ABSTRACT
Colonic luminal aromatic amines have been historically considered to be derived from dietary source, especially fermented foods; however, recent studies indicate that the gut microbiota serves as an alternative source of these amines. Herein, we show that five prominent genera of Firmicutes (Blautia, Clostridium, Enterococcus, Ruminococcus, and Tyzzerella) have the ability to abundantly produce aromatic amines through the action of aromatic amino acid decarboxylase (AADC). In vitro cultivation of human fecal samples revealed that a significant positive correlation between aadc copy number of Ruminococcus gnavus and phenylethylamine (PEA) production. Furthermore, using genetically engineered Enterococcus faecalis-colonized BALB/cCrSlc mouse model, we showed that the gut bacterial aadc stimulates the production of colonic serotonin, which is reportedly involved in osteoporosis and irritable bowel syndrome. Finally, we showed that human AADC inhibitors carbidopa and benserazide inhibit PEA production in En. faecalis.
Introduction
The effects of gut bacterial metabolites on host health have been demonstrated in rodent models. For example, acetic acid protects the host against pathogen infectionCitation1, butyric acid induces the colonic regulatory T cell differentiation,Citation2 deoxycholic acid increases liver cancer incidence,Citation3 lithocholic acid ameliorates inflammation in colitis,Citation4 and polyamines extend host longevity,Citation5,Citation6 and maintain mucosal homeostasis.Citation7 The importance of gut bacterial metabolites is also recognized in humans has also been reported, e.g., de novo biosynthesis of vitamin K (menaquinones), which humans cannot biosynthesize, in gut.Citation8 Moreover, polyamines produced by gut bacteria contribute to atherosclerosis prevention.Citation9 Considering these findings, the regulation of gut bacterial metabolite production in the human is crucial for the maintenance of nutritional homeostasis and diseases prevention in humans. The molecular mechanism of gut bacterial metabolites production and their physiological function in the host have been gradually elucidated. For example, aromatic lactic acid produced by aromatic lactate dehydrogenase of Bifidobacterium affects the host immune system,Citation10 and isoallolithocholic acid produced by 5α-reductase and 3β-hydroxysteroid dehydrogenase of Parabacteroides merdae St3 through shows bactericidal activity against gram-positive pathogens.Citation11 However, our knowledge about the sources of gut bacterial metabolites and their production mechanism and physiological effects on host remains fragmentary.
Aromatic amines are among the compounds detected in the colon;Citation12,Citation13 tryptamine, tyramine, and phenylethylamine (PEA) are typically known as the trace amines that can affect neurotransmission even in small amounts.Citation14,Citation15 Diet and gut microbiota have been identified as sources of colonic luminal aromatic amines. Aromatic amines are found in fermented foods,Citation16 nuts, and citrus fruitsCitation17 as well as in roasted coffee and cacao, where they are generated by Maillard- and Strecker reactions.Citation17–19 Several previous studies have reported that some gut bacteria possess aromatic amine-producing capabilitiesCitation20 and contribute to colonic aromatic amine level using germ-free animal models such as rats, chickens, and mice.Citation21–24 Previous in vitro studies have revealed that gut bacteria produce aromatic amines from aromatic amino acids through decarboxylation catalyzed by aromatic amino acid decarboxylase (AADC).Citation23,Citation25 The kinetic parameters of AADC from several bacterial species, including gut bacteria (such as Enterococcus faecalis, Clostridium sporogenes, and Ruminococcus gnavus), have been determined using purified recombinant enzymes.Citation25,Citation26 Almost all characterized AADC has shown relatively broad substrate specificity,Citation25,Citation27 catalyzing the decarboxylation of not only proteinogenic aromatic amino acids (Phe, Tyr, and Trp), but also non-proteinogenic aromatic amino acids such as 3,4-dihydroxyphenylalanine (L-DOPA)Citation28–31 and 5-hydroxytryptophan.Citation27 Most previous studies on aromatic amino acid decarboxylation and aromatic amine production during the cultivation were focused on aromatic amine production from a non-proteinogenic aromatic amino acid L-DOPA.Citation29,Citation32 Gut bacteria produce biogenic amines, including aromatic amines, from proteinogenic amino acids abundant in the diet,Citation33,Citation34 such as tryptamine synthesis through the decarboxylation of tryptophan by gut bacteria.Citation25 However, few studies have elucidated at the genetic level how gut bacteria contribute to the production of trace amines from proteinogenic aromatic amino acids in the colon, and further research is needed to elucidate it.
PEA has been shown to induce efflux of neurotransmitters (dopamine, norepinephrine, and serotonin) and inhibit uptake of these neurotransmitters in human cell lines and in brain synaptosomes from mouse, juvenile rhesus, and tamarin.Citation35 PEA activates polymorphonuclear leukocytes and induces allergic reactions.Citation36 These physiological effects of PEA are mediated by trace amine-associated receptor 1 (TAAR1) activation.Citation35,Citation36 Because TAAR1 is also expressed in the colonic epithelium,Citation37 PEA from gut bacteria is expected to exert physiological functions through TAAR1.Citation38 However, no studies have been conducted to investigate this possibility.
Serotonin (5-hydroxytryptamine) is a monoamine neurotransmitter distributed in the central and peripheral nervous systems, and plays different roles depending on location. Serotonin in the central nervous system is a neurotransmitter in the brain and affects sleepCitation39 and appetite,Citation40 whereas peripheral serotonin is a regulatory factor in different organs, regulating bone development,Citation41,Citation42 immune response,Citation43 and brown adipose tissue thermogenesis.Citation44 Peripheral serotonin, which accounts for 90% of serotonin in the body, is produced by enterochromaffin (EC) cells in the gastrointestinal tract.Citation45 Bhattarai et al. demonstrated that Bacteroides thetaiotaomicron heterologously expressing aadc of R. gnavus produced tryptamine in the mouse gut, and the produced tryptamine increased anion and fluid secretion in the proximal colon via Serotonin receptor-4, one of the G protein-coupled receptors (GPCRs).Citation23 Gut bacteria modulate gastrointestinal motility and platelet function by promoting peripheral serotonin production from EC cells, mediated by tyramine and other gut bacterial metabolites.Citation24 Therefore, aromatic amines produced by gut bacteria have a significant effect on host physiology, by way of serotoninCitation24 or serotonin signaling pathways.Citation23,Citation46 However, the serotonin-mediated relationship between the host physiology and PEA, another natural aromatic amine, remains to be studied.
In this study, we identified five species of PEA-producing gut bacteria among 32 species of dominant human gut bacteria and verified that PEA production depends on aadc. The effects of aadc on colonic luminal aromatic amine and colonic serotonin production in the host were evaluated using mouse model. In addition, the PEA production by En. faecalis was successfully inhibited using established inhibitors.
Results
Discovery of the PEA-producing bacteria in the most predominant species in the human indigenous gut microbiota
Recently, we reported that 32 of the most predominant species of human indigenous gut microbiota were culturable in Gifu anaerobic medium (GAM)Citation47 and evaluated polyamine biosynthesis and transport using this system.Citation48 Reanalysis of the high-performance liquid chromatography (HPLC) chromatograms obtained in the polyamine study revealed that Blautia hansenii, Clostridium asparagiforme, Tyzzerella nexilis, En. faecalis, and R. gnavus produced unidentified biogenic amine in the culture supernatant ( and Supplementary Figure S1). The retention time of the unidentified biogenic amine did not correspond to that of polyamines (putrescine, cadaverine, spermidine, spermine, and agmatine) (). To identify the unidentified biogenic amine, we purified this compound using an ion exchange chromatography from the culture supernatant of T. nexilis. The MS/MS spectra of the purified biogenic amine corresponded to that of PEA (). The retention time of the purified unidentified biogenic amine corresponded to PEA standard sample, in two different HPLC systems (). These results indicated that the unidentified biogenic amine is PEA.
Figure 1. Identification of PEA and PEA-producing gut bacteria. (a) Unidentified biogenic amine (x) observed in B. hansenii, C. asparagiforme, T. nexilis, En. faecalis, and R. gnavus. The upper panel shows the HPLC chromatogram of culture supernatant of T. nexilis as representative data, and the lower and middle panels are chromatograms of polyamines (PAs) standard (Put, putrescine; Cad, cadaverine; Spd, spermidine; Spm, spermine) and agmatine (Agm) standard, respectively. *Trace amounts of putrescine were present as a contaminant in the agmatine standard reagent. (b) Comparison of MS/MS spectra of PEA standard and unidentified biogenic amine purified from culture supernatant of T. nexilis. (c and d) Comparison of chromatograms of PEA standard and unidentified biogenic amine purified from culture supernatant of T. nexilis: (c) Chromatograms obtained by reverse-phase HPLC (d) Chromatograms obtained by cation exchange HPLC. (e) PEA concentration in the culture supernatant of 32 species of the GAM culturable, dominant human gut bacteria.Citation47 White and gray bars indicate PEA concentration in the culture supernatant in the growing and stationary phase, respectively. Data represent the mean ± SD of three individual experiments. See also Supplementary Figure S1.
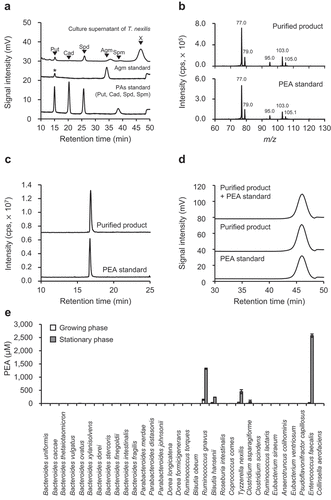
Five species of the human dominant gut bacteria (B. hansenii, C. asparagiforme, T. nexilis, En. faecalis, and R. gnavus) produced PEA in their culture supernatants. The PEA concentrations in the culture supernatants in stationary phase were 240 μM for B. hansenii, 74 μM for C. asparagiforme, 447 μM for T. nexilis, 2,572 μM for En. faecalis, and 1,317 μM for R. gnavus ().
