studies, genome editing experiments aimed at engineering other intestinal bacteria are still scarce. For example, Bacteroides is among the most abundant genera in the human gut, and reduced relative abundances of this genera have been observed in IBD patients.Citation234 However, engineering Bacteroides is challenging due to their high intraspecies genetic diversity and natural resistance to antibiotics that are used for genetic selection in the lab, which can result in low efficiency in genome editing experiments.Citation235–237 In recent work by Zheng and colleagues, different CRISPR-Cas methods were used to edit the genome of several Bacteroides species.Citation227 Results showed that FnCas12a (a nuclease from the Cas12a family) had the highest genome-editing efficiency across Bacteroides species. The efficacy of FnCas12a-engineered Bacteroides strains as therapeutics for IBD remains to be explored.
While most IBD therapies are aimed at modulating inflammatory pathways, approaches that restore intestinal barrier function and promote mucosal healing have been less explored. During acute flares the intestinal epithelial barrier is disrupted, exposing the gut lining to bacteria and exogenous antigens which can exacerbate inflammation and lead to systemic infections. In one study, the bacterium L. lactis was engineered to produce trefoil factors (TFF), which are known to promote intestinal barrier function and epithelial restitution.Citation238 Orally administered TFF-producing L. lactis (LL-mTFF) was able to secrete TFF in the colon and prevent and heal colitis in a DSS-induced acute colitis murine model. Rectal administration of purified TFF was also able to reduce colitis symptoms, but much higher concentrations were needed and could not achieve the same results obtained with LL-mTFF. More recently, E. coli Nissle1917 (EcN), a well-studied probiotic strain, was engineered to secrete therapeutic matrices to promote gut epithelial integrity.Citation239 Matrices, composed of nanofibers displaying TFFs, were able to protect against DSS-induced colitis in mice, by promoting mucosal healing and immunomodulation.
A limitation of using engineered bacteria as therapeutics is that they do not always necessarily reach the site of inflammation. To overcome this issue, targeting mechanisms for localized delivery of biotherapeutics at disease sites are being explored. Recently, a research group designed an invasive L. lactis strain containing a DNA vector, pValac:il-10, that was used to deliver the DNA into intestinal eukaryotic cells and treat mice with induced colitis. Once in the host cells, the eukaryotic cellular machinery expressed the ORF of interest, and translated and synthesized IL-10 in the intestine of the animals, which experienced a reduction of symptom severity and a delay in disease onset and development.Citation240–242 Similarly, an EcN strain was engineered to invade the colon epithelium (and degrade later in the phagosome) and deliver a plasmid to express small hairpin RNA (shRNA) targeting tumor necrosis factor (TNF).Citation243 This method reduced levels of TNF as well as inflammatory mediators. Finally, McKay and colleagues elegantly engineered commensal EcN expressing a therapeutic biologic directed toward nitric oxide (NO), a biomarker of IBD that is greatly increased in the intestine of patients compared to healthy controls,Citation244 exploiting a phenomenon known as pseudotaxis. In this phenomenon, cell motility is dictated by the concentration of a chemical. Mimicking this natural process, authors engineered EcN to sense and accumulate at sites with high content of NO. At the same time, EcN were engineered to selectively synthesize a biologic, granulocyte macrophage‐colony stimulating factor (GM‐CSF), co-expressed with a signal molecule that would trigger the synthesis of the biologic in response to the presence of NO.Citation245 This study opens new research and therapeutic possibilities by using “smart probiotic” bacteria specifically directed toward a known biomarker, which can elicit a genetic response from the engineered bacteria, triggering the synthesis of the biotherapeutic of interest at the precise site of disease or inflammation.
Despite the development of such innovative solutions and some promising results, most of the documented benefits of engineered bacteria producing therapeutic molecules are from studies based on animal models. There are some safety concerns about the use of modified bacteria in therapeutic settings, mainly due to potential exchange of genetic material with other bacteria, which may result in unexpected consequences. Therefore, the use of safe and genetically stable strains, and designs that ensure containment of strains in the intestinal environment or self-removal mechanisms should be a prioritized area of research. Ferenczi and collaborators removed more than 20% of the genome of the EcN strain used in their study, including transposons, and eliminated the recombination and conjugation abilities of the bacteria, minimizing the risk of horizontal gene transfer.Citation243 Moreover, the ability of engineered bacteria to “escape” the intestinal ecosystem and invade other sites could be reduced by strategies that introduce mechanisms that block their growth or lead to bacterial self-destruction within a few days of synthesizing the therapeutic of interest.Citation230,Citation231,Citation243 Finally, an additional limitation that remains elusive, even to engineered bacteria is the ability to control the dose effect, a problem that researchers have not yet been able to solve and which will require further research efforts.
Besides engineering individual bacteria, efforts are being made to selectively edit the intestinal microbiome in situ as treatments for intestinal inflammation. For example, Hughes and colleagues found that molybdenum-cofactor-dependent metabolic pathways (such as nitrate reduction and formate oxidation) are a signature of inflammation-associated dysbiosis and contribute to the expansion of microorganisms, such as members of the Enterobacteriaceae family, which are dependent on such pathways.Citation246 In subsequent work by the same group, authors were able to prevent expansion of Enterobacteriaceae during gut inflammation by tungsten treatment, which selectively inhibits molybdenum-cofactor-dependent microbial respiratory pathways.Citation247 As such pathways are operational only during episodes of inflammation, tungsten treatment acts only on the enterobacterial population in the disease state, and does not affect Enterobacteriaceae during homeostatic conditions. This tungsten-mediated precision editing of the microbiota can therefore ameliorate the severity of intestinal inflammation in mouse models of colitis, and represents a promising and innovative avenue of research.
Phage therapy: phages as a treatment vector
The use of phages as curative agents to treat bacterial infections has been applied for almost a century.Citation248–251 However, due to variation in effectiveness and the discovery of antibiotics, the study of phages as therapeutics suffered a slow-down.Citation252 More recently, with the rise of antibiotic resistance in bacteria, the need for alternative treatment solutions has re-focused the attention on phages. Phage therapy consists of administering lytic phages with the purpose of selectively removing specific bacteria that are thought to cause a disease, also known as “decolonization”.Citation253–256 One of the main benefits of phage therapy is the ability to precisely target highly conserved species-specific membrane proteins and kill bacterial pathogens without disrupting the other members of the gut microbiota or attacking human cells.Citation254 In addition, phage enzymes are able to disrupt bacterial biofilms, penetrating areas and infecting bacteria that are frequently difficult to reach by antibiotics.Citation257,Citation258 Some phage therapies have already been used in humans, including the use of phage preparations to treat Pseudomonas aeruginosa infections in burn wounds and chronic otitis,Citation251,Citation254,Citation259 to treat gastrointestinal distress in donors with no diagnosed gastrointestinal condition,Citation260 and as a last resource treatment in some cases of multidrug-resistant infections.Citation261–263 Phage-based therapies are also being explored as treatment for UC and CD, and studies in mouse models of colitis have yielded encouraging results. In one study, mice infected with AIEC and administered with a cocktail of three AIEC-specific bacteriophages isolated from wastewater benefited from a significant decrease in fecal AIEC and reduced induced-colitis symptoms. In addition, the same phage cocktail was able to target AIEC in homogenates of ileal biopsies taken from CD patients.Citation264 Similarly, another study identified a phage from the Myoviridae family, also isolated from wastewater that could reduce the abundance of diarrheagenic E. coli in a mouse model of intestinal colonization, while microbiome alpha and beta diversity remained unaltered.Citation265 Efficacy of this phage’s E. coli-killing capabilities were also confirmed in vitro. Moreover, in a different study, three AIEC-specific Caudovirales phages isolated from an IBD patient reduced colonization of E. coli in germ-free mice.Citation164 Using the same phage cocktail in a mouse model of colitis, however, authors observed that expansion in phage numbers exacerbated inflammation within the intestine and contributed to colitis symptoms, suggesting that, as seen in IBD, increased phage abundances may contribute to disease severity in chronic inflammation settings.
Despite some promising results obtained from animal models, the evidence of phage therapy efficacy on human IBD patients is still in its infancy. The safety and efficacy of EcoActive, a collection of bacteriophages against AEIC, is currently under investigation in a phase 2 double-blind, placebo-controlled trial with CD patients (www.clinicaltrials.gov: NCT03808103). Another phage cocktail therapy, BX002, is being evaluated as a treatment to target Klebsiella pneumonia in IBD/Primary Sclerosing Cholangitis (PSC) patients. The two diseases are thought to be related since a great proportion of PSC patients also suffer from IBD and previous studies have shown that K. pneumonia strains isolated from IBD/PSC patients are associated with the onset or exacerbation of both diseases in murine models.Citation266,Citation267 In a phase 1a single-blind, placebo-controlled trial, the safety, tolerability, and feasibility of delivering the phage cocktail BX002 to the recipients’ gut was demonstrated in healthy participants (NCT04737876). Based on these results, the efficacy of BX002 at reducing the levels of the target bacteria will be tested in a 1b/2a clinical trial. On the other hand, FMT could also represent an indirect tool of phage therapy, as previous work has shown transference of phages from donors to their recipients.Citation268,Citation269 In the study by Chehoud and colleagues, authors studied the viral fraction in stool samples after FMT from an adult donor to three pediatric patients with UC, and confirmed the transference of phages to the recipients.Citation269 Similar results have been observed after FMT in the context of Clostridium difficile infections (CDI). Along with bacteria, viral populations were transferred from FMT donors to their recipients.Citation268 Importantly, it has been suggested that the “phageome” alone may be sufficient to treat CDI. In a recent trial, the transference of fecal filtrates (devoid of whole bacteria but containing viral populations and bacterial products) from healthy donors to CDI patients with chronic relapse was able to restore normal stool habits and eliminated symptoms in recipients for at least six months.Citation270 These studies support the theory that the effect of FMT could be mediated not exclusively by bacteria, and opens new phage applications to be explored to treat IBD and other intestinal diseases with microbial-dysbiosis components.