Production of aromatic amines by PEA-producing gut bacteria in aromatic-amino-acids-defined medium
It was reported that PEA was biosynthesized from Phe in the reaction catalyzed by AADC, a pyridoxal-5′-phosphate-dependent decarboxylase.Citation25,Citation27 AADC decarboxylates not only Phe, but also other proteinogenic aromatic amino acids (Tyr and Trp)Citation25,Citation27 (). The PEA-producing gut bacteria (B. hansenii, C. asparagiforme, T. nexilis, En. faecalis, and R. gnavus) possess the potential for the production of tyramine and tryptamine (). As the concentrations of the substrates of AADC: Phe, Tyr, and Trp, in GAM were different from each other (Supplementary Table S1), the production ability of each aromatic amine could not be quantitatively compared based on the concentration of PEA, tyramine, and tryptamine in the culture supernatant. Therefore, an aromatic-amino-acid-defined (AAAD) medium, where the concentrations of aromatic amino acids were adjusted to 1 mM, was prepared (Supplementary Table S2). The concentrations of the corresponding aromatic amines, PEA, tyramine, and tryptamine, in the culture supernatant of PEA-producing gut bacteria were analyzed.
Figure 2. Aromatic amine production profile of identified PEA-producing gut bacteria. (a) Aromatic amino acid decarboxylase (AADC) decarboxylates proteinogenic aromatic amino acids (Tyr, Phe, and Trp) and generates corresponding aromatic amines (tyramine, PEA, and tryptamine). (b-f) Aromatic amine concentration in the culture supernatants of PEA-producing gut bacteria grown in AAAD medium: (b) Blautia hansenii (c) Clostridium asparagiforme (d) Enterococcus faecalis (e) Ruminococcus gnavus (f) Tyzzerlla nexilis. Green, red, blue circles indicate tyramine, PEA, and tryptamine concentrations in the culture supernatants, respectively. The “Production” value on the vertical axis was calculated by subtracting the amount of each aromatic amine originally contained in AAAD medium from the amount of aromatic amine at each time point. Data represent the mean ± SD of three or four individual experiments. The statistical significance of the PEA, tyramine, and tryptamine production at 72 h was determined using the one-way ANOVA post-hoc Tukey-Kramer test, and the p-values for the Tukey-Kramer test are shown. See also Supplementary Figure S2 and Supplementary Table S3.
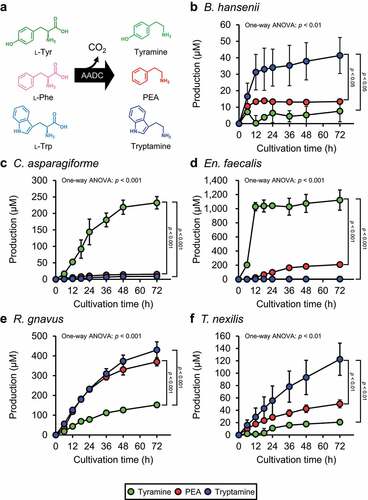
All tested PEA-producing gut bacteria could grow in the AAAD medium (Supplementary Figure S2) and PEA was detected in the culture supernatants of four species (B. hansenii, En. faecalis, R. gnavus, and T. nexilis) (). Aromatic amine concentrations at all measured cultivation times were analyzed by repeated measures one-way ANOVA (Supplementary Table S3). For those aromatic amines with significantly different concentrations, values at 0 h and 72 h after inoculation were subjected to a post-hoc Tukey-Kramer test to verify whether they were significantly increased by the cultivation of the bacteria. Statistically significant aromatic amine production was observed, except for tyramine production by B. hasenii and tryptamine production by En. faecalis and C. asparagiforme ( and Supplementary Table S3). Production of PEA in the culture supernatants of PEA-producing gut bacteria grown in AAAD medium was reached the maximum at 72 h after inoculation; the concentrations were 13 μM for B. hansenii, 15 μM for C. asparagiforme, 208 μM for En. faecalis, 370 μM for R. gnavus, and 50 μM for T. nexilis, (). Then, one-way ANOVA and post-hoc Tukey-Kramer test () were performed to compare the concentrations of the three aromatic amines in each species after 72 h of cultivation. In B. hansenii, the concentration of tryptamine was significantly higher than that of tyramine and PEA, with no significant difference between the concentrations of PEA and tyramine (). In C. asparagiforme, the concentration of tyramine was significantly higher than that of PEA and tryptamine, with no significant difference between the concentrations of PEA and tryptamine (). In En. faecalis, the concentration of tyramine was significantly higher than that of PEA and tryptamine, with no significant difference between the concentrations of PEA and tryptamine (). Although there was no significant difference between the production of PEA and tryptamine in En. faecalis, the p-value obtained in the Tukey-Kramer test was 0.109, indicating a tendency toward higher PEA production than tryptamine (). In R. gnavus, the concentration of tryptamine and PEA were significantly higher than that of tyramine, with no significant difference between the concentrations of PEA and tryptamine (). In T. nexilis, the concentration of tryptamine was significantly higher than that of PEA and tyramine, with no significant difference between the concentrations of PEA and tyramine ().
Heterologous expression of aadc and aadc homologs
Recombinant AADC proteins of R. gnavus (AADCRg) and En. faecalis (AADCEf) synthesize aromatic amines from aromatic amino acids, in vitro.Citation25,Citation26 The AADC of other three PEA-producing gut bacteria (B. hansenii, C. asparagiforme, and T. nexilis) were not identified experimentally; however, BLASTP analysisCitation49 using AADCRg as the query protein showed that AADC homologs were present in B. hansenii (BLAHAN_06497, hereafter referred to as aadcBh), C. asparagiforme (CLOSTASPAR_05940, hereafter referred to as aadcCa), and T. nexilis (CLONEX_01451, hereafter referred to as aadcTn). To determine the productivity of aromatic amine by AADC and AADC homologs of PEA-producing gut bacteria, the genes or its homologs encoding AADCs: aadcRg, aadcEf, aadcBh, aadcCa, and aadcTn were cloned into overexpression vectors and introduced into E. coli and the concentration of aromatic amines in the culture supernatants of E. coli strains were measured.
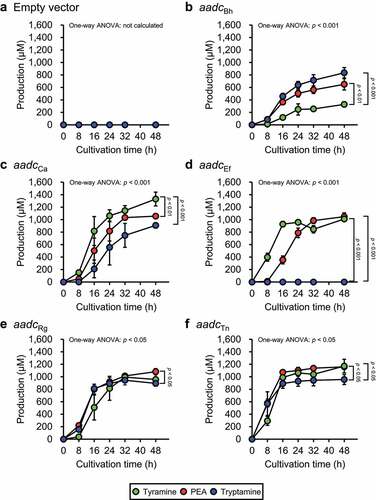
E. coli harboring the empty vector (YS297) did not produce any of the aromatic amines (), whereas the E. coli strains heterologously expressing the aadc and aadc homologs produced more than 600 μM PEA, 800 μM tryptamine, and 300 μM tyramine at 48 h after inoculation (), confirming that the production of aromatic amines was due to the heterologous expression of the aadc and aadc homologs. Aromatic amine concentrations at all measured cultivation times were analyzed by repeated measures one-way ANOVA (Supplementary Table S4). For the aromatic amines with significantly different concentrations, values at 0 h and 48 h after inoculation were subjected to a post-hoc Tukey-Kramer test to verify whether they were significantly increased by the cultivation of the bacteria. Statistically significant aromatic amine production was observed, except for tryptamine production by YS317 harboring aadcEf ( and Supplementary Table S4). One-way ANOVA and post-hoc Tukey-Kramer test () were performed to compare the concentrations of the three aromatic amines in the culture supernatant of each E. coli strain after 48 h of cultivation. In YS389 harboring aadcBh, the concentration of tryptamine and PEA was significantly higher than that of tyramine (). In contrast to the results obtained from B. hansenii cultured in AAAD medium (), the results obtained from YS389 showed a significantly higher concentration of PEA than tyramine at the end of cultivation (). In YS300 harboring aadcCa, the concentration of tyramine was significantly higher than that of PEA and tryptamine (). Similar results were obtained when C. asparagiforme was cultured in AAAD medium (). YS389, YS300, YS298, and YS299 harboring aadcBh (), aadcCa (), aadcRg (), and aadcTn (), respectively, produced tryptamine, whereas YS317 harboring aadcEf did not produce tryptamine during the cultivation period. In contrast to the culture supernatant of En. faecalis in AAAD medium (), the culture supernatant of YS317 harboring aadcEf showed no significant difference in the final concentrations of PEA and tyramine, although the concentration of PEA reached its maximum after tyramine (). In YS298 harboring aadcRg, the concentration of PEA was significantly higher than that of tryptamine (). The concentrations of tryptamine and tyramine were significantly different in R. gnavus cultivated in AAAD medium () but not in YS298 (). In contrast to T. nexilis cultivated in AAAD medium (), the culture supernatant of YS299 harboring aadcTn contained a significantly higher concentration of PEA and tyramine than tryptamine ().
Figure 3. Aromatic amine concentration in the culture supernatants of E. coli harboring aadc or aadc candidate gene of PEA-producing gut bacteria. E. coli transformants were grown in M9AAA-medium and the aromatic amine concentrations in the culture supernatants were measured: (a) YS297 (empty vector), (b) YS389 (expressing aadcBh), (c) YS300 (expressing aadcCa), (d) YS317 (expressing aadcEf), (e) YS298 (expressing aadcRg), and (f) YS299 (expressing aadcTn). Green, red, and blue circles indicate tyramine, PEA, and tryptamine concentrations in the culture supernatants, respectively. Data represent the mean ± SD of three individual experiments. The statistical significance of the PEA, tyramine, and tryptamine production at 48 h was determined using the one-way ANOVA post-hoc Tukey-Kramer test, and the p-values for the Tukey-Kramer test are shown. See also Supplementary Table S4.
Correlation of gut bacterial aadc copy number and PEA production ability in human feces
Heterologous expression experiments indicated that aadc candidates: aadcBh, aadcCa, aadcTn, were involved in PEA production (), in addition to aadcEf and aadcRg, whose role in PEA production was already reported. However, it was still unclear whether aadc of PEA-producing gut bacteria produce PEA in the human colonic lumen. Therefore, the correlation between the copy number of aadcEf, aadcRg, aadcBh, and aadcTn and the amount of PEA produced when incubated with 1 mM Phe was analyzed, using human fecal samples. C. asparagiforme produced 254 μM of tyramine in AAAD medium; however, PEA production was extremely low (15 μM, ), and therefore, we excluded aadcCa from the analysis.
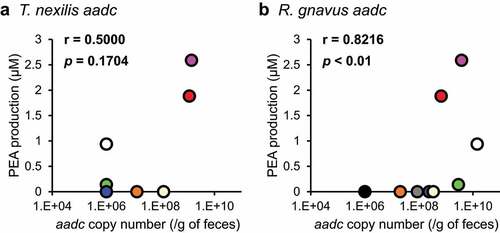
The concentrations of PEA produced in the fecal culture ranged from 0 to 2.6 μM (). aadcTn was detected in 44% of donors (4/9); aadcRg, in 88% (8/9), while aadcEf and aadcBh were not detected by the qPCR assay. A significant correlation was observed between PEA production and aadcRg (r = 0.8216 and p= 0.0066, ), but not between PEA production and aadcTn (r = 0.5000 and p= 0.1704, ).
Figure 4. PEA production in feces is associated with aadc of R. gnavus. Nine human feces were separately incubated with or without 1 mM Phe. PEA production was calculated from the difference between PEA concentration when incubated with 1 mM Phe and that without 1 mM Phe. Copy number of aadc of T. nexilis and R. gnavus were determined using qPCR. (a) Correlation between PEA production and copy number of aadc of T. nexilis. (b) Correlation between PEA production and copy number of aadc of R. gnavus. Spearman’s rank correlation test was used for the correlation analysis (r = correlation coefficient). The copy number of the aadc gene in samples where the aadc gene was below the detection limit in our system was set as 106 copies/g of feces. Donors are distinguished by color; the same color in (a) and (b) indicates the same donor.
Influence of gut bacterial aadc on colonic aromatic amine production in colon and serotonin level
We hypothesized that colonic luminal PEA, as with tyramine,Citation24 induces serotonin production from EC cells via TAAR1. To confirm this hypothesis in experiments with BLAB/cCrSlc mice, we purchased and used the colons of BLAB/cCrSlc mice to evaluate Taar1 expression in colon. RNA was extracted from the purchased colons of BALB/cCrSlc mice, PCR was performed using the reverse transcription product as a template with Taar1-specific primers,Citation50 and the PCR product was sequenced. As a result, the expression of Taar1 in the BALB/cCrSlc mouse colon was confirmed (Supplementary Figure S3). Next, to analyze the effects of indigenous bacterial PEA production in the intestinal lumen on the colonic serotonin production in the host at gut bacterial gene level, we selected En. faecalis, which can be genetically engineered, among the PEA-producing gut bacteria and generated aadc-deletion (SK981) and complementation (SK982) strains of En. faecalis. The production of aromatic amines was completely lost by the deletion of aadc, as previously describedCitation29 and the production of aromatic amines was recovered by the complementation of aadc (). The colonic serotonin levels of mice colonized with wild-type En. faecalis (SK947), Δaadc (SK981), or aadc-complemented strain (SK982) were measured.
Figure 5. aadc modulates colonic serotonin levels. (a) Effects of deletion and complementation of aadc on the aromatic amine production of En. faecalis in vitro. En. faecalis WT (SK947), Δaadc (SK981), and aadc complemented (SK982) strains were pre-cultured at 37°C in GAM, containing chloramphenicol (10 μg/mL) for 18 h in an anaerobic chamber. Each bacterial preculture was inoculated at a final optical density (OD600) of 0.03 in GAM. Strains were cultured at 37°C in GAM with chloramphenicol (10 μg/mL) for 72 h in an anaerobic chamber. Aromatic amine concentration in the culture supernatants was quantified using HPLC. Data represent the mean ± SD of three individual experiments. One-way ANOVA was performed to assess for significant differences in the aromatic amine concentrations between the groups, and the p-values for one-way ANOVA are indicated. Statistical significance between the strains was further analyzed by Tukey-Kramer test. The groups indicated by different letters were statistically different to each other, and the p-values between the groups indicated by the different letters are shown in each panel. (b) The feeding schedule for mice. Six-weeks-old female BALB/cCrSlc mice were given antibiotics in drinking water for two weeks to remove the indigenous bacteria. Mice were fed a standard diet for 13 d, then a Phe-rich diet for 5-days. En. faecalis (WT, Δaadc, aadc complementation) (1 × 108 cfu) was inoculated into mice on day 15, indicated by the red arrow (1 day after stopping antibiotics treatment). (c) En. faecalis colonization of mouse colon confirmed using qPCR. Statistical analysis was performed using Steel-Dwass test. No significant differences were observed between the groups, for total bacterial 16S rRNA gene and En. faecalis 16S rRNA gene. Two independent experiments were performed (n = 4 and 6 in each group, respectively), and data is shown in box plots with the median ± interquartile range. Statistical significance for each gene copy number between the groups was assessed by the Kruskal-Wallis test post-hoc Steel-Dwass test. The p-values for the Kruskal-Wallis test and Steel-Dwass test are shown. (d) Colonic serotonin levels in mice colonized with En. faecalis. Serotonin concentrations were measured using ELISA. The amount of serotonin in each mouse was normalized by the mean value obtained for WT colonized mice. Two independent experiments were performed (n = 4 and 6 in each group, respectively). The amounts are expressed as serotonin (a.u.). The data are shown by box plots, in which the horizontal line inside the box is the median. Statistical significance was assessed by the Kruskal-Wallis test post-hoc Steel-Dwass test. The p-values for the Kruskal-Wallis test and Steel-Dwass test are shown. See also Supplementary Figure S4.
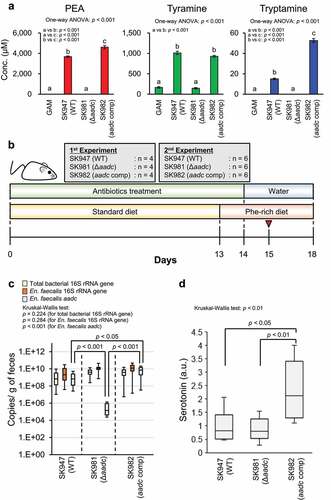
A schematic overview of the mice experiment is shown in . The number of En. faecalis was almost the same in the feces of mice colonized with wild type, ∆aadc, and aadc-complemented En. faecalis, suggesting that the colonization efficiency was not influenced by aadc deletion or complementation (). The fecal aadc copy number was significantly higher in mice colonized with aadc-complemented strain than in those colonized with wild-type En. faecalis. The fecal aadc copy number of mice colonized with ∆aadc En. faecalis was in the order of 1/3,000, compared to that of mice colonized with wild type En. faecalis (). Serotonin levels were significantly higher in the colon tissue of mice colonized with aadc-complemented strain than those colonized with wild-type En. faecalis or ∆aadc En. faecalis. However, no significant difference in the serotonin levels in the colon tissue was observed between mice colonized with wild-type En. faecalis and mice colonized with ∆aadc En. faecalis (). These data suggest that a high copy number of aadc (~1010 copy number/g of feces), which was also observed in some samples during our human fecal assays (), induces colonic serotonin production (). However, PEA was not detected in the feces and the cecal contents of any of the mice. A small amount of tyramine was detected in the feces of mice colonized with wild-type and aadc-complemented En. faecalis, while no tyramine was detected in the feces of mice colonized with ∆aadc En. faecalis (Supplementary Figure S4A). These results indicate that gut bacterial aadc contributes to aromatic amine production in the host colonic lumen. However, no significant correlation between the fecal tyramine amounts and colonic serotonin amounts was observed in the groups (Supplementary Figure S4B).
Human AADC inhibitor decreases PEA production in En. faecalis
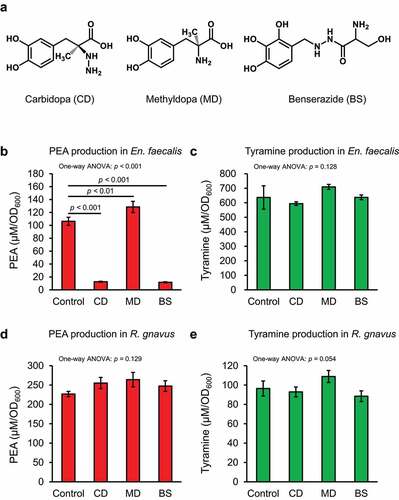
Several human AADC inhibitors have been clinically used for the treatment of Parkinson’s disease,Citation51 and these inhibitors may be candidates for drug repositioning. We evaluated whether human AADC inhibitors (carbidopa, methyldopa, and benserazide []) could inhibit PEA production of En. faecalis and R. gnavus, which produced PEA in vitro (). Severe growth deficiency was not observed with any tested AADC inhibitors (Supplementary Figure S5A and S5B). PEA production in En. faecalis was strongly inhibited by carbidopa and benserazide, however methyldopa did not inhibit PEA production ( and Supplementary Figure S5C). Surprisingly, tyramine concentration in the culture supernatant of En. faecalis did not change when treated with carbidopa and benserazide ( and Supplementary Figure S5D). None of the tested human AADC inhibitors inhibited PEA production by R. gnavus ( and Supplementary Figure S5E). Although carbidopa and benserazide significantly reduced tyramine production by R. gnavus (Supplementary Figure S5F), they also significantly reduced R. gnavus growth (Supplementary Figure S5B). Therefore, the tested inhibitors exerted no significant effects after the tyramine concentration of R. gnavus was normalized to bacterial growth (OD600) ().