This approach, however, has some limitations. Extensive steps of purification of phage preparations are essential to eliminate bacterial residues, such as membrane components and endotoxins that may be present from propagating the phage in its host.Citation271 The high specificity of phages is simultaneously an advantage and a limitation: while phages do not disturb microbial communities, and therefore are exempt of antibiotic-related side effects,Citation272 target pathogens need to be identified beforehand, which narrows the spectrum of action and demands etiological understanding of the disease a priori.Citation254 To avoid introducing harmful genes (i.e. antibiotic resistance or virulence factors) into the bacterial communities, only strictly lytic and fully sequenced phages should be utilized. However, lytic phage genomes contain many hypothetical genes with no known function. Additionally, the function and pathways of encoding proteins that alter bacterial physiology are not fully understood. As with antibiotic treatment, phage effectiveness can be hampered by bacterial resistance.Citation256 Lytic phages exert strong antimicrobial selective pressures on their hosts, which possess a battery of anti-phage defense systems, including changes in receptor proteins that phages need to enter the cells, or CRISPR-Cas systems that recognize and degrade previously encountered foreign viral DNA.Citation273 Simultaneously, phages continually evolve and adapt to maintain their infective capabilities, in an evolutionary race with their hosts.Citation256,Citation274 In the study by Goghokia and colleagues, E. coli strains developed resistance to phage therapy, but continuous administration of phage treatment was able to suppress bacterial growth.Citation164 However, this process may not be sufficient to counter the emergence of phage-resistance when “mono-phage” therapies are being applied. Instead, the use of phage cocktails is a commonly used strategy that increases the spectrum of action.Citation254 Nevertheless, use of such cocktails requires longer preparation and purification times, and have reduced pharmacokinetic predictability. Combination therapy of phages and lower concentration of antibiotics (enough to prevent cell division but not to cause cell death), known as “phage-antibiotic synergy” treatment has been suggested to be more effective against bacterial-associated diseases and at preventing the emergence of resistance.Citation254,Citation275 In addition, combining phage therapy with FMT may enhance microbiota engraftment and improve treatment efficacy. Overall, scientific evidence on phage therapy efficacy is still scarce, but there is a growing interest in the field and a strong need for novel therapeutics. Although some phage therapies are already in use in the food industry and agriculture, their use in humans has not been yet approved in the European Union or the United States.Citation256 Adequate double-blind, randomized placebo-controlled trials are essential to prove their safety and efficacy to treat human infections and diseases, including IBD.
Conclusions
In this review, we have provided an overview of recent technologies, both experimental and computational, used to analyze how the microbiome interacts with the host in IBD, and described their major advantages and limitations. Other technologies that have still not been extensively applied to the study of host–microbiome interactions in IBD include single-cell profiling and in situ characterization. Resolving heterogeneity of microbial species at the single cell level in complex communities is an important problem given the potentially large number of phenotypes arising from similar genotypes. Current approaches are limited to “bulk” estimates of composition and function, and approaches that can break down differences at higher resolution are necessary.Citation276 Techniques for fluorescent hybridization or fluorescent spectral imaging can also be applied to visualize spatial structure of microbial communities, which can provide novel knowledge to understand community function and interaction with the host.Citation277,Citation278 There is also a pressing need to improve analytical techniques of microbiome data, including accurate approaches to reduce false correlations,Citation191 methods to identify strain diversity,Citation279,Citation280 and characterize the increasing amount of genes and proteins of unknown function in microbial genomes.Citation281,Citation282
Finally, characterizing microbiome processes at the community level will be fundamental to better understand its role in IBD. Although many efforts are now directed toward characterization of specific isolates, consortia of microbes often interact in a synergistic manner to synthesize products that cannot be obtained in isolation.Citation283 This is particularly relevant in IBD, as it has been previously demonstrated that the effect of SCFA to induce anti-inflammatory Tregs in the gut is not achievable by individual bacteria but require a defined set of bacteria.Citation284 Further, microbes do not exist as isolated cells, but often form aggregates and biofilms, which are also highly relevant for IBD.Citation54 Technologies that can better dissect such structures and can characterize bacterial signals at the community level are therefore necessary next steps to provide novel insights into IBD.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Cleynen I, Boucher G, Jostins L, Schumm LP, Zeissig S, Ahmad T, Andersen V, Andrews JM, Annese V, Brand S, et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2016;387(10014):156–31. doi:10.1016/S0140-6736(15)00465-1.
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Philip Schumm L, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi:10.1038/nature11582.
- Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi:10.1016/j.chom.2014.02.005.
- Clemente JC, Manasson J, Scher JU. The role of the gut microbiome in systemic inflammatory disease. BMJ. 2018;360:j5145. doi:10.1136/bmj.j5145.
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–656. doi:10.1038/s41586-019-1237-9.
- D’Auria G, Peris-Bondia F, Džunková M, Mira A, Collado MC, Latorre A, Moya A, et al. Active and secreted IgA-coated bacterial fractions from the human gut reveal an under-represented microbiota core. Sci Rep. 2013;3(1):3515. doi:10.1038/srep03515.
- Palm NW, De zoete M, Cullen T, Barry N, Stefanowski J, Hao L, Degnan P, Hu J, Peter I, Zhang W, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158(5):1000–1010. doi:10.1016/j.cell.2014.08.006.
- Bunker JJ, Flynn T, Koval J, Shaw D, Meisel M, McDonald B, Ishizuka I, Dent A, Wilson P, Jabri B, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity. 2015;43(3):541–553. doi:10.1016/j.immuni.2015.08.007.
- Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, Manary MJ, Liu T-C, Stappenbeck TS, Maleta KM, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med. 2015;7(276):276ra24. doi:10.1126/scitranslmed.aaa4877.
- Jackson MA, Pearson C, Ilott NE, Huus KE, Hegazy AN, Webber J, Finlay BB, Macpherson AJ, Powrie F, Lam LH, et al. Accurate identification and quantification of commensal microbiota bound by host immunoglobulins. Microbiome. 2021;9(1):33. doi:10.1186/s40168-020-00992-w.
- Rengarajan S, Vivio EE, Parkes M, Peterson DA, Roberson ED, Newberry RD, Ciorba MA, Hsieh CS, et al. Dynamic immunoglobulin responses to gut bacteria during inflammatory bowel disease. Gut Microbes. 2020;11(3):405–420. doi:10.1080/19490976.2019.1626683.
- Shapiro JM, de Zoete MR, Palm NW, Laenen Y, Bright R, Mallette M, Bu K, Bielecka AA, Xu F, Hurtado-Lorenzo A, et al. Immunoglobulin A targets a unique subset of the microbiota in inflammatory bowel disease. Cell Host Microbe. 2021;29(1):83–93 e3. doi:10.1016/j.chom.2020.12.003.
- Viladomiu M, Kivolowitz C, Abdulhamid A, Dogan B, Victorio D, Castellanos JG, Woo V, Teng F, Tran NL, Sczesnak A, Chai C, et al. IgA-coated E.coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med. 2017;9(376). doi:10.1126/scitranslmed.aaf9655.
- Doron I, Mesko M, Li XV, Kusakabe T, Leonardi I, Shaw DG, Fiers WD, Lin W-Y, Bialt-DeCelie M, Román E, et al. Mycobiota-induced IgA antibodies regulate fungal commensalism in the gut and are dysregulated in Crohn’s disease. Nat Microbiol. 2021;6(12):1493–1504. doi:10.1038/s41564-021-00983-z.
- Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4(2):293–305. doi:10.1038/s41564-018-0306-4.
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Doré J, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15(8):1183–1189. doi:10.1002/ibd.20903.
- Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, et al. A decrease of the butyrate-producing species Roseburiahominis and Faecalibacterium prausnitzi defines dysbiosis in patients with ulcerativecolitis. Gut. 2014;63(8):1275–1283. doi:10.1136/gutjnl-2013-304833.
- Pascal V, Pozuelo M, Borruel N, Casellas F, Campos D, Santiago A, Martinez X, Varela E, Sarrabayrouse G, Machiels K, et al. A microbial signature for Crohn’s disease. Gut. 2017;66(5):813–822. doi:10.1136/gutjnl-2016-313235.
- Aden K, Rehman A, Waschina S, Pan W-H, Walker A, Lucio M, Nunez AM, Bharti R, Zimmerman J, Bethge J, et al. Metabolic functions of gut microbes associate with efficacy of tumor necrosis factor antagonists in patients with inflammatory bowel diseases. Gastroenterology. 2019;157(5):1279–1292 e11. doi:10.1053/j.gastro.2019.07.025.
- Sofia MA, Ciorba MA, Meckel K, Lim CK, Guillemin GJ, Weber CR, Bissonnette M, Pekow JR, et al. Tryptophan metabolism through the kynurenine pathway is associated with endoscopic inflammation in ulcerative colitis. Inflamm Bowel Dis. 2018;24(7):1471–1480. doi:10.1093/ibd/izy103.
- Sugihara K, Morhardt TL, Kamada N. The role of dietary nutrients in inflammatory bowel disease. Front Immunol. 2019;9:3183. doi:10.3389/fimmu.2018.03183.
- Palego L, Betti L, Rossi A, Giannaccini G, et al. Tryptophan biochemistry: structural, nutritional, metabolic, and medical aspects in humans. J Amino Acids. 2016;2016:8952520. doi:10.1155/2016/8952520.
- Jahnel J, Fickert P, Hauer AC, Högenauer C, Avian A, Trauner M, et al. Inflammatory bowel disease alters intestinal bile acid transporter expression. Drug Metab Dispos. 2014;42(9):1423–1431. doi:10.1124/dmd.114.058065.
- Sinha SR, Haileselassie Y, Nguyen LP, Tropini C, Wang M, Becker LS, Sim D, Jarr K, Spear ET, Singh G, et al. Dysbiosis-Induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. 2020;27(4):659–670.e5. doi:10.1016/j.chom.2020.01.021.
- Bauermeister A, Mannochio-Russo H, Costa-Lotufo LV, Jarmusch AK, Dorrestein PC, et al. Mass spectrometry-based metabolomics in microbiome investigations. Nat Rev Microbiol. 2022;20(3):143–160. doi:10.1038/s41579-021-00621-9.
- Melnik AV, da Silva RR, Hyde ER, Aksenov AA, Vargas F, Bouslimani A, Protsyuk I, Jarmusch AK, Tripathi A, Alexandrov T, et al. Coupling targeted and untargeted mass spectrometry for metabolome-Microbiome-wide association studies of human fecal samples. Anal Chem. 2017;89(14):7549–7559. doi:10.1021/acs.analchem.7b01381.
- Han J, Lin K, Sequeira C, Borchers CH, et al. An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography-tandem mass spectrometry. Anal Chim Acta. 2015;854:86–94. doi:10.1016/j.aca.2014.11.015.
- Zheng X, Qiu Y, Zhong W, Baxter S, Su M, Li Q, Xie G, Ore BM, Qiao S, Spencer MD, et al. A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids. Metabolomics. 2013;9(4):818–827. doi:10.1007/s11306-013-0500-6.
- Song HE, Lee HY, Kim SJ, Back SH, Yoo HJ, et al. A facile profiling method of short chain fatty acids using liquid chromatography-Mass spectrometry. Metabolites. 2019;9(9):173. doi:10.3390/metabo9090173.