Figure 6. Human AADC inhibitors inhibit PEA production in En. faecalis. En. faecalis and R. gnavus were cultured with 1.5 mM human AADC inhibitor in AAAD medium for 24 h, and the concentration of PEA and tyramine in the culture supernatants were quantified using HPLC. PEA and tyramine concentrations were normalized to the OD600 values and shown as μM/OD600. (a) Structure of the tested human AADC inhibitors. (b) and (c) Effect of the human AADC inhibitors on PEA and tyramine production in En. faecalis, respectively. (d) and (e) Effect of the human AADC inhibitors on PEA and tyramine production in R. gnavus, respectively. Data represent the mean ± SD of three individual experiments. Statistical significance was assessed by one-way ANOVA post-hoc Dunnett’s test. The p-values for one-way ANOVA and Dunnett’s test are shown. See also Supplementary Figure S5.
Discussion
Gut microbiota produce various metabolites in the colonic lumen, affecting the host physiology.Citation52 Identification of gut bacterial species and genes responsible for metabolite production is essential for the optimization of the state of the intestinal environment. Some molecular mechanisms of metabolite production in the colonic lumen have been established at the genetic level,Citation53–55 however, even for well-known metabolites, there are unidentified mechanisms modulating the production. Few studies have investigated between intestinal bacterial genes and the host physiology. In this study, we identified five PEA-producing bacteria from the GAM-culturable human dominant gut bacteria () and revealed that aadc is indispensable for the production of PEA (). Our results suggested that gut bacterial AADC and its reaction product, PEA, participate in the peripheral serotonin production in the host. Furthermore, fecal culture and mouse experiments indicated that gut bacterial aadc contributes to aromatic amine production in the colon ( and Supplementary Figure S4A).
AADC of R. gnavus was recently identified as a Trp decarboxylase, and studies using the recombinant AADC indicated that the catalytic efficiency (kcat/Km) for Trp is 1,000-fold higher than that for Phe.Citation25 However, in our experiment using the AAAD medium containing 1 mM of Trp, Tyr, and Phe, R. gnavus produced statistically significantly more tryptamine than PEA, but the difference was 1.2-fold, which was much smaller than the difference expected from kcat/Km value (). B. hansenii () and T. nexilis (), which were considered tryptamine-producing gut bacteriaCitation25 produced not only tryptamine, but also PEA and/or tyramine in the AAAD medium. Aromatic amine productivity of E. coli transformants expressing each aadc from PEA-producing gut bacteria in M9AAA-medium were partially but not entirely consistent with the PEA-producing gut bacteria in AAAD medium (). These differences were hypothesized to be due to the different abilities of E. coli and PEA-producing gut bacteria to uptake aromatic amino acids or to release aromatic amines. These results suggested that the production of metabolites secreted by bacterial cells is influenced not only by the activity of the enzymes, but also by that of transporters. A significant correlation was observed between aadcRg and the production of PEA in fecal culture (). Therefore, our results suggest that PEA production was not a side reaction, but rather the main reaction catalyzed by AADC in the intestinal lumen. On the other hand, no significant correlation was observed between aadcTn and the PEA production (). This could possibly be because of the lower number of the fecal samples possessing aadcTn. In addition, in vitro analysis showed that R. gnavus produced more PEA than T. nexilis (), suggesting that the contribution of aadcRg is significant compared to that of aadcTn, in the PEA production in colonic lumen. We also observed that while possessing the same level of aadcRg in the feces, the fecal samples differ in PEA production (). Gut bacteria reductively metabolize aromatic amino acids to aromatic lactate, aromatic acrylate, and aromatic propionate by aromatic lactate dehydrogenase, phenyl lactate dehydrogenase, phenyl lactate dehydratase, or acyl-CoA dehydrogenase after deamination.Citation10,Citation55 Therefore, we speculate that the amount of the genes related to reductive aromatic amino acid metabolism pathway affects PEA production.
Peripheral serotonin is produced by EC cellsCitation56 and its production heavily relies on gut bacteria.Citation24 Physiologically active metabolites derived from gut bacteria, such as short-chain fatty acids (acetic acid and butyric acid),Citation57 and deoxycholateCitation24 stimulate peripheral serotonin production. Intrarectally injected tyramine induces peripheral serotonin secretion in mouse.Citation24 EC cells express a variety of receptors, including TAAR1;Citation58 like tyramine, PEA is also a ligand of TAAR1.Citation35 Colonic Taar1 expression has been reported in humans,Citation37 and BALB/cCrSlc mice of the same strain as used in the experiment to assess the effect of gut bacterial PEA on the colonic serotonin, was also expressing Taar1 in the colon (Supplementary Figure S3). Furthermore, activation of TAAR1 induced serotonin production in a cell model.Citation35 In this study, colonic serotonin levels of mice colonized with aadc-complemented En. faecalis were higher than those in mice colonized with wild-type En. faecalis and Δaadc En. faecalis (). There are two possible sources of phenylalanine in the mouse colon: diet and metabolic activity of gut bacteria. Phe, a substrate for AADC, has been detected in mice colon and its amount increase in response to feeding a high-fat diet.Citation59 Therefore, it is possible that PEA is generated in the murine colonic lumen by the reaction catalyzed by AADC. However, in our experiments, PEA was detected neither in the feces nor in the cecal contents of mice. A possible reason for this is that PEA produced by gut bacteria in the intestinal tract after inducing serotonin production in mice was rapidly degraded by a reaction catalyzed by monoamine oxidase-B (MAO-B)Citation60 and was undetectable by the time of analysis. Actually, the expression of Mao-b in the colon of BALB/cCrSlc mice of the same strain as used in the experiment to assess the effect of gut bacterial PEA on the colonic serotonin was confirmed by PCR (Supplementary Figure S3). The reason why tyramine, but not PEA, was detected (Supplementary Figure S4A) in the colonic lumen could be that the Km of MAO-B, which reportedly is expressed in the intestinal tract,Citation24 to tyramine is described to be 290-fold higher than that to PEA.Citation61 Therefore, it is likely that only PEA was preferentially degraded by MAO-B. In the present study, a maximum of 41.8 pmol of tyramine per mg of feces was detected (Supplementary Figure S4A), but tyramine concentration in the feces did not correlate significantly with serotonin production in the intestinal tract (Supplementary Figure S4B). On the contrary, Yano et al. demonstrated that injection of about 20 μmol of tyramine into the mouse colonic lumen activated colonic serotonin production.Citation61 Given a mouse fecal mass of 100 to 200 mg,Citation62,Citation63 the amount of tyramine detected in the colon in the present study was 4.2 to 8.4 nmol, much less than the dose given by Yano et al.Citation24 This could explain why tyramine amounts in the intestinal lumen did not correlate with serotonin production in the intestinal tract; thus, this study does not exclude the theory that tyramine promotes serotonin productionCitation24 in the colon.
Peripheral serotonin is an important regulator and is implicated in several diseases, such as osteoporosis,Citation64 diarrhea-predominant irritable bowel syndrome,Citation65 celiac disease,Citation66 inflammatory bowel disease,Citation67 and obesity.Citation68 Marketed drugs: Carbidopa and benserazide, developed as human AADC inhibitor, significantly inhibited PEA production in En. faecalis (). However, none of these inhibitors inhibited tyramine production by En. faecalis and PEA and tyramine production by R. gnavus (). AADCEf is capable of decarboxylating L-DOPA, while AADCRg cannot.Citation29 The human AADC inhibitor is an analogue of L-DOPA, and therefore likely fits into the substrate pocket of AADCEf to inhibit PEA production. However, the low affinity of AADCRg for L-DOPA might have abrogated the inhibitory effect. (S) -α-fluoromethyl tryptophan inhibits AADC of R. gnavus,Citation25 while (S)-α-fluoromethyl tyrosine inhibits AADC of En. faecalis . Therefore, (S)-α-fluoromethyl derivatives, such as (S)-α-fluoromethyl phenylalanine may be promising as inhibitors of PEA synthesis by AADC of gut bacteria.
Here, we showed that gut bacterial AADC contributes colonic serotonin production, and that two commercial AADC inhibitors can inhibit PEA production by En. faecalis. However, our study had some limitations. First, we used BALB/cCrSlc mice as a model. This strain possesses a mutation in the tryptophan hydroxylase-2 gene, which is responsible for central serotonin biosynthesis.Citation69,Citation70 BALB/cCrSlc mice has not been reported to exhibit a mutation in tryptophan hydroxylase-1, which is responsible for the peripheral serotonin biosynthesis evaluated in the present study. The determined colonic serotonin level (0.49 to 6.8 μg/g of colonic tissue) was comparable to that in previous studies using C57BL/6 mice.Citation71 However, other models are needed to assess PEA effects on serotonin, including in the central nervous system. Second, we evaluated the effects of human AADC inhibitors on gut bacterial aromatic amine production with 1.5 mM. This dosage has technical and toxicological limitations, e.g., to reach 1.5 mM of carbidopa in the intestinal tract, a minimum of 1 g must be ingested.Citation72 To solve this problem, it is necessary to develop efficient drug delivery methods to the intestinal tract; for example, the administration of carbidopa in enteric capsules and the development of effective gut bacterial AADC inhibitors that are not absorbed in the intestine should be considered. In the future, selective and efficient inhibition of gut bacterial AADC could lead to the prevention and treatment of diseases involving peripheral serotonin.
Conclusion
The present study presented five PEA-producing gut bacteria species and determined that aromatic amine production from proteinogenic aromatic amino acids depends on AADC activity. Human fecal culture experiments revealed a significant positive correlation between aadc of R. gnavus and PEA production, suggesting that R. gnavus contributes to PEA production in the human colonic lumen. Furthermore, gut bacterial aadc activity upregulated colonic serotonin levels in a mice model treated with genetically modified En. faecalis. Finally, we revealed that the human AADC inhibitors carbidopa and benserazide prevented PEA production by En. faecalis. This study shows that AADC in gut bacteria may be a potential target in the prevention and treatment of diseases involving peripheral serotonin.
Materials and Methods
Chemicals
Phenylethylamine hydrochloride (Cat# P0086), tryptamine (Cat# T0890), tyramine hydrochloride (Cat# A0303), L-tyrosine (Cat# T0550), carbidopa monohydrate (Cat# C2450), methyldopa sesquihydrate (Cat# D1817), and benserazide hydrochloride (Cat# B4108) were purchased from Tokyo Chemical Industry (Tokyo, Japan). L-Phenylalanine (Cat# 169–01303), Amino Acids Mixture Standard Solution, Type H (Cat# 013–08391) were obtained from FUJIFILM Wako Pure Chemical (Osaka, Japan). L-Tryptophan (Cat# 35607–74) was obtained from Nacalai Tesque (Kyoto, Japan). Other reagents of analytical grade were from FUJIFILM Wako Pure Chemical, Nacalai Tesque (Kyoto, Japan), and Sigma-Aldrich (St. Louis, MO, USA).
Bacterial strains
The strains used in this study are listed in . Bacteria were obtained from the Japan Collection of Microorganisms (JCM), the American Type Culture Collection (ATCC), and the German Collection of Microorganisms and Cultures (DSMZ).
Table 1. Bacterial strains, plasmids, and primers used in this study.
High-performance liquid chromatography (HPLC)
Quantification of aromatic amines and aromatic amino acids were carried out using HPLC. PEA was quantified by the post-column labeling method as describe previously.Citation82 Briefly, PEA was separated using a cation-exchange column (#2619PH, Hitachi, Tokyo, Japan) in normal-phase mode, and derivatized with o-phthalaldehyde and detected using fluorescence detector (λex: 340 nm and λem: 435 nm). Tyramine and tryptamine were separated in reverse-phase mode using a Discovery HS-F5 column (4.6 × 250 mm, 5 μm, SUPECLCO, Bellefonte, PA, USA) at 35°C. The elution was carried out at a flow rate of 0.4 mL/min with 10 mM ammonium formate (pH 3.0) and acetonitrile using the following gradient program: the concentration of acetonitrile was linearly increased from 3 to 27% during 0‒22 min, and linearly increased from 27 to 66% during 22–80 min, increased to 100% during 80‒81 min, maintained at 100% during 81‒86 min, returned to 3% during 86‒87 min, and maintained at 3% during 87‒102 min. In this analytical system, the retention time for Tyr, tyramine, Trp, and tryptamine were 18.5, 28, 29.3, and 51 min, respectively. The elution was monitored based on the fluorescence with excitation at 280 nm and emission at 325 nm, using a Waters 2475 Multi-wavelength Fluorescence detector (Waters, Milford, MA, USA). Phe concentration was quantified using HPLC equipped with a cation-exchange column (#2619, Hitachi). The temperature of the column oven was maintained at 70°C. Buffer-A (23 mM sodium citrate, 96.8 mM sodium chloride, 84.7 mM citric acid, 13% ethanol, and 0.0001% caprylic acid), buffer-B (26.3 mM sodium citrate, 121 mM sodium chloride, 105 mM citric acid, 0.02% ethanol, and 0.0001% caprylic acid), buffer-C (90 mM sodium citrate, 930 mM sodium chloride, 104 mM citric acid, and 0.0001% caprylic acid), and buffer-D (200 mM sodium hydroxide, 10% ethanol, and 0.0001% caprylic acid) were used as the mobile phase, at a flow rate of 0.4 mL/min. The gradient program was as follows: buffer-A was kept at 100% during 0–13 min. The buffer was changed to buffer-B and the proportion was kept at 100% from 13–18 min. From 18–26 min, buffer-B was linearly decreased from 100 to 75% and buffer-C was linearly increased from 0 to 25%. Buffer-C was linearly increased to 100% during 26–56 min and kept at 100% during 56–67 min. The buffer was changed to buffer-D and the proportion of buffer-D was kept at 100% during 67–74 min. The buffer was changed to buffer-A and the proportion of buffer-A was kept at 100% during 74–105 min. Detection of Phe was carried out using the same method as that for PEA. A standard sample was always analyzed within each batch to quantify aromatic amines and aromatic amino acids in the sample. Aromatic amine and aromatic amino acid species in the samples were identified by comparison with the retention times of the standards, and each compound concentration was determined using a standard curve generated from the peak areas of the standards at known concentrations.
Purification of unidentified biogenic amine from culture supernatant of T. nexilis
The unidentified biogenic amine was purified from culture supernatant of T. nexilis using a cation exchange resin (Dowex 50 W X8, [H+-form; 100–200 mm], FUJIFILM Wako Pure Chemicals). Ten milliliters of resin was packed into a column. T. nexilis was anaerobically pre-cultured overnight in 1 mL of GAM at 37°C. Pre-cultures (500 μL) were inoculated into 500 mL of GAM and cultured overnight at 37°C in an anaerobic chamber (INVIVO2 400; Ruskinn Technology, Bridgend, UK) until stationary phase.Citation47 The culture was centrifuged (12,680 × g, 4°C, 20 min), and the supernatant was applied to the resin-packed column. After loading, the unidentified biogenic amine was purified using the following procedures: the column was washed with 400 mL of deionized water. Polyamines (spermidine, cadaverine, and spermine) were eluted with 120 mL of 1 M HCl. Residual HCl was removed from the column by washing the column with 200 mL of deionized water. Agmatine was eluted with 115 mL of 500 mM of NH3 and the column was washed with 100 mL of deionized water. The unidentified biogenic amine was eluted with 100 mM HCl. The eluted unidentified amine was again absorbed onto the same column, washed with 2 L of deionized water, and then eluted by increasing the NH3 concentration in steps of 2, 4, and 8 M in 80 mL increments. Every 1 mL was recovered, and the concentration and purity of the unidentified biogenic amine were tested using HPLC. The fraction eluted with 8 M NH3 contained the highest amount of the unidentified biogenic amine, and was therefore subjected to further analysis.
Determination of molecular mass of the purified unidentified biogenic amine using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS)
The mass of the purified product was determined using a 3200 QTRAP MS system (Applied Biosystems, Foster City, CA, USA). The sample was directly injected to the electrospray ionization-MS detector in scan mode with positive ionization. The molecular ion peak with m/z of 122 was further analyzed by MS/MS fragmentation. The purified sample was separated using an Agilent HPLC system (Agilent) as follows: column, TSKgel ODS-80Ts (4.6 × 250 mm2, 5 μm particles; Tosoh); solvent system, A: 0.1% acetic acid in water, B: 0.1% acetic acid in acetonitrile; gradient modes: 90% A (0–5 min), 90–60% A (5–20 min), and 10% A (20–30 min); flow rate, 0.5 mL/min at 40°C. The separated sample was analyzed by LC-MS/MS, using select ion mode at m/z = 122. Standard PEA was used for comparison. The biogenic amine was confirmed as PEA by comparing its retention time and daughter ions with that of standard PEA in LC-MS/MS analysis.
Aromatic amine production profile of PEA-producing gut bacteria
PEA-producing gut bacteria were anaerobically pre-cultured in GAM at 37°C for 18 h in an anaerobic chamber (INVIVO2 400). After pre-cultivation, cells in the stationary phaseCitation47 were washed and resuspended with 30 mL of the AAAD medium (Supplementary Table S2) at an initial OD600 of 0.03 and cultured at 37°C in an anaerobic chamber. One milliliter of the culture was collected at the indicated times and centrifuged (18,900 × g, 4°C, 10 min) to obtain the culture supernatant. The culture supernatant was filtrated using a Cosmonice filter W (Nacalai Tesque) after protein precipitation by trichloroacetic acid, as described previously.Citation82 The filtrated sample was analyzed using HPLC.
Aromatic amine production profile of En. faecalis and R. gnavus in the presence of human AADC inhibitors
Human AADC inhibitors: carbidopa,Citation83 methyldopa,Citation84 and benserazideCitation85 were dissolved in MilliQ water and added to the AAAD medium at a final concentration of 1.5 mM. The other culture conditions were the same as described above. Following 24 h cultivation in the presence of inhibitors, PEA and tyramine concentrations in the culture supernatants and OD600 were measured.