- Zhang J, Yang S, Wang J, Xu Y, Zhao H, Lei J, Zhou Y, Chen Y, Wu L, Zhou M, et al. Integrated LC-MS metabolomics with dual derivatization for quantification of FFAs in fecal samples of hepatocellular carcinoma patients. J Lipid Res. 2021;62:100143. doi:10.1016/j.jlr.2021.100143.
- Tumanov S, Bulusu V, Kamphorst JJ. Analysis of fatty acid metabolism using stable isotope tracers and mass spectrometry. Methods Enzymol. 2015;561:197–217.
- Chan JC, Kioh DY, Yap GC, Lee BW, Chan EC, et al. A novel LCMSMS method for quantitative measurement of short-chain fatty acids in human stool derivatized with (12)C- and (13)C-labelled aniline. J Pharm Biomed Anal. 2017;138:43–53. doi:10.1016/j.jpba.2017.01.044.
- Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, Harmsen HJ, Faber KN, Hermoso MA, et al. Short chain fatty acids (SCFAs)-Mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019;10(277):1–16. doi:10.3389/fimmu.2019.0027.
- Vernia P, Monteleone G, Grandinetti G, Villotti G, Di Giulio E, Frieri G, Marcheggiano A, Pallone F, Caprilli R, Torsoli A, et al. Combined oral sodium butyrate and mesalazine treatment compared to oral mesalazine alone in ulcerative colitis: randomized, double-blind, placebo-controlled pilot study. Dig Dis Sci. 2000;45(5):976–981. doi:10.1023/A:1005537411244.
- Vernia P, Annese V, Bresci G, D’Albasio G, D’Incà R, Giaccari S, Ingrosso M, Mansi C, Riegler G, Valpiani D, et al. Topical butyrate improves efficacy of 5-ASA in refractory distal ulcerative colitis: results of a multicentre trial. Eur J Clin Invest. 2003;33(3):244–248. doi:10.1046/j.1365-2362.2003.01130.x.
- Di Sabatino A, Morera R, Ciccocioppo R, Cazzola P, Gotti S, Tinozzi FP, Tinozzi S, Corazza GR, et al. Oral butyrate for mildly to moderately active Crohn’s disease. Aliment Pharmacol Ther. 2005;22(9):789–794. doi:10.1111/j.1365-2036.2005.02639.x.
- Whiley L, Nye LC, Grant I, Andreas N, Chappell KE, Sarafian MH, Misra R, Plumb RS, Lewis MR, Nicholson JK, et al. Ultrahigh-performance liquid chromatography tandem mass spectrometry with electrospray ionization quantification of tryptophan metabolites and markers of gut health in serum and plasma—Application to clinical and epidemiology cohorts. Anal Chem. 2019;91(8):5207–5216. doi:10.1021/acs.analchem.8b05884.
- Nikolaus S, Schulte B, Al-Massad N, Thieme F, Schulte DM, Bethge J, Rehman A, Tran F, Aden K, Häsler R, et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology. 2017;153(6):1504–1516.e2. doi:10.1053/j.gastro.2017.08.028.
- Beghini F, McIver LJ, Blanco-Míguez A, Dubois L, Asnicar F, Maharjan S, Mailyan A, Manghi P, Scholz M, Thomas AM, et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife. 2021;10:1–42. doi:10.7554/eLife.65088.
- Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12(10):902–903. doi:10.1038/nmeth.3589.
- Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257. doi:10.1186/s13059-019-1891-0.
- Aggarwala V, Mogno I, Li Z, Yang C, Britton GJ, Chen-Liaw A, Mitcham J, Bongers G, Gevers D, Clemente JC, Colombel JF, et al. Precise quantification of bacterial strains after fecal microbiota transplantation delineates long-term engraftment and explains outcomes. Nat Microbiol. 2021;6(10):1309–1318. doi:10.1038/s41564-021-00966-0.
- Smillie CS, Sauk J, Gevers D, Friedman J, Sung J, Youngster I, Hohmann EL, Staley C, Khoruts A, Sadowsky MJ, et al. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe. 2018;23(2):229–240 e5. doi:10.1016/j.chom.2018.01.003.
- Sunagawa S, Mende DR, Zeller G, Izquierdo-Carrasco F, Berger SA, Kultima JR, Coelho LP,Arumugam M, Tap J, Nielsen HB, Rasmussen S, et al. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods. 2013;10(12):1196–1199. doi:10.1038/nmeth.2693.
- Olm MR, Crits-Christoph A, Bouma-Gregson K, Firek BA, Morowitz MJ, Banfield JF, et al. inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat Biotechnol. 2021;39(6):727–736. doi:10.1038/s41587-020-00797-0.
- Brand EC, Klaassen MAY, Gacesa R, Vich Vila A, Ghosh H, de Zoete MR, Boomsma DI, Hoentjen F, Horjus Talabur Horje CS, van de Meeberg PC, et al. Healthy cotwins share gut microbiome signatures with their inflammatory bowel disease twins and unrelated patients. Gastroenterology. 2021;160(6):1970–1985. doi:10.1053/j.gastro.2021.01.030.
- Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9(1):103. doi:10.1186/s13073-017-0490-5.
- Xia Y, Wang J, Fang X, Dou T, Han L, Yang C, et al. Combined analysis of metagenomic data revealed consistent changes of gut microbiome structure and function in inflammatory bowel disease. J Appl Microbiol. 2021;131(6):3018–3031. doi:10.1111/jam.15154.
- Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, Cleland T, Xavier RJ, et al. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe. 2017;21(5):603–610 e3. doi:10.1016/j.chom.2017.04.010.
- Donovan PD, Gonzalez G, Higgins DG, Butler G, Ito K, et al. Identification of fungi in shotgun metagenomics datasets. PLoS One. 2018;13(2):e0192898. doi:10.1371/journal.pone.0192898.
- Soverini M, Turroni S, Biagi E, Brigidi P, Candela M, Rampelli S, et al. HumanMycobiomeScan: a new bioinformatics tool for the characterization of the fungal fraction in metagenomic samples. BMC Genomics. 2019;20(1):496. doi:10.1186/s12864-019-5883-y.
- Marcelino VR, Clausen PT, Buchmann JP, Wille M, Iredell JR, Meyer W, Lund O, Sorrell TC, Holmes EC. CC Metagen, et al. CCMetagen: comprehensive and accurate identification of eukaryotes and prokaryotes in metagenomic data. Genome Biol. 2020;21(1):103. doi:10.1186/s13059-020-02014-2.
- Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, Wu GD, et al. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2015;21(8):1948–1956. doi:10.1097/MIB.0000000000000454.
- Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio. 2016;7(5). doi:10.1128/mBio.01250-16.
- Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66(6):1039–1048. doi:10.1136/gutjnl-2015-310746.
- Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, et al. Malassezia is associated with crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe. 2019;25(3):377–388.e6. doi:10.1016/j.chom.2019.01.007.
- Akhter S, Aziz RK, Edwards RA. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012;40(16):e126. doi:10.1093/nar/gks406.
- Roux S,Enault F, Hurwitz BL, Sullivan MB, et al. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi:10.7717/peerj.985.
- Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):W16–21. doi:10.1093/nar/gkw387.
- Park H, Yeo S, Kang S, Huh CS, et al. Longitudinal microbiome analysis in a dextran sulfate sodium-Induced colitis mouse model. Microorganisms. 2021;9(2):370. doi:10.3390/microorganisms9020370.
- Mars RAT, Yang Y, Ward T, Houtti M, Priya S, Lekatz HR, Tang X, Sun Z, Kalari KR, Korem T, et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell. 2020;182(6):1460–1473 e17. doi:10.1016/j.cell.2020.08.007.
- Paramsothy S, Nielsen S, Kamm MA, Deshpande NP, Faith JJ, Clemente JC, Paramsothy R, Walsh AJ, van den Bogaerde J, Samuel D, et al. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology. 2019;156(5):1440–1454 e2. doi:10.1053/j.gastro.2018.12.001.
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-Type lectin receptor dectin-1 influence colitis. Science. 2012;336(6086):1314–1317. doi:10.1126/science.1221789.
- Liang G, Conrad MA, Kelsen JR, Kessler LR, Breton J, Albenberg LG, Marakos S, Galgano A, Devas N, Erlichman J, Zhang H, et al. Dynamics of the stool virome in very early-Onset inflammatory bowel disease. Journal of Crohn’s and Colitis. 2020;14(11):1600–1610. doi:10.1093/ecco-jcc/jjaa094.
- Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD, et al. Rapid evolution of the human gut virome. Proc Natl Acad Sci U S A. 2013;110(30):12450–12455. doi:10.1073/pnas.1300833110.
- Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, et al. The gut mycobiome of the human microbiome project healthy cohort. Microbiome. 2017;5(1):153. doi:10.1186/s40168-017-0373-4.
- Tang J, Iliev ID, Brown J, Underhill DM, Funari VA, et al. Mycobiome: approaches to analysis of intestinal fungi. J Immunol Methods. 2015;421:112–121. doi:10.1016/j.jim.2015.04.004.
- Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8(9):e1002687. doi:10.1371/journal.pcbi.1002687.
- Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA, et al. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol. 2015;11(5):e1004226. doi:10.1371/journal.pcbi.1004226.
- Deng Y,Jiang YH, Yang Y, He Z, Luo F, Zhou J, et al. Molecular ecological network analyses. BMC Bioinform. 2012;13(1):113. doi:10.1186/1471-2105-13-113.
- Bucci V, Tzen B, Li N, Simmons M, Tanoue T, Bogart E, Deng L, Yeliseyev V, Delaney ML, Liu Q, Olle B, et al. MDSINE: microbial dynamical systems inference engine for microbiome time-series analyses. Genome Biol. 2016;17(1):121. doi:10.1186/s13059-016-0980-6.
- Chen L, Collij V, Jaeger M, van den Munckhof ICL, Vich Vila A, Kurilshikov A, Gacesa R, Sinha T, Oosting M, Joosten LAB, et al. Gut microbial co-abundance networks show specificity in inflammatory bowel disease and obesity. Nat Commun. 2020;11(1):4018. doi:10.1038/s41467-020-17840-y.
- Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012;12(12):821–832. doi:10.1038/nri3322.
- Wilmore JR, Gaudette BT, Gomez Atria D, Hashemi T, Jones DD, Gardner CA, Cole SD, Misic AM, Beiting DP, Allman D, et al. Commensal microbes induce serum IgA responses that protect against polymicrobial sepsis. Cell Host Microbe. 2018;23(3):302–311.e3. doi:10.1016/j.chom.2018.01.005.