Plasmid construction
Cells of B. hansenii, C. asparagiforme, T. nexilis, En. faecalis, and R. gnavus in 0.5 mL of overnight culture, were centrifuged and stored at −20°C until the genomic DNA (gDNA) extraction. Cells of T. nexilis and R. gnavus were suspended in 100 μL of TE buffer and disrupted using zirconia beads (Thermo Fisher Scientific, Waltham, MA, USA) in a SHAKE MASTER ver. 1.2 (Bio Medical Science, Tokyo, Japan). The disrupted cells were centrifuged (21,500 × g, 4°C, 10 min) and gDNA in the resulting supernatant fractions were used as templates in PCR reactions. B. hansenii and C. asparagiforme gDNA were extracted using the phenol-chloroform methods.Citation47 Enterococcus faecalis gDNA was extracted using a Wizard Genomic DNA purification kit (Promega, Madison, WI), according to the manufacturer’s protocol. The genes of putative aadc (BLAHAN_06497 [aadcBh], CLONEX_01451 [aadcTn], and CLOSTASPAR_05940 [aadcCa]) and aadc (EF_0634 [aadcEf] and RUMGNA_01526 [aadcRg]) were amplified using KOD-plus- Neo (Toyobo, Osaka, Japan) or PrimeSTAR Max (TaKaRa Bio, Shiga, Japan) from the respective gDNA using the primers listed in . Amplified DNA was cloned between the NdeI and XhoI sites of the expression vector, pCDF23Citation74 using an In-Fusion HD cloning kit (Clontech Laboratories Inc., Mountain View, CA, USA). The aadc in the resulting plasmid was sequenced to ensure that there were no PCR-introduced errors. Plasmids were used to transform Escherichia coli BL21 (DE3) for heterologous expression.
Plasmids used for generating a standard curve of 16S rRNA genes and aadcEf in qPCR were constructed as follows: 16S rRNA gene and aadcEf were amplified from gDNA of En. faecalis using the primers listed in . Amplified DNA was cloned into SmaI site of pUC19 using DNA ligation kit Mighty Mix (TaKaRa Bio).
Heterologous expression of aadc in Escherichia coli
Escherichia coli BL21 (DE3) strains harboring aadc expression plasmids or an empty plasmid were pre-cultured in 5 mL of Luria-Bertani medium containing 75 μg/mL spectinomycin in 100 mL Erlenmeyer flask, at 37°C with reciprocal shaking at 140 rpm for 17 h. M9 mediumCitation86 containing 0.2% of glucose and 1 mM of aromatic amino acids: Phe, Tyr, and Trp (M9AAA-medium) was used for the main culture. Pre-cultured cells were washed with M9 medium containing 0.2% glucose and suspended in M9AAA-medium. The suspension was inoculated into 10 mL of M9AAA-medium, supplemented with 10 μM isopropyl β-D-thiogalactopyranoside in a 100 mL Erlenmeyer flask at an initial OD600 of 0.03 and cultured at 37°C with reciprocal shaking at 140 rpm. An aliquot of the culture was collected every 6 h and centrifuged for 10 min at 21,500 × g at 4°C. The resulting culture supernatant was treated with 10% (w/v) trichloroacetic acid, and was subjected to HPLC analysis, as previously reported.Citation87
Generation of deletion mutant of aadc in En. faecalis V583
The aadc gene of En. faecalis V583 was deleted using pLT06 as described previously.Citation53 Primers and plasmids used for aadc deletion and complementation are listed in . Complementation plasmid pLZ12-aadc+ was constructed as follows: aadc and its upstream 500 bp were amplified using PCR with PrimeSTAR Max DNA polymerase (TaKaRa Bio) with C_ddc+0.5K_F_pLZ_Bam and C_ddc+0.5K_R_pLZ_Bam as primers. The product was cloned into BamHI site of pLZ12. The resulting plasmid was introduced into En. faecalis by electroporation, as described previously.Citation53
Conversion of phenylalanine to phenylethylamine by human fecal sample
Fecal samples from nine healthy Japanese donors (eight male and one female; age: 21.8 ± 10.2, range 4–40 years) were analyzed. Feces were collected using stool collecting kit (LSI medience Co., Tokyo, Japan) and stored under anaerobic condition using Anaero Pack system (Mitsubishi Gas Chemical Co., Inc., Tokyo, Japan) at −80°C until use. Fecal samples were suspended in a 4-fold volume of phosphate-buffered saline (PBS) or PBS containing 1 mM of Phe and were incubated anaerobically at 37°C. After 8 h incubation, the culture supernatants were harvested by centrifugation (21,500 × g, 4°C, 10 min) and subjected to HPLC analysis, as described in the “Aromatic amine producing profile of PEA-producing gut bacteria” section. The net PEA production was calculated by subtracting PEA amount formed in the absence of Phe from that formed in the presence of 1 mM Phe in the fecal suspension.
Extraction of gDNA from fecal samples
Fecal samples (10–20 mg) were suspended in 95 μL of TE buffer. Five microliters of 300 mg/mL lysozyme (Sigma-aldrich, MO. USA) and 11 μL of 1,000 U/μL achromopeptidase (FUJIFILM Wako Pure Chemicals) were added to the suspension and incubated for 30 min at 37°C. Twelve microliters of 20% sodium dodecyl sulfate solution was added to the suspension and incubated at 60°C for 20 min.Citation88 The bacterial DNA was extracted from the incubated mixture using QIAamp Fast DNA Stool Mini Kit (QIAGEN, Hilden, Germany).
Quantification of DNA copy numbers in fecal sample
The copy number of the targeted gene was determined by quantitative PCR (qPCR) using a thermal cycler (StepOne Real-time PCR system, Applied Biosystems) and TB Green Premix Ex Taq II (Tli RNaseH Plus) (TaKaRa Bio). Primers used for qPCR are listed in . The Reaction mixture (20 μL) consisted of 10 μL of 2× TB Green II mix, 9.2 ng of gDNA, and 0.7 μM (for aadc gene) or 0.35 μM (for 16S rDNA) of primers. PCR cycling conditions for the amplification of aadcTn were 95°C for 30s, followed by 40 cycles of 95°C for 5 s and 64°C for 45s. PCR cycling conditions for the amplification of the other genes were 95°C for 30s, followed by 40 cycles of 95°C for 5 s and 60°C for 1 min. Gene copy numbers were calculated based on the standard curve generated using varying concentrations of gDNA or plasmids containing the target gene.
Animal experiments
Six-week-old female BALB/cCrSlc mice were purchased from Japan SLC (Hamamatsu, Japan). All mice were maintained in a 12 h light-dark cycle and housed in group cages with 2‒3 animals per cage with free access to water and diet and bred based on the regulations regarding the protection of laboratory animals at the Kanazawa University. All animal experiments were performed according to the Guideline for the Care and Use of Laboratory Animals at Kanazawa University (Approval number: AP-163778). Mice were fed a standard diet (CE-2, CLEA Japan Inc., Tokyo, Japan) for 13 days. Tyr was removed from the diet and Phe content was increased from 0.87% (w/w) in the L-Amino Acid Defined AIN-93 G (Dyets Inc., Bethlehem, PA, USA) (Supplementary Table S5) to 8.7% (w/w) in the Phe-rich diet (#511379, Dyets Inc.) (Supplementary Table S5). The mice were fed a Phe-rich diet from for 5 d, beginning on day 13 (). Drinking water and the diet were sterilized by autoclave or γ-irradiation, respectively. Antibiotics (0.25 mg/mL of doripenem and 0.5 mg/mL of vancomycin) were provided ad libitum in the drinking waterCitation89 for 14 days until one day before En. faecalis administration.
The En. faecalis strains, WT, Δaadc, and aadc complemented En. faecalis were grown anaerobically in GAM containing 10 μg/mL chloramphenicol at 37°C for 12 h. Cells were collected from 1 mL of culture by centrifugation (6,000 × g, 25°C, 5 min) and washed twice with 1 mL of PBS containing 10 μg/mL chloramphenicol. Cells were suspended in 5 mL of PBS and the colony forming unit (cfu) was measured on GAM agar plate containing 10 μg/mL chloramphenicol. Cells were prepared to 1 × 108 cfu/200 μL and administered to mice. Three days after En. faecalis administration, serotonin content in the colon tissue was evaluated. These experiments were independently performed twice (n = 4 and 6 in each group, respectively), with a resulting n = 10 per group.
Quantification of serotonin concentration in the colon tissue
Mice were euthanized by cervical dislocation under the anesthetization with Dormicum (Meiji Seika Pharma Co., Ltd., Tokyo, Japan), Vetorphale (Astellas Pharma, Inc., Tokyo, Japan), and Domitor (Nippon Zenyaku Kogyo Co., Ltd., Fukushima, Japan). The entire length of the colon with full-thickness was washed with PBS to remove the luminal contents, sonicated in 10 mL of PBS using Branson model 250 (BRANSON, St. Louis, MO, USA), and stored at −25°C, until use. The colon lysate was centrifuged (21,500 × g, 4°C, 5 min) to remove tissue debris. Serotonin concentration was determined using Serotonin ELISA Kit (Enzo Lifescience, Farmingdale, NY, USA), according to the manufacturer’s protocol. The serotonin ELISA Kit (Enzo Lifescience, Farmingdale, NY, USA) was used because the Serotonin ELISA Kit (Eagle Biosciences) used by Yano et al.Citation24 could not be purchased in Japan due to legal restrictions. Serotonin amounts were normalized based on the weight of the colonic tissue. Animal experiments were independently performed twice (n = 4 and 6 in each group, respectively), and for each experiment, the amount of serotonin was normalized by the average amount of serotonin in mice colonized with wild-type En. faecalis. The normalized values were assigned an arbitrary unit (a.u.) and the serotonin levels in the colonic tissue were expressed as serotonin (a.u.).
Analysis of Taar1 and Mao-b expression in mouse colon
Colon tissue from six-week-old female BALB/cCrSlc mice was purchased from Japan SLC. The tissue was treated with RNAlater-ICE (Thermo Fisher Scientific) for 20 h at −20°C and used for RNA extraction. The frozen colon was placed into a tube with a stainless bead and disrupted using a Multi-Beads shocker (Yasui Kikai Co., Osaka, Japan). RNA extraction and on-column genomic DNA digestion were then performed using ISOSPIN Cell & Tissue RNA Kit (Nippon Gene, Tokyo, Japan) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized from 0.2 or 1 μg of RNA using PrimeScript™ RT reagent Kit (Perfect Real Time) (TaKaRa Bio). TB Green® Premix Ex Taq™ (Tli RNaseH Plus) (TaKaRa Bio) was used for PCR, which was performed using 1 µL cDNA solution for samples that used 0.2 μg of RNA for cDNA synthesis and 0.2 μL cDNA solution for samples that used 1 μg of RNA for cDNA synthesis as a template. The PCR cycling conditions were 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. The PCR products were electrophoresed with 3% agarose gel and visualized using ethidium bromide under ultraviolet light.
Statistical analysis
Statistical analyses were performed using SPSS software version 21 (IBM, Armonk, NY) and BellCurve for Excel (Social Survey Research Information Co., LTD.). Correlation between PEA production and aadc copy number was analyzed by Spearman’s rank correlation test. Repeated measures one-way ANOVA was employed to evaluate the significance of change in aromatic amine concentration with cultivation time. The Tukey-Kramer and Dunnett’s tests were used for multiple comparisons of aromatic amine concentrations. aadcEf and 16S rRNA gene copy numbers in mouse feces and colonic serotonin level were statistically analyzed using Kruskal-Wallis test followed by Steel-Dwass test. A p < .05 was considered statistically significant.
Ethical approval
This study was approved by the Ethics Committee of Ishikawa prefectural university (2016–2) and was performed in accordance with the Declaration of Helsinki. Informed consent was obtained from all donors or their parents.
Data and materials availability
All data needed to evaluate the conclusions in this work are present in the paper and/or the Supplementary Materials. Additional data related to this paper will provided by the authors upon reasonable request.
Author contributions
Y.S., S.O., and S.K. conceived the project and designed the experiments. M.N., A.N., and H.M. purified PEA and carried out LC-MS analyses. Y.S., M.N., H.K., and H.S. cultured gut bacteria and measured aromatic amines in the culture supernatant. T.Katayama retrieved aadc candidates of PEA producing-gut bacteria from the genomic information and supervised heterologous expression of aadc in E. coli transformants. Y.S., A.N., H.M., and T.Koyanagi generated E. coli transformants and measured aromatic amines in the culture supernatants. Y.S., Y.M., Y.K., M.K., N.I., R.H., and S.O. were responsible for animal experiments. S.K. genetically engineered En. faecalis. Y.S., A.G., M.S. contributed to collect human fecal samples and analyze human fecal DNA. Y.S. and S.K. performed fecal culture experiments. E.N. and S.K. evaluated the effects of human AADC inhibitors on the gut bacterial aromatic amine production. Y.S. and R. H. performed statistical analyses. Y.S., S.O. and S.K. wrote the draft manuscript. M.S., N.I., T.Katayama, and S.K. edited the manuscript. All authors discussed the data and contributed to the completion of the final manuscript.
Data availability
All necessary data are included in the manuscript since no omics analysis or determination of new genomes has been undertaken in this study.
Supplemental Material
Download PDF (812.5 KB)Acknowledgments
We are grateful to Dr. B. A. Duerkop, and Professor L. Hooper from the University of Texas Southwestern Medical Center, Dallas for technical advice and for the gift of pLZ12. We acknowledge Dr. L. E. Hancock, University of Kansas, for providing pLT06. We thank Dr. Kohei Ogura (Kanazawa University) and Dr. Yuhei Araiso (Kanazawa University) for their advice on RNA purification from mouse colon samples, and Ms. Aya Kitakata (Ishikawa Prefectural University) for technical support.
Disclosure statement
The authors report no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2022.2128605
Additional information
Funding
References
- Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–24. doi:10.1038/nature09646.
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi:10.1038/nature12721.
- Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499(7456):97–101. doi:10.1038/nature12347.
- Sinha SR, Haileselassie Y, Nguyen LP, Tropini C, Wang M, Becker LS, Sim D, Jarr K, Spear ET, Singh G, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. 2020;27:659–70 e5. doi:10.1016/j.chom.2020.01.021.
- Kibe R, Kurihara S, Sakai Y, Suzuki H, Ooga T, Sawaki E, Muramatsu K, Nakamura A, Yamashita A, Kitada Y, et al. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci Rep. 2014;4(1):4548. doi:10.1038/srep04548.
- Matsumoto M, Kurihara S, Kibe R, Ashida H, Benno Y. Longevity in mice is promoted by probiotic-induced suppression of colonic senescence dependent on upregulation of gut bacterial polyamine production. PLoS One. 2011;6:e23652. doi:10.1371/journal.pone.0023652.
- Nakamura A, Kurihara S, Takahashi D, Ohashi W, Nakamura Y, Kimura S, Onuki M, Kume A, Sasazawa Y, Furusawa Y, et al. Symbiotic polyamine metabolism regulates epithelial proliferation and macrophage differentiation in the colon. Nat Commun. 2021;12:2105. doi:10.1038/s41467-021-22212-1.
- LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160–168. doi:10.1016/j.copbio.2012.08.005.
- Matsumoto M, Kitada Y, Naito Y. Endothelial Function is improved by Inducing Microbial Polyamine Production in the Gut: a Randomized Placebo-Controlled Trial. Nutrients. 2019;11(5):1188.
- Laursen MF, Sakanaka M, von Burg N, Morbe U, Andersen D, Moll JM, Pekmez CT, Rivollier A, Michaelsen KF, Mølgaard C, et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat Microbiol. 2021;6(11):1367–1382. doi:10.1038/s41564-021-00970-4.
- Sato Y, Atarashi K, Plichta DR, Arai Y, Sasajima S, Kearney SM, Suda W, Takeshita K, Sasaki T, Okamoto S, et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature. 2021;599(7885):458–464. doi:10.1038/s41586-021-03832-5.
- Santoru ML, Piras C, Murgia A, Palmas V, Camboni T, Liggi S, Ibba I, Lai MA, Orrù S, Blois S, et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep. 2017;7(1):9523. doi:10.1038/s41598-017-10034-5.
- Dong F, Hao F, Murray IA, Smith PB, Koo I, Tindall AM, Kris-Etherton PM, Gowda K, Amin SG, Patterson AD, et al. Intestinal microbiota-derived tryptophan metabolites are predictive of Ah receptor activity. Gut Microbes. 2020;12(1):1–24. doi:10.1080/19490976.2020.1788899.
- Ryu IS, Kim OH, Kim JS, Sohn S, Choe ES, Lim RN, Kim TW, Seo JW, Jang EY. Effects of β-Phenylethylamine on Psychomotor, Rewarding, and Reinforcing Behaviors and Affective State: The Role of Dopamine D1 Receptors. Int J Mol Sci. 2021;22(17):9485. doi:10.3390/ijms23010022.
- Khan MZ, Nawaz W. The emerging roles of human trace amines and human trace amine-associated receptors (hTAARs) in central nervous system. Biomed Pharmacother. 2016;83:439–449. doi:10.1016/j.biopha.2016.07.002.
- Doeun D, Davaatseren M, Chung MS. Biogenic amines in foods. Food Sci Biotechnol. 2017;26:1463–1474. doi:10.1007/s10068-017-0239-3.
- Sanchez-Perez S, Comas-Baste O, Rabell-Gonzalez J, Veciana-Nogues MT, Latorre-Moratalla ML, Vidal-Carou MC. Biogenic Amines in Plant-Origin Foods: are They Frequently Underestimated in Low-Histamine Diets? Foods. 2018;7(12):205.
- Macheiner L, Schmidt A, Wagner M, Mayer HK. Thermogenic formation of biogenic amines during commercial coffee roasting processes. Lwt. 2022;154:112664. doi:10.1016/j.lwt.2021.112664.
- Granvogl M, Bugan S, Schieberle P. Formation of amines and aldehydes from parent amino acids during thermal processing of cocoa and model systems: new insights into pathways of the strecker reaction. J Agric Food Chem. 2006;54:1730–1739. doi:10.1021/jf0525939.
- Pugin B, Barcik W, Westermann P, Heider A, Wawrzyniak M, Hellings P, Akdis CA, O'Mahony L. A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Microb Ecol Health Dis. 2017;28:1353881. doi:10.1080/16512235.2017.1353881.
- Phillips AW, Newcomb HR, Smith JE. Lachapeller. Serotonin in the small intestine of conventional and germfree chicks. Nature. 1961;192:380. doi:10.1038/192380a0.
- Beaver MH, Wostmann BS. Histamine and 5-hydroxytryptamine in the intestinal tract of germ-free animals, animals harbouring one microbial species and conventional animals. Br J Pharmacol Chemother 1962; 19:385.
- Bhattarai Y, Williams BB, Battaglioli EJ, Whitaker WR, Till L, Grover M, Linden DR, Akiba Y, Kandimalla KK, Zachos NC, et al. Gut microbiota-produced tryptamine activates an epithelial G-protein-coupled receptor to increase colonic secretion. Cell Host Microbe. 2018;23:775–85 e5. doi:10.1016/j.chom.2018.05.004.
- Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler C, Ismagilov R, Mazmanian S, Hsiao E, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161(2):264–276. doi:10.1016/j.cell.2015.02.047.
- Williams BB, Van Benschoten AH, Cimermancic P, Donia MS, Zimmermann M, Taketani M, Ishihara A, Kashyap P, Fraser J, Fischbach M, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe. 2014;16(4):495–503. doi:10.1016/j.chom.2014.09.001.
- Liu F, Xu W, Du L, Wang D, Zhu Y, Geng Z, Zhang M, Xu W. Heterologous expression and characterization of tyrosine decarboxylase from Enterococcus faecalis R612Z1 and Enterococcus faecium R615Z1. J Food Prot. 2014;77:592–598. doi:10.4315/0362-028X.JFP-13-326.