- Cullender TC,Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA, Walter J, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14(5):571–581. doi:10.1016/j.chom.2013.10.009.
- Kubinak JL, Petersen C, Stephens WZ, Soto R, Bake E, O’Connell RM, Round JL, et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe. 2015;17(2):153–163. doi:10.1016/j.chom.2014.12.009.
- Moor K, Diard M, Sellin ME, Felmy B, Wotzka SY, Toska A, Bakkeren E, Arnoldini M, Bansept F, Co AD, et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature. 2017;544(7651):498–502. doi:10.1038/nature22058.
- Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou W-C, Conner ME, Earl AM, Knight R, Bjorkman PJ, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. 2018;360(6390):795–800. doi:10.1126/science.aaq0926.
- Viladomiu M, Kivolowitz C, Abdulhamid A, Dogan B, Victorio D, Castellanos JG, Woo V, Teng F, Tran NL, Sczesnak A, Chai C, et al. IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote T(H)17-dependent inflammation. Sci Transl Med. 2017;9(376). doi:10.1126/scitranslmed.aaf9655.
- Shapiro JM, de Zoete MR, Palm NW, Laenen Y, Bright R, Mallette M, Bu K, Bielecka AA, Xu F, Hurtado-Lorenzo A, et al. Immunoglobulin a targets a unique subset of the microbiota in inflammatory bowel disease. Cell Host Microbe. 2021;29(1):83–93.e3. doi:10.1016/j.chom.2020.12.003.
- Yang C, Mogno I, Contijoch EJ, Borgerding JN, Aggarwala V, Li Z, Siu S, Grasset EK, Helmus DS, Dubinsky MC, et al. Fecal IgA levels are determined by strain-level differences in bacteroides ovatus and are modifiable by gut microbiota manipulation. Cell Host Microbe. 2020;27(3):467–475.e6. doi:10.1016/j.chom.2020.01.016.
- Kabbert J, Benckert J, Rollenske T, Hitch TC, Clavel T, Cerovic V, Wardemann H, Pabst O, et al. High microbiota reactivity of adult human intestinal IgA requires somatic mutations. J Exp Med. 2020;217(11). doi:10.1084/jem.20200275.
- Bunker JJ, Erickson SA, Flynn TM, Henry C, Koval JC, Meisel M, Jabri B, Antonopoulos DA, Wilson PC, Bendelac A, et al. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science. 2017;358(6361). doi:10.1126/science.aan6619.
- Magri G, Comerma L, Pybus M, Sintes J, Lligé D, Segura-Garzón D, Bascones S, Yeste A, Grasset EK, Gutzeit C, et al. Human secretory IgM emerges from plasma cells clonally related to gut memory B cells and targets highly diverse commensals. Immunity. 2017;47(1):118–134.e8. doi:10.1016/j.immuni.2017.06.013.
- Fadlallah J, Sterlin D, Fieschi C, Parizot C, Dorgham K, El Kafsi H, Autaa G, Ghillani-Dalbin P, Juste C, Lepage P, et al. Synergistic convergence of microbiota-specific systemic IgG and secretory IgA. J Allergy Clin Immunol. 2019;143(4):1575–1585.e4. doi:10.1016/j.jaci.2018.09.036.
- Lin R, Chen H, Shu W, Sun M, Fang L, Shi Y, Pang Z, Wu W, Liu Z, et al. Clinical significance of soluble immunoglobulins A and G and their coated bacteria in feces of patients with inflammatory bowel disease. J Transl Med. 2018;16(1):359. doi:10.1186/s12967-018-1723-0.
- Ost KS, O’Meara TR, Stephens WZ, Chiaro T, Zhou H, Penman J, Bell R, Catanzaro JR, Song D, Singh S, et al. Adaptive immunity induces mutualism between commensal eukaryotes. Nature. 2021;596(7870):114–118. doi:10.1038/s41586-021-03722-w.
- Doron I, Leonardi I, Li XV, Fiers WD, Semon A, Bialt-DeCelie M, Migaud M, Gao IH, Lin W-Y, Kusakabe T, et al. Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell. 2021;184(4):1017–1031.e14. doi:10.1016/j.cell.2021.01.016.
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi:10.1038/s41586-019-1237-9.
- Plichta DR, Graham DB, Subramanian S, Xavier RJ, et al. Therapeutic opportunities in inflammatory bowel disease: mechanistic dissection of host-Microbiome relationships. Cell. 2019;178(5):1041–1056. doi:10.1016/j.cell.2019.07.045.
- Aden K, Rehman A, Waschina S, Pan W-H, Walker A, Lucio M, Nunez AM, Bharti R, Zimmerman J, Bethge J, et al. Metabolic functions of gut microbes associate with efficacy of tumor necrosis Factor antagonists in patients with inflammatory bowel diseases. Gastroenterology. 2019;157(5):1279–1292.e11. doi:10.1053/j.gastro.2019.07.025.
- Segain JP, Segain JP, Kornprobst M, et al. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut. 2000;47(3):397–403. doi:10.1136/gut.47.3.397.
- Lührs H, Gerke T, Müller JG, Melcher R, Schauber J, Boxberger F, Scheppach W, Menzel T, et al. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol. 2002;37(4):458–466. doi:10.1080/003655202317316105.
- Vernero M, De Blasio F, Ribaldone DG, Bugianesi E, Pellicano R, Saracco GM, Astegiano M, Caviglia GP, et al. The usefulness of microencapsulated sodium butyrate add-on therapy in maintaining remission in patients with ulcerative colitis: a prospective observational study. J Clin Med. 2020;9(12):3941. doi:10.3390/jcm9123941.
- Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487(7408):477–481. doi:10.1038/nature11228.
- Islam J, Sato S, Watanabe K, Watanabe T, Hirahara K, Aoyama Y, Tomita S, Aso H, Komai M, Shirakawa H, et al. Dietary tryptophan alleviates dextran sodium sulfate-induced colitis through aryl hydrocarbon receptor in mice. J Nutr Biochem. 2017;42:43–50. doi:10.1016/j.jnutbio.2016.12.019.
- Bettenworth D, Nowacki TM, Ross M, Kyme P, Schwammbach D, Kerstiens L, Thoennissen GB, Bokemeyer C, Hengst K, Berdel WE, Heidemann J, et al. Nicotinamide treatment ameliorates the course of experimental colitis mediated by enhanced neutrophil-specific antibacterial clearance. Mol Nutr Food Res. 2014;58(7):1474–1490. doi:10.1002/mnfr.201300818.
- Kim CJ, Kovacs-Nolan JA, Yang C, Archbold T, Fan MZ, Mine Y, et al. l-Tryptophan exhibits therapeutic function in a porcine model of dextran sodium sulfate (DSS)-induced colitis. J Nutr Biochem. 2010;21(6):468–475. doi:10.1016/j.jnutbio.2009.01.019.
- Scott SA, Fu J, Chang PV. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proceedings of the National Academy of Sciences. 2020;117(32):19376–19387. doi:10.1073/pnas.2000047117.
- Lanis JM, Alexeev EE, Curtis VF, Kitzenberg DA, Kao DJ, Battista KD, Gerich ME, Glover LE, Kominsky DJ, Colgan SP, et al. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol. 2017;10(5):1133–1144. doi:10.1038/mi.2016.133.
- Marcos J, Renau N, Valverde O, Aznar-Lain G, Gracia-Rubio I, Gonzalez-Sepulveda M, Pérez-Jurado LA, Ventura R, Segura J, Pozo OJ, et al. Targeting tryptophan and tyrosine metabolism by liquid chromatography tandem mass spectrometry. J Chromatogr A. 2016;1434:91–101. doi:10.1016/j.chroma.2016.01.023.
- Akobeng AK, Miller V, Stanton J, Elbadri AM, Thomas AG, et al. Double-blind randomized controlled trial of glutamine-enriched polymeric diet in the treatment of active Crohn’s disease. J Pediatr Gastroenterol Nutr. 2000;30(1):78–84. doi:10.1097/00005176-200001000-00022.
- Ockenga J, Borchert K, Stüber E, Lochs H, Manns MP, Bischoff SC, et al. Glutamine-enriched total parenteral nutrition in patients with inflammatory bowel disease. Eur J Clin Nutr. 2005;59(11):1302–1309. doi:10.1038/sj.ejcn.1602243.
- Ticho AL, Malhotra P, Dudeja PK, Gill RK, Alrefai WA, et al. Intestinal absorption of bile acids in health and disease. Compr Physiol. 2019;10(1):21–56.
- Guzior DV, Quinn RA. Review: microbial transformations of human bile acids. Microbiome. 2021;9(1):140. doi:10.1186/s40168-021-01101-1.
- Joyce SA, Gahan CG. Disease-Associated changes in bile acid profiles and links to altered Gut microbiota. Dig Dis. 2017;35(3):169–177. doi:10.1159/000450907.
- Song Z, Cai Y, Lao X, Wang X, Lin X, Cui Y, Kalavagunta PK, Liao J, Jin L, Shang J, et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome. 2019;7(1):9. doi:10.1186/s40168-019-0628-3.
- Weingarden AR, Chen C, Zhang N, Graiziger CT, Dosa PI, Steer CJ, Shaughnessy MK, Johnson JR, Sadowsky MJ, Khoruts A, et al. Ursodeoxycholic acid inhibits clostridium difficile spore germination and vegetative growth, and prevents the recurrence of ileal pouchitis associated with the infection. J Clin Gastroenterol. 2016;50(8):624–630. doi:10.1097/MCG.0000000000000427.
- Pearson T, Caporaso JG, Yellowhair M, Bokulich NA, Padi M, Roe DJ, Wertheim BC, Linhart M, Martinez JA, Bilagody C, et al. Effects of ursodeoxycholic acid on the gut microbiome and colorectal adenoma development. Cancer Med. 2019;8(2):617–628. doi:10.1002/cam4.1965.
- Fiorucci S, Carino A, Baldoni M, Santucci L, Costanzi E, Graziosi L, Distrutti E, Biagioli M, et al. Bile acid signaling in inflammatory bowel diseases. Dig Dis Sci. 2021;66(3):674–693. doi:10.1007/s10620-020-06715-3.
- Hou RG, Fan L, Liu JJ, Cheng Y, Chang ZP, Wu B, Shao YY, et al. Bile acid malabsorption is associated with diarrhea in acute phase of colitis. Can J Physiol Pharmacol. 2018;96(12):1328–1336. doi:10.1139/cjpp-2018-0017.
- Jung D, Fantin AC, Scheurer U, Fried M, Kullak-Ublick GA, et al. Human ileal bile acid transporter gene ASBT (SLC10A2) is transactivated by the glucocorticoid receptor. Gut. 2004;53(1):78–84. doi:10.1136/gut.53.1.78.
- Barkun AN, Love J, Gould M, Pluta H, Steinhart AH, et al. Bile acid malabsorption in chronic diarrhea: pathophysiology and treatment. Can J Gastroenterol. 2013;27(11):653–659. doi:10.1155/2013/485631.
- Devarakonda A, Arnott I, Satsangi J. P700 The efficacy of colesevelam to treat bile acid malabsorption in Crohn’s disease: data from TOPPIC trial. Journal of Crohn’s and Colitis. 2019;13(Supplement_1):S470–S471. doi:10.1093/ecco-jcc/jjy222.824.
- Beigel F, Teich N, Howaldt S, Lammert F, Maul J, Breiteneicher S, Rust C, Göke B, Brand S, Ochsenkühn T, et al. Colesevelam for the treatment of bile acid malabsorption-associated diarrhea in patients with Crohn’s disease: a randomized, double-blind, placebo-controlled study. Journal of Crohn’s and Colitis. 2014;8(11):1471–1479. doi:10.1016/j.crohns.2014.05.009.
- Keely SJ, Steer CJ, Lajczak-McGinley NK. Ursodeoxycholic acid: a promising therapeutic target for inflammatory bowel diseases? Am J Physiol Gastrointest Liver Physiol. 2019;317(6):G872–g881. doi:10.1152/ajpgi.00163.2019.
- Martínez-Moya P, Romero-Calvo I, Requena P, Hernández-Chirlaque C, Aranda CJ, González R, Zarzuelo A, Suárez MD, Martínez-Augustin O, Marín JJG, et al. Dose-dependent antiinflammatory effect of ursodeoxycholic acid in experimental colitis. Int Immunopharmacol. 2013;15(2):372–380. doi:10.1016/j.intimp.2012.11.017.
- Troy EB, Kasper DL, Kasper DL, Kasper DL. Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front Biosci (Landmark Ed). 2010;15(1):25–34. doi:10.2741/3603.
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–625. doi:10.1038/nature07008.
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–12209. doi:10.1073/pnas.0909122107.
- Zheng L, Luo M, Kuang G, Liu Y, Liang D, Huang H, Yi X, Wang C, Wang Y, Xie Q, et al. Capsular polysaccharide from bacteroides fragilis protects against ulcerative colitis in an undegraded form. Front Pharmacol. 2020;11(1835). doi:10.3389/fphar.2020.570476.
- Brito IL, Alm EJ. Tracking strains in the microbiome: insights from metagenomics and models. Front Microbiol. 2016;7:712. doi:10.3389/fmicb.2016.00712.
- Faith JJ, Colombel JF, Gordon JI. Identifying strains that contribute to complex diseases through the study of microbial inheritance. Proc Natl Acad Sci U S A. 2015;112(3):633–640. doi:10.1073/pnas.1418781112.
- Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel J-F, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with crohn’s disease. Gastroenterology. 1998;115(6):1405–1413. doi:10.1016/S0016-5085(98)70019-8.
- Fang X, Monk JM, Nurk S, Akseshina M, Zhu Q, Gemmell C, Gianetto-Hill C, Leung N, Szubin R, Sanders J, Beck PL, et al. Metagenomics-Based, strain-level analysis of Escherichia coli from a time-series of microbiome samples from a crohn’s disease patient. Front Microbiol. 2018;9:2559. doi:10.3389/fmicb.2018.02559.
- Miquel S, Peyretaillade E, Claret L, de Vallée A, Dossat C, Vacherie B, Zineb EH, Segurens B, Barbe V, Sauvanet P, et al. Complete genome sequence of Crohn’s disease-associated adherent-invasive E.coli strain LF82. PLoS One. 2010;5(9):e12714. doi:10.1371/journal.pone.0012714.
- Nagayama M, Yano T, Atarashi K, Tanoue T, Sekiya M, Kobayashi Y, Sakamoto H, Miura K, Sunada K, Kawaguchi T, et al. TH1 cell-inducing Escherichia coli strain identified from the small intestinal mucosa of patients with crohn’s disease. Gut Microbes. 2020;12(1):1788898. doi:10.1080/19490976.2020.1788898.
- Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, Alpern JA, Akyildiz A, Theiss AL, Nusrat A, et al. Invasive Escherichia coli are a feature of Crohn’s disease. Lab Invest. 2007;87(10):1042–1054. doi:10.1038/labinvest.3700661.
- Liu YX, Qin Y, Chen T, Lu M, Qian X, Guo X, Bai Y, et al. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell. 2020. 12.
- Miossec MJ, Valenzuela SL, Pérez-Losada M, Johnson WE, Crandall KA, Castro-Nallar E, et al. Evaluation of computational methods for human microbiome analysis using simulated data. PeerJ. 2020;8:e9688. doi:10.7717/peerj.9688.
- Ye SH, Siddle KJ, Park DJ, Sabeti PC, et al. Benchmarking metagenomics tools for taxonomic classification. Cell. 2019;178(4):779–794. doi:10.1016/j.cell.2019.07.010.
- Gwak HJ, Lee SJ, Rho M. Application of computational approaches to analyze metagenomic data. J Microbiol. 2021;59(3):233–241. doi:10.1007/s12275-021-0632-8.
- Haas A, Zimmermann K, Graw F, Slack E, Rusert P, Ledergerber B, Bossart W, Weber R, Thurnheer MC, Battegay M, et al. Systemic antibody responses to gut commensal bacteria during chronic HIV-1 infection. Gut. 2011;60(11):1506–1519. doi:10.1136/gut.2010.224774.
- Halfvarson J, Brislawn CJ, Lamendella R, Vázquez-Baeza Y, Walters WA, Bramer LM, D’Amato M, Bonfiglio F, McDonald D, Gonzalez A, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2(5):17004. doi:10.1038/nmicrobiol.2017.4.
- Gevers D, Kugathasan S, Denson L, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song S, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi:10.1016/j.chom.2014.02.005.
- Hashash JG, Chintamaneni P, Ramos Rivers CM, Koutroubakis IE, Regueiro MD, Baidoo L, Swoger JM, Barrie A, Schwartz M, Dunn MA, et al. Patterns of antibiotic exposure and clinical disease activity in inflammatory bowel disease: a 4-year prospective study. Inflamm Bowel Dis. 2015;21(11):2576–2582. doi:10.1097/MIB.0000000000000534.
- Lewis JD, Chen E, Baldassano R, Otley A, Griffiths A, Lee D, Bittinger K, Bailey A, Friedman E, Hoffmann C, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric crohn’s disease. Cell Host Microbe. 2015;18(4):489–500. doi:10.1016/j.chom.2015.09.008.
- Rogler G, Zeitz J, Biedermann L. The search for causative environmental factors in inflammatory bowel disease. Dig Dis. 2016;34(1):48–55. doi:10.1159/000447283.
- Shaw KA, Bertha M, Hofmekler T, Chopra P, Vatanen T, Srivatsa A, Prince J, Kumar A, Sauer C, Zwick ME, et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med. 2016;8(1):75. doi:10.1186/s13073-016-0331-y.
- Ho SM, Lewis JD, Mayer EA, Bernstein CN, Plevy SE, Chuang E, Rappaport SM, Croitoru K, Korzenik JR, Krischer J, et al. Challenges in IBD research: environmental triggers. Inflamm Bowel Dis. 2019;25(Suppl 2):S13–S23. doi:10.1093/ibd/izz076.
- Mars RAT, Frith M, Kashyap PC. Functional gastrointestinal disorders and the microbiome-What is the best strategy for moving microbiome-based therapies for functional gastrointestinal disorders into the clinic? Gastroenterology. 2021;160(2):538–555. doi:10.1053/j.gastro.2020.10.058.
- Fite A, Macfarlane S, Furrie E, Bahrami B, Cummings JH, Steinke DT, Macfarlane GT, et al. Longitudinal analyses of gut mucosal microbiotas in ulcerative colitis in relation to patient age and disease severity and duration. J Clin Microbiol. 2013;51(3):849–856. doi:10.1128/JCM.02574-12.
- Xun Z, Zhang Q, Xu T, Chen N, Chen F, et al. Dysbiosis and ecotypes of the salivary microbiome associated with inflammatory bowel diseases and the assistance in diagnosis of diseases using oral bacterial profiles. Front Microbiol. 2018;9:1136. doi:10.3389/fmicb.2018.01136.
- De Fazio L, Cavazza E, Spisni E, Strillacci A, Centanni M, Candela M, Praticò C, Campieri M, Ricci C, Valerii MC, et al. Longitudinal analysis of inflammation and microbiota dynamics in a model of mild chronic dextran sulfate sodium-induced colitis in mice. World J Gastroenterol. 2014;20(8):2051–2061. doi:10.3748/wjg.v20.i8.2051.
- Sharpton T, Lyalina S, Luong J, Pham J, Deal EM, Armour C, Gaulke C, Sanjabi S, Pollard KS, et al. Development of Inflammatory Bowel Disease Is Linked To A Longitudinal Restructuring Of The Gut Metagenome in mice. mSystems. 2017;2(5). doi:10.1128/mSystems.00036-17.
- Ward MA, Pierre JF, Leal RF, Huang Y, Shogan B, Dalal SR, Weber CR, Leone VA, Musch MW, An GC, et al. Insights into the pathogenesis of ulcerative colitis from a murine model of stasis-induced dysbiosis, colonic metaplasia, and genetic susceptibility. Am J Physiol Gastrointest Liver Physiol. 2016;310(11):G973–88. doi:10.1152/ajpgi.00017.2016.
- Wardill HR, Choo JM, Dmochowska N, Mavrangelos C, Campaniello MA, Bowen JM, Rogers GB, Hughes PA. Acute colitis drives tolerance by persistently altering the epithelial barrier and innate and adaptive immunity. Inflamm Bowel Dis. 2019;25(7):1196–1207. doi:10.1093/ibd/izz011.
- Galazzo G, Tedjo DI, Wintjens DS, Savelkoul PH, Masclee AA, Bodelier AG, Pierik MJ, Jonkers DM, Penders J, et al. Faecal microbiota dynamics and their relation to disease course in crohn’s disease. J Crohns Colitis. 2019;13(10):1273–1282. doi:10.1093/ecco-jcc/jjz049.
- Ashton JJ, Colquhoun CM, Cleary DW, Coelho T, Haggarty R, Mulder I, Batra A, Afzal NA, Beattie RM, Scott KP, et al. 16S sequencing and functional analysis of the fecal microbiome during treatment of newly diagnosed pediatric inflammatory bowel disease. Medicine (Baltimore). 2017;96(26):e7347. doi:10.1097/MD.0000000000007347.
- Yin W, Ludvigsson JF, Liu Z, Roosaar A, Axéll T, Ye W, et al. Inverse association between poor oral health and inflammatory bowel diseases. Clin Gastroenterol Hepatol. 2017;15(4):525–531. doi:10.1016/j.cgh.2016.06.024.
- Schaffler H, Herlemann DP, Klinitzke P, Berlin P, Kreikemeyer B, Jaster R, Lamprecht G, et al. Vitamin D administration leads to a shift of the intestinal bacterial composition in Crohn’s disease patients, but not in healthy controls. J Dig Dis. 2018;19(4):225–234. doi:10.1111/1751-2980.12591.
- Vazquez-Baeza Y, Gonzalez A, Xu ZZ, Washburne A, Herfarth HH, Sartor RB, Knight R. Guiding longitudinal sampling in IBD cohorts. Gut. 2018;67(9):1743–1745. doi:10.1136/gutjnl-2017-315352.
- Clooney AG, Eckenberger J, Laserna-Mendieta E, Sexton KA, Bernstein MT, Vagianos K, Sargent M, Ryan FJ, Moran C, Sheehan D, et al. Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut. 2021;70(3):499–510. doi:10.1136/gutjnl-2020-321106.
- Effenberger M, Reider S, Waschina S, Bronowski C, Enrich B, Adolph TE, Koch R, Moschen AR, Rosenstiel P, Aden K, et al. Microbial butyrate synthesis indicates therapeutic efficacy of azathioprine in IBD patients. J Crohns Colitis. 2021;15(1):88–98. doi:10.1093/ecco-jcc/jjaa152.
- Dollive S, Peterfreund GL, Sherrill-Mix S, Bittinger K, Sinha R, Hoffmann C, Nabel CS, Hill DA, Artis D, Bachman MA, et al. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol. 2012;13(7):R60. doi:10.1186/gb-2012-13-7-r60.
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Bolchacova E, Voigt K, Crous PW, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences. 2012;109(16):6241–6246. doi:10.1073/pnas.1117018109.
- Frau A, Kenny JG, Lenzi L, Campbell BJ, Ijaz UZ, Duckworth CA, Burkitt MD, Hall N, Anson J, Darby AC, et al. DNA extraction and amplicon production strategies deeply inf luence the outcome of gut mycobiome studies. Sci Rep. 2019;9(1):9328. doi:10.1038/s41598-019-44974-x.
- Blaalid R, Kumar S, Nilsson RH, Abarenkov K, Kirk PM, Kauserud H. ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol Ecol Resour. 2013;13:218–224. doi:10.1111/1755-0998.12065.
- Duerkop BA, Clements CV, Rollins D, Rodrigues JLM, Hooper LV. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proceedings of the National Academy of Sciences. 2012;109(43):17621–17626. doi:10.1073/pnas.1206136109.
- Norman JM, Handley S, Baldridge M, Droit L, Liu C, Keller B, Kambal A, Monaco C, Zhao G, Fleshner P, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160(3):447–460. doi:10.1016/j.cell.2015.01.002.
- Pérez-Brocal V, García-López R, Vázquez-Castellanos JF, Nos P, Beltrán B, Latorre A, Moya A. Study of the viral and microbial communities associated with Crohn’s disease: a metagenomic approach. Clin Transl Gastroenterol. 2013;4(6):e36. doi:10.1038/ctg.2013.9.
- Wagner J, Maksimovic J, Farries G, Sim WH, Bishop RF, Cameron DJ, Catto-Smith AG, Kirkwood CD. Bacteriophages in gut samples from pediatric crohn’s disease patients: metagenomic analysis using 454 pyrosequencing. Inflamm Bowel Dis. 2013;19(8):1598–1608. doi:10.1097/MIB.0b013e318292477c.
- Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, O’Regan O, Ryan FJ, Draper LA, Plevy SE, Ross RP, et al. Whole-Virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe. 2019;26(6):764–778.e5. doi:10.1016/j.chom.2019.10.009.
- Gogokhia L, Buhrke K, Bell R, Hoffman B, Brown DG, Hanke-Gogokhia C, Ajami NJ, Wong MC, Ghazaryan A, Valentine JF, et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe. 2019;25(2):285–299.e8. doi:10.1016/j.chom.2019.01.008.
- Zuo T, Lu X-J, Zhang Y, Cheung CP, Lam S, Zhang F, Tang W, Ching JYL, Zhao R, Chan PKS, et al. Gut mucosal virome alterations in ulcerative colitis. Gut. 2019;68(7):1169–1179. doi:10.1136/gutjnl-2018-318131.
- Shkoporov AN, Hill C. Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe. 2019;25(2):195–209. doi:10.1016/j.chom.2019.01.017.
- Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, et al. Bacteriophage adhering to mucus provide a non–host-derived immunity. Proceedings of the National Academy of Sciences. 2013;110(26):10771–10776. doi:10.1073/pnas.1305923110.
- Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. Healthy human gut phageome. Proceedings of the National Academy of Sciences. 2016;113(37):10400–10405. doi:10.1073/pnas.1601060113.
- Baltimore D. Expression of animal virus genomes. Bacteriol Rev. 1971;35(3):235–241. doi:10.1128/br.35.3.235-241.1971.
- Liang G, Bushman FD. The human virome: assembly, composition and host interactions. Nat Rev Microbiol. 2021;19(8):514–527. doi:10.1038/s41579-021-00536-5.
- Krishnamurthy SR, Wang D. Origins and challenges of viral dark matter. Virus Res. 2017;239:136–142. doi:10.1016/j.virusres.2017.02.002.
- Reyes A, Semenkovich NP, Whiteson K, Rohwer F,Gordon JI. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol. 2012;10(9):607–617. doi:10.1038/nrmicro2853.
- Thurber RV, Haynes M, Breitbart M, Weglei L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4(4):470–483. doi:10.1038/nprot.2009.10.
- Foster SJ, Monahan BJ. Amplification of fungal genomes using multiple displacement amplification. In: Sharon A, editor. Molecular and cell biology methods for fungi. Totowa (NJ): Humana Press; 2010. p. 175–185.
- Reyes A, Haynes M, Hanson N, Angley FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–338. doi:10.1038/nature09199.
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi:10.1038/nmeth.1923.
- Schmieder R, Edwards R, Rodriguez-Valera F. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One. 2011;6(3):e17288. doi:10.1371/journal.pone.0017288.
- Zolfo M, Pinto F, Asnicar F, Manghi P, Tett A, Bushman FD, Segata N. Detecting contamination in viromes using ViromeQC. Nat Biotechnol. 2019;37(12):1408–1412. doi:10.1038/s41587-019-0334-5.
- White JR, Maddox C, White O, Angiuoli SV, Fricke WF. CloVR-ITS: automated internal transcribed spacer amplicon sequence analysis pipeline for the characterization of fungal microbiota. Microbiome. 2013;1(1):6. doi:10.1186/2049-2618-1-6.
- Gweon HS, Oliver A, Taylor J, Booth T, Gibbs M, Read DS, Griffiths RI, Schonrogge K. PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol Evol. 2015;6(8):973–980. doi:10.1111/2041-210X.12399.
- Wylie TN, Wylie KM. ViroMatch: a computational pipeline for the detection of viral sequences from complex metagenomic data. Microbiol Resour Announc. 2021;10(9). doi:10.1128/MRA.01468-20.
- Clarke EL, Taylor LJ, ZhaoC, Connell A, Lee JJ,Fett B, Bushman FD, Bittinger K. Sunbeam: an extensible pipeline for analyzing metagenomic sequencing experiments. Microbiome. 2019;7(1):46. doi:10.1186/s40168-019-0658-x.
- Zhao G, Wu G, Lim ES, DroitL, KrishnamurthyS, Barouch DH, Virgin HW, Wang D. VirusSeeker, a computational pipeline for virus discovery and virome composition analysis. Virology. 2017;503:21–30. doi:10.1016/j.virol.2017.01.005.
- Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350(6261):663–666. doi:10.1126/science.aad2602.
- Layeghifard M, Hwang DM, Guttman DS. Disentangling Interactions in the microbiome: a network perspective. Trends Microbiol. 2017;25(3):217–228. doi:10.1016/j.tim.2016.11.008.
- Aitchison J. The statistical-Analysis of compositional data. Journal of the Royal Statistical Society Series B-Methodological. 1982;44:139–177.
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi:10.1038/nmeth.f.303.
- McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10(4):e1003531. doi:10.1371/journal.pcbi.1003531.
- Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5(1):27. doi:10.1186/s40168-017-0237-y.
- Lovell D, Pawlowsky-Glahn V, Egozcue JJ, Marguerat S, Bähler J. Proportionality: a valid alternative to correlation for relative data. PLoS Comput Biol. 2015;11(3):e1004075. doi:10.1371/journal.pcbi.1004075.
- Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. Microbiome datasets are compositional: and this is not optional. Front Microbiol. 2017;8:8. doi:10.3389/fmicb.2017.00008.
- Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, Zengler K, Knight R. Establishing microbial composition measurement standards with reference frames. Nat Commun. 2019;10(1):2719. doi:10.1038/s41467-019-10656-5.
- Bu K, Wallach DS, Wilson Z, Shen N, Segal LN, Bagiella E, Clemente JJ. Identifying correlations driven by influential observations in large datasets. Brief Bioinform. 2021;23(1).
- Faust K, Raes J. CoNet app: inference of biological association networks using Cytoscape. F1000Res. 2016;5:1519. doi:10.12688/f1000research.9050.1.
- Ban Y, An L, Jiang H. Investigating microbial co-occurrence patterns based on metagenomic compositional data. Bioinformatics. 2015;31(20):3322–3329. doi:10.1093/bioinformatics/btv364.
- Fang H, et al. CCLasso: correlation inference for compositional data through Lasso. Bioinformatics. 2015;31(19):3172–3180. doi:10.1093/bioinformatics/btv349.
- Biswas S Mcdonald M, Lundberg DS, Dangl JL, Jojic V. Learning microbial interaction networks from metagenomic count data. J Comput Biol. 2016;23(6):526–535. doi:10.1089/cmb.2016.0061.
- Fang H, Huang C, Zhao H, Deng M. gCoda: conditional dependence network inference for compositional data. J Comput Biol. 2017;24(7):699–708. doi:10.1089/cmb.2017.0054.
- Morton JT, Aksenov AA, Nothias LF, Foulds JR, Quinn RA, Badri MH, Swenson TL, Van Goethem MW, Northen TR, Vazquez-Baeza Y, et al. Learning representations of microbe-metabolite interactions. Nat Methods. 2019;16(12):1306–1314. doi:10.1038/s41592-019-0616-3.
- Stein RR, Bucci V, Toussaint NC, Buffie CG, Rätsch G, Pamer EG, Sander C, Xavier JB. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput Biol. 2013;9(12):e1003388. doi:10.1371/journal.pcbi.1003388.
- Schluter J, Peled JU, Taylor BP, Markey KA, Smith M, Taur Y, Niehus R, Staffas A, Dai A, Fontana E, et al. The gut microbiota is associated with immune cell dynamics in humans. Nature. 2020;588(7837):303–307. doi:10.1038/s41586-020-2971-8.
- Rao C, Coyte KZ, Bainter W, Geha RS, Martin CR, Rakoff-Nahoum S. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature. 2021;591(7851):633–638. doi:10.1038/s41586-021-03241-8.
- Erhard F. Estimating pseudocounts and fold changes for digital expression measurements. Bioinformatics. 2018;34(23):4054–4063. doi:10.1093/bioinformatics/bty471.
- Brotons P, Bassat Q, Lanaspa M, Henares D, Perez-Arguello A, Madrid L, Balcells R, Acacio S, Andres-Franch M, Marcos MA, et al. Nasopharyngeal bacterial load as a marker for rapid and easy diagnosis of invasive pneumococcal disease in children from Mozambique. PLoS One. 2017;12(9):e0184762. doi:10.1371/journal.pone.0184762.
- Vandeputte D, Kathagen G, D’hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551(7681):507–511. doi:10.1038/nature24460.
- Hirten RP, Grinspan A, Fu S-C, Luo Y, Suarez-Farinas M, Rowland J, Contijoch EJ, Mogno I, Yang N, Luong T, et al. Microbial engraftment and efficacy of fecal microbiota transplant for clostridium difficile in patients with and without inflammatory bowel disease. Inflamm Bowel Dis. 2019;25(6):969–979. doi:10.1093/ibd/izy398.
- Grinspan A. FMT for severe C.difficile infection: if at first you do no harm, the second time’s a charm. Dig Dis Sci. 2020;66:5–6.
- Eiseman B, Silen W, Bascom GS, Kauvar AJ. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery. 1958;44(5):854–859.
- Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2012;9(2):88–96. doi:10.1038/nrgastro.2011.244.
- Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Armstrong D, Marshall JK, Kassam Z, Reinisch W, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149(1):102–109.e6. doi:10.1053/j.gastro.2015.04.001.
- Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, Leong RWL, Connor S, Ng W, Paramsothy R, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389(10075):1218–1228. doi:10.1016/S0140-6736(17)30182-4.
- DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, Turbett S, Chung RT, Chen Y-B, Hohmann EL, et al. Drug-resistant E.coli bacteremia transmitted by fecal microbiota transplant. New England Journal of Medicine. 2019;381(21):2043–2050. doi:10.1056/NEJMoa1910437.
- Kelly CR, Yen EF, Grinspan AM, Kahn SA, Atreja A, Lewis JD, Moore TA, Rubin DT, Kim AM, Serra S, et al. Fecal microbiota transplantation is highly effective in Real-world practice: initial results from the FMT national registry. Gastroenterology. 2021;160(1):183–+. doi:10.1053/j.gastro.2020.09.038.
- Youngster I, Russell GH, Pindar C, Ziv-Baran T, Sauk J, Hohmann EL. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA. 2014;312(17):1772–1778. doi:10.1001/jama.2014.13875.
- Du C, Luo Y, Walsh S, Grinspan A. Oral fecal microbiota transplant capsules are safe and effective for recurrent Clostridioides difficile infection a systematic review and meta-analysis. J Clin Gastroenterol. 2021;55(4):300–308. doi:10.1097/MCG.0000000000001495.
- Henn MR, O’Brien EJ, Diao L, Feagan BG, Sandborn WJ, Huttenhower C, Wortman JR, McGovern BH, Wang-Weigand S, Lichter DI, et al. A phase 1b safety study of SER-287, a spore-based microbiome therapeutic, for active mild to moderate ulcerative colitis. Gastroenterology. 2021;160(1):115–127.e30. doi:10.1053/j.gastro.2020.07.048.
- Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, Leong RWL, Connor S, Ng W, Paramsothy R, Xuan W, et al. Multi-donor intense faecal microbiota transplantation is an effective treatment for resistant ulcerative colitis: a randomised placebo-controlled trial. Journal of Crohns & Colitis. 2016;10:S14–S14.
- Stein RR, Tanoue T, Szabady RL, BhattaraiSK, Olle B, Norman JM, Suda W, Oshima K, Hattori M, Gerber GK, et al. Computer-guided design of optimal microbial consortia for immune system modulation. eLife. 2018;7:e30916.
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–208. doi:10.1038/nature13828.
- van der Lelie D, Oka A, Taghavi S, Umeno J, Fan T-J, Merrell KE, Watson SD, Ouellette L, Liu B, Awoniyi M, et al. Rationally designed bacterial consortia to treat chronic immune-mediated colitis and restore intestinal homeostasis. Nat Commun. 2021;12(1):3105. doi:10.1038/s41467-021-23460-x.
- Cook DP, Gysemans C, Mathieu C. Lactococcus lactis as a versatile Vehicle for Tolerogenic Immunotherapy. Front Immunol. 2018;8(1961). doi:10.3389/fimmu.2017.01961.
- Ramachandran G, Bikard D. Editing the microbiome the CRISPR way. Philos Trans R Soc Lond B Biol Sci. 2019;374(1772):20180103. doi:10.1098/rstb.2018.0103.
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini, LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–239. doi:10.1038/nbt.2508.
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41(7):4336–4343. doi:10.1093/nar/gkt135.
- Pires DP, Cleto S, Sillankorva S, Azeredo J, Lu, TK. Genetically engineered phages: a review of advances over the last decade. Microbiology and Molecular Biology Reviews. 2016;80(3):523–543. doi:10.1128/MMBR.00069-15.
- Yan MY, Yan HQ, Ren GX, Zhao JP, Guo XP, Sun YC . .CRISPR-Cas12a-Assisted Recombineering in Bacteria. Appl Environ Microbiol. 2017 ;83(17):e00947–17.doi:10.1128/AEM.00947-17.
- Zheng L, Tan Y, Hu Y, Shen J, Qu Z, Chen X, Ho CL, Leung ELH, Zhao W, Dai L, et al. CRISPR/Cas-Based genome editing for human gut commensal Bacteroides species. ACS Synth Biol. 2022;11(1):464–472. doi:10.1021/acssynbio.1c00543.
- Roda G, Jharap B, Neeraj N, Colombel JF .Loss of response to anti-TNFs: definition, epidemiology, and management. Clin Transl Gastroenterol. 2016;7(1):e135. doi:10.1038/ctg.2015.63.
- Yanai H, Hanauer SB. Assessing response and loss of response to biological therapies in IBD. Am J Gastroenterol. 2011;106(4):685–698. doi:10.1038/ajg.2011.103.
- Steidler L, Neirynck S, Huyghebaert N, Snoeck V, Vermeire A, Goddeeris B, Cox E, Remon JP, Remaut E. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol. 2003;21(7):785–789. doi:10.1038/nbt840.
- Braat H, Rottiers P, Hommes DW, Huyghebaert N, Remaut E, Remon J, van Deventer SJH, Neirynck S, Peppelenbosch MP, Steidler L, et al. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin Gastroenterol Hepatol. 2006;4(6):754–759. doi:10.1016/j.cgh.2006.03.028.
- Foligne B, Dessein R, Marceau M, Poiret S, Chamaillard M, Pot B, Simonet M, Daniel C . Prevention and treatment of colitis with Lactococcus lactis secreting the immunomodulatory Yersinia LcrV protein. Gastroenterology. 2007;133(3):862–874. doi:10.1053/j.gastro.2007.06.018.
- Vandenbroucke K, de Haard H, Beirnaert E, Dreier T, Lauwereys M, Huyck L, Van Huysse J, Demetter P, Steidler L, Remaut E, et al. Orally administered L.lactis secreting an anti-TNF nanobody demonstrate efficacy in chronic colitis. Mucosal Immunol. 2010;3(1):49–56. doi:10.1038/mi.2009.116.
- Zhou Y, Zhi F. Lower level of bacteroides in the gut microbiota is associated with inflammatory bowel disease: a meta-Analysis. Biomed Res Int. 2016;2016:5828959. doi:10.1155/2016/5828959.
- Kino Y, Nakayama-Imaohji H, Fujita M, Tada A, Yoneda S, Murakami K, Hashimoto M, Hayashi T, Okazaki K, Kuwahara T, et al. Counterselection employing mutated pheS for markerless genetic deletion in Bacteroides species. Anaerobe. 2016;42:81–88. doi:10.1016/j.anaerobe.2016.09.004.
- García-Bayona L, Comstock LE. Streamlined genetic manipulation of diverse Bacteroides and Parabacteroides isolates from the human gut microbiota. mBio. 2019;10(4). doi:10.1128/mBio.01762-19.
- Bencivenga-Barry NA, Lim B, Herrera C, Trent MS, Goodman AL. Genetic manipulation of wild human gut Bacteroides. J Bacteriol. 2020;202(3). doi:10.1128/JB.00544-19.
- Vandenbroucke K, Hans W, Van Huysse J, Neirynck S, Demetter P, Remaut E, Rottiers P, Steidler L . Active delivery of trefoil factors by genetically modified Lactococcus lactis prevents and heals acute colitis in mice. Gastroenterology. 2004;127(2):502–513. doi:10.1053/j.gastro.2004.05.020.
- Praveschotinunt P, Duraj-Thatte AM, Gelfat I, Bahl F, Chou DB, Joshi NS . Engineered E.coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat Commun. 2019;10(1):5580. doi:10.1038/s41467-019-13336-6.
- Del Carmen S, Zurita-Turk M, Lima FA, Dos Santos JSC, Leclercq SY, Chatel J-M, Azevedo V, De Moreno De Leblanc A, Miyoshi A, Leblanc JG, et al. A novel interleukin-10 DNA mucosal delivery system attenuates intestinal inflammation in a mouse model. European Journal of Inflammation. 2013;11(3):641–654. doi:10.1177/1721727X1301100308.
- Zurita-Turk M, Del Carmen S, Santos AC, Pereira VB, Cara DC, Leclercq SY, de Leblanc AD, Azevedo V, Chatel J-M, LeBlanc JG, et al. Lactococcus lactis carrying the pValac DNA expression vector coding for IL-10 reduces inflammation in a murine model of experimental colitis. BMC Biotechnol. 2014;14(1):73. doi:10.1186/1472-6750-14-73.
- Zurita-Turk M, Souza BM, de Castro CP, Pereira VB, da Cunha VP, Preisser TM, de Faria AMC, Machado DCC, Miyoshi A . Attenuation of intestinal inflammation in IL-10 deficient mice by a plasmid carrying Lactococcus lactis strain. BMC Biotechnol. 2020;20(1):38. doi:10.1186/s12896-020-00631-0.
- Ferenczi S, Solymosi N, Horváth I, Szeőcs N, Grózer Z, Kuti D, Juhász B, Winkler Z, Pankotai T, Sükösd F. Efficient treatment of a preclinical inflammatory bowel disease model with engineered bacteria. Mol Ther Methods Clin Dev. 2021;20:218–226. doi:10.1016/j.omtm.2020.11.010.
- Reinders CI, Herulf M, Ljung T, Hollenberg J, Weitzberg E, Lundberg JO, Hellström PM . Rectal mucosal nitric oxide in differentiation of inflammatory bowel disease and irritable bowel syndrome. Clin Gastroenterol Hepatol. 2005;3(8):777–783. doi:10.1016/S1542-3565(05)00182-5.
- McKay R, Hauk P, Quan D, Bentley WE. Development of cell-based sentinels for nitric oxide: ensuring marker expression and unimodality. ACS Synth Biol. 2018;7(7):1694–1701. doi:10.1021/acssynbio.8b00146.
- Hughes ER, Winter MG, Duerkop BA, Spiga L, Furtado de Carvalho T, Zhu W, Gillis CC, Büttner L, Smoot MP, Behrendt CL, et al. Microbial respiration and formate oxidation as metabolic signatures of inflammation-Associated dysbiosis. Cell Host Microbe. 2017;21(2):208–219. doi:10.1016/j.chom.2017.01.005.
- Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Büttner L, de Lima Romão E, Behrendt CL, Lopez CA, et al. Precision editing of the gut microbiota ameliorates colitis. Nature. 2018;553(7687):208–211. doi:10.1038/nature25172.
- Dubos RJ, Straus JH, Pierce C. The multiplication of bacteriophage in vivo and its protective effect against an experimental infection with Shigella dysenteriae. J Exp Med. 1943;78(3):161–168. doi:10.1084/jem.78.3.161.
- Kutter E, De Vos D, Gvasalia G, Alavidze Z, Gogokhia L, Kuhl S, Abedon ST. Phage therapy in clinical practice: treatment of human infections. Curr Pharm Biotechnol. 2010;11(1):69–86. doi:10.2174/138920110790725401.
- Sulakvelidze A, Alavidze Z, Morris JG. Bacteriophage Therapy. Antimicrob Agents Chemother. 2001;45(3):649–659. doi:10.1128/AAC.45.3.649-659.2001.
- Wright A, Hawkins CH, Änggård EE, Harper DR. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin Otolaryngol. 2009;34(4):349–357. doi:10.1111/j.1749-4486.2009.01973.x.
- Tsonos J, Vandenheuvel D, Briers Y, De Greve H, Hernalsteens J-P, Lavigne R. Hurdles in bacteriophage therapy: deconstructing the parameters. Vet Microbiol. 2014;171(3–4):460–469. doi:10.1016/j.vetmic.2013.11.001.
- Bragg R, van der Westhuizen W, Lee JY, Coetsee E, Boucher C. Bacteriophages as potential treatment option for antibiotic resistant bacteria. Adv Exp Med Biol. 2014;807:97–110 doi:10.1007/978-81-322-1777-0_7.
- Gordillo Altamirano FL, Barr JJ. Phage therapy in the postantibiotic era. Clin Microbiol Rev. 2019;32(2). doi:10.1128/CMR.00066-18.
- Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013;13(12):1057–1098. doi:10.1016/S1473-3099(13)70318-9.
- Sabino J, Hirten RP, Colombel JF. Review article: bacteriophages in gastroenterology-from biology to clinical applications. Aliment Pharmacol Ther. 2020;51(1):53–63. doi:10.1111/apt.15557.
- Gabisoniya TG, Loladze MZh, Nadiradze, MM, Chakhunashvili NK, Alibegashvili MG, Tamarashvili NG, Pushkina VA . [Effects of bacteriophages on biofilm formation by strains of Pseudomonas aeruginosa]. Prikl Biokhim Mikrobiol. 2016;52(3):312–317. 29509387
- Motlagh AM, Bhattacharjee AS, Goel R. Biofilm control with natural and genetically-modified phages. World J Microbiol Biotechnol. 2016;32(4):67. doi:10.1007/s11274-016-2009-4.
- Jault P, Leclerc T, Jennes S, Pirnay JP, Que Y-A, Resch G, Rousseau AF, Ravat F, Carsin H, Le Floch R, et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect Dis. 2019;19(1):35–45. doi:10.1016/S1473-3099(18)30482-1.
- Gindin M, Febvre HP, Rao S, Wallace TC, Weir TL . Bacteriophage for gastrointestinal health (PHAGE) study: evaluating the safety and tolerability of supplemental bacteriophage consumption. J Am Coll Nutr. 2019;38(1):68–75. doi:10.1080/07315724.2018.1483783.
- LaVergne S, Hamilton T, Biswas B, Kumaraswamy M, Schooley RT, Wooten D . Phage therapy for a multidrug-Resistant Acinetobacter baumannii craniectomy site infection. Open Forum Infect Dis. 2018;5(4):ofy064. doi:10.1093/ofid/ofy064.
- Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, Barr J, Reed S, Rohwer F, Benler S . Development and use of personalized bacteriophage-Based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii. Infection Antimicrobial Agents and Chemotherapy. 2017;61(10):e00954–17 doi:10.1128/AAC.00954-17.
- Aslam S, Pretorius V, Lehman SM, Morales S, Schooley RT . Novel bacteriophage therapy for treatment of left ventricular assist device infection. The Journal of Heart and Lung Transplantation. 2019;38(4):475–476. doi:10.1016/j.healun.2019.01.001.
- Galtier M, De Sordi L, Sivignon A, de Vallée A, Maura D, Neut C, Rahmouni O, Wannerberger K, Darfeuille-Michaud A, Desreumaux P, et al. Bacteriophages targeting adherent invasive Escherichia coli strains as a promising new treatment for crohn’s disease. J Crohns Colitis. 2017;11(7):840–847. doi:10.1093/ecco-jcc/jjw224.
- Cepko LCS, Garling EE, Dinsdale MJ, Scott WP, Bandy L, Nice T, Faber-Hammond J, Mellies JL . Myoviridae phage PDX kills enteroaggregative Escherichia coli without human microbiome dysbiosis. J Med Microbiol. 2020;69(2):309–323. doi:10.1099/jmm.0.001162.
- Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, et al. Ectopic colonization of oral bacteria in the intestine drives T 1 cell induction and inflammation. Science. 2017;358(6361):359–365. doi:10.1126/science.aan4526.
- Nakamoto N, Sasaki N, Aoki R, Miyamoto K, Suda W, Teratani T, Suzuki T, Koda Y, Chu P-S, Taniki N, et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nature Microbiology. 2019;4(3):492–503. doi:10.1038/s41564-018-0333-1.
- Broecker F, Klumpp J, Moelling K. Long-term microbiota and virome in a Zürich patient after fecal transplantation against Clostridium difficile infection. Ann N Y Acad Sci. 2016;1372(1):29–41. doi:10.1111/nyas.13100.
- Chehoud C, Dryga A, Hwang Y, Nagy-Szakal D, Hollister EB, Luna RA, Versalovic J, Kellermayer R, Bushman FD. Transfer of viral communities between human individuals during fecal microbiota transplantation. mBio. 2016;7(2):e00322–16. doi:10.1128/mBio.00322-16.
- Ott SJ, Waetzig GH, Rehman A, Moltzau-Anderson J, Bharti R, Grasis JA, Cassidy L, Tholey A, Fickenscher H, Seegert D, et al. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology. 2017;152(4):799–811.e7. doi:10.1053/j.gastro.2016.11.010.
- Szermer-Olearnik B, Boratyński J, Skurnik M. Removal of endotoxins from bacteriophage preparations by extraction with organic solvents. PLoS One. 2015;10(3):e0122672. doi:10.1371/journal.pone.0122672.
- Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016;8(1):39. doi:10.1186/s13073-016-0294-z.
- Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R. Systematic discovery of antiphage defense systems in the microbial pangenome. Science. 2018;359(6379):eaar4120. doi:10.1126/science.aar4120.
- Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2011;33(1):43–51. doi:10.1002/bies.201000071.
- Torres-Barceló C, Hochberg ME. Evolutionary rationale for phages as complements of antibiotics. Trends Microbiol. 2016;24(4):249–256. doi:10.1016/j.tim.2015.12.011.
- Hatzenpichler R, Krukenberg V, Spietz RL, Jay ZJ. Next-generation physiology approaches to study microbiome function at single cell level. Nat Rev Microbiol. 2020;18(4):241–256. doi:10.1038/s41579-020-0323-1.
- Welch JLM, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A. 2016;113(6):E791–E800. doi:10.1073/pnas.1522149113.
- Kim D, Barraza JP, Arthur RA, Hara A, Lewis K, Liu Y, Scisci EL, Hajishengallis E, Whiteley M, Koo H, et al. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc Natl Acad Sci U S A. 2020;117(22):12375–12386. doi:10.1073/pnas.1919099117.
- Carrow HC, Batachari LE, Chu HT. Strain diversity in the microbiome: lessons from bacteroides fragilis. PLoS Pathog. 2020;16(12):e1009056. doi:10.1371/journal.ppat.1009056.
- Yan Y, Nguyen LH, Franzosa EA, Huttenhower C. Strain-level epidemiology of microbial communities and the human microbiome. Genome Med. 2020;12(1). doi:10.1186/s13073-020-00765-y.
- Thomas AM, Segata N. Multiple levels of the unknown in microbiome research. BMC Biol. 2019;17(1):17. doi:10.1186/s12915-019-0636-6.
- Zou YQ, Xue W, Luo G, Deng Z, Qin P, Guo R, Sun H, Xia Y, Liang S, Dai Y, et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol. 2019;37(2):179–+. doi:10.1038/s41587-018-0008-8.
- Stierle AA, Stierle D, Decato D, Priestley N, Alverson J, Hoody J, McGrath K, Klepacki D. The berkeleylactones, antibiotic macrolides from fungal coculture. J Nat Prod. 2017;80(4):1150–1160. doi:10.1021/acs.jnatprod.7b00133.
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi:10.1038/nature12331.