- Luqman A, Nega M, Nguyen MT, Ebner P, Gotz F. SadA-expressing staphylococci in the human gut show increased cell adherence and internalization. Cell Rep. 2018;22:535–545. doi:10.1016/j.celrep.2017.12.058.
- Koyanagi T, Nakagawa A, Sakurama H, Yamamoto K, Sakurai N, Takagi Y, Minami H, Katayama T, Kumagai H. Eukaryotic-type aromatic amino acid decarboxylase from the root colonizer Pseudomonas putida is highly specific for 3,4-dihydroxyphenyl-L-alanine, an allelochemical in the rhizosphere. Microbiology (Reading). 2012;158:2965–2974. doi:10.1099/mic.0.062463-0.
- van Kessel SP, Frye AK, El-Gendy AO, Castejon M, Keshavarzian A, van Dijk G, El Aidy S. Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson’s disease. Nat Commun. 2019;10:310. doi:10.1038/s41467-019-08294-y.
- Anderson WA, Moo-Young M, Legge RL. Development of a multienzyme reactor for dopamine synthesis: II. Reactor engineering and simulation. Biotechnol Bioeng. 1992;40(3):388–395. doi:10.1002/bit.260400308.
- Lyte M. Microbial endocrinology as a basis for improved L-DOPA bioavailability in Parkinson’s patients treated for Helicobacter pylori. Med Hypotheses. 2010;74(5):895–897. doi:10.1016/j.mehy.2009.11.001.
- Maini Rekdal V, Bess EN, Bisanz JE, Turnbaugh PJ, Balskus EP. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science. 2019;364. doi:10.1126/science.aay2204.
- Mou Z, Yang Y, Hall AB, Jiang X. The taxonomic distribution of histamine-secreting bacteria in the human gut microbiome. BMC Genomics. 2021;22(1):695. doi:10.1186/s12864-021-08004-3.
- Lyte JM. Eating for 3.8 x 10 (13):Examining the Impact of Diet and Nutrition on the Microbiota-Gut-Brain Axis Through the Lens of Microbial Endocrinology. Front Endocrinol (Lausanne). 2018;9:796. doi:10.3389/fendo.2018.00796.
- Xie Z, Miller GM. Beta-phenylethylamine alters monoamine transporter function via trace amine-associated receptor 1: implication for modulatory roles of trace amines in brain. J Pharmacol Exp Ther. 2008;325:617–628. doi:10.1124/jpet.107.134247.
- Babusyte A, Kotthoff M, Fiedler J, Krautwurst D. Biogenic amines activate blood leukocytes via trace amine-associated receptors TAAR1 and TAAR2. J Leukoc Biol. 2013;93:387–394. doi:10.1189/jlb.0912433.
- Ohta H, Takebe Y, Murakami Y, Takahama Y, Morimura S. Tyramine and beta-phenylethylamine, from fermented food products, as agonists for the human trace amine-associated receptor 1 (hTAAR1) in the stomach. Biosci Biotechnol Biochem. 2017;81:1002–1006. doi:10.1080/09168451.2016.1274640.
- Bugda Gwilt K, Gonzalez DP, Olliffe N, Oller H, Hoffing R, Puzan M, El Aidy S, Miller GM. Actions of Trace Amines in the Brain-Gut-Microbiome Axis via Trace Amine-Associated Receptor-1 (TAAR1). Cell Mol Neurobiol. 2020;40:191–201. doi:10.1007/s10571-019-00772-7.
- Monti JM. Serotonin control of sleep-wake behavior. Sleep Med Rev. 2011;15:269–281. doi:10.1016/j.smrv.2010.11.003.
- Blundell JE. Serotonin and appetite. Neuropharmacology. 1984;23:1537–1551. doi:10.1016/0028-3908(84)90098-4.
- Yun HM, Park KR, Hong JT, Kim EC. Peripheral serotonin-mediated system suppresses bone development and regeneration via serotonin 6 G-protein-coupled receptor. Sci Rep. 2016;6:30985. doi:10.1038/srep30985.
- Nam SS, Lee JC, Kim HJ, Park JW, Lee JM, Suh JY, Um HS, Kim JY, Lee Y, Kim YG, et al. Serotonin inhibits osteoblast differentiation and bone regeneration in rats. J Periodontol. 2016;87:461–469. doi:10.1902/jop.2015.150302.
- Shajib MS, Khan WI. The role of serotonin and its receptors in activation of immune responses and inflammation. Acta Physiol (Oxf). 2015;213:561–574. doi:10.1111/apha.12430.
- Crane JD, Palanivel R, Mottillo EP, Bujak AL, Wang H, Ford RJ, Collins A, Blümer RM, Fullerton MD, Yabut JM, et al. Inhibiting peripheral serotonin synthesis reduces obesity and metabolic dysfunction by promoting brown adipose tissue thermogenesis. Nat Med. 2015;21(2):166–172. doi:10.1038/nm.3766.
- Gershon MD, Tack J. The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology. 2007;132:397–414. doi:10.1053/j.gastro.2006.11.002.
- Nzakizwanayo J, Dedi C, Standen G, Macfarlane WM, Patel BA, Jones BV. Escherichia coli Nissle 1917 enhances bioavailability of serotonin in gut tissues through modulation of synthesis and clearance. Sci Rep. 2015;5:17324. doi:10.1038/srep17324.
- Gotoh A, Nara M, Sugiyama Y, Sakanaka M, Yachi H, Kitakata A, Nakagawa A, Minami H, Okuda S, Katoh T, et al. Use of Gifu Anaerobic Medium for culturing 32 dominant species of human gut microbes and its evaluation based on short-chain fatty acids fermentation profiles. Biosci Biotechnol Biochem. 2017;81:2009–2017. doi:10.1080/09168451.2017.1359486.
- Sugiyama Y, Nara M, Sakanaka M, Gotoh A, Kitakata A, Okuda S, Kurihara S. Comprehensive analysis of polyamine transport and biosynthesis in the dominant human gut bacteria: potential presence of novel polyamine metabolism and transport genes. Int J Biochem Cell Biol. 2017;93:52–61. doi:10.1016/j.biocel.2017.10.015.
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi:10.1186/1471-2105-10-421.
- Nelson DA, Tolbert MD, Singh SJ, Bost KL. Expression of neuronal trace amine-associated receptor (Taar) mRNAs in leukocytes. J Neuroimmunol. 2007;192(1–2):21–30. doi:10.1016/j.jneuroim.2007.08.006.
- Deleu D, Northway MG, Hanssens Y. Clinical pharmacokinetic and pharmacodynamic properties of drugs used in the treatment of Parkinson’s disease. Clin Pharmacokinet. 2002;41:261–309. doi:10.2165/00003088-200241040-00003.
- Schroeder BO, Backhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22:1079–1089. doi:10.1038/nm.4185.
- Kitada Y, Muramatsu K, Toju H, Kibe R, Benno Y, Kurihara S, Matsumoto M. Bioactive polyamine production by a novel hybrid system comprising multiple indigenous gut bacterial strategies. Sci Adv. 2018;4:eaat0062. doi:10.1126/sciadv.aat0062.
- Liou CS, Sirk SJ, Diaz CAC, Klein AP, Fischer CR, Higginbottom SK, Erez A, Donia MS, Sonnenburg JL, Sattely ES, et al. A metabolic pathway for activation of dietary glucosinolates by a human gut symbiont. Cell. 2020;180:717–28 e19. doi:10.1016/j.cell.2020.01.023.
- Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551:648–652. doi:10.1038/nature24661.
- Penttila A, Lempinen M. Enterochromaffin cells and 5-hydroxytryptamine in the human intestinal tract. Gastroenterology. 1968;54:375–381. doi:10.1016/S0016-5085(68)80098-8.
- Reigstad CS, Salmonson CE, Rainey JF 3rd, Szurszewski JH, Linden DR, Sonnenburg JL, Farrugia G, Kashyap PC. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015;29:1395–1403. doi:10.1096/fj.14-259598.
- Kidd M, Modlin IM, Gustafsson BI, Drozdov I, Hauso O, Pfragner R. Luminal regulation of normal and neoplastic human EC cell serotonin release is mediated by bile salts, amines, tastants, and olfactants. Am J Physiol Gastrointest Liver Physiol. 2008;295:G260–72. doi:10.1152/ajpgi.00056.2008.
- Zeng SL, Li SZ, Xiao PT, Cai YY, Chu C, Chen BZ, Li P, Li J, Liu EH. Citrus polymethoxyflavones attenuate metabolic syndrome by regulating gut microbiome and amino acid metabolism. Sci Adv. 2020;6:eaax6208. doi:10.1126/sciadv.aax6208.
- Chen H, Nwe PK, Yang Y, Rosen CE, Bielecka AA, Kuchroo M, Cline GW, Kruse AC, Ring AM, Crawford JM, et al. A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell. 2019;177:1217–31 e18. doi:10.1016/j.cell.2019.03.036.
- Tsugeno Y, Ito A. A key amino acid responsible for substrate selectivity of monoamine oxidase A and B. J Biol Chem. 1997;272:14033–14036. doi:10.1074/jbc.272.22.14033.
- Mishima E, Fukuda S, Shima H, Hirayama A, Akiyama Y, Takeuchi Y, Fukuda NN, Suzuki T, Suzuki C, Yuri A, et al. Alteration of the Intestinal Environment by Lubiprostone Is Associated with Amelioration of Adenine-Induced CKD. J Am Soc Nephrol. 2015;26(8):1787–1794. doi:10.1681/ASN.2014060530.
- Fujimura T, Shibata S, Shimojima N, Morikawa Y, Okano H, Kuroda T. Fluorescence Visualization of the Enteric Nervous Network in a Chemically Induced Aganglionosis Model. PLoS One. 2016;11:e0150579. doi:10.1371/journal.pone.0150579.
- Yadav VK, Balaji S, Suresh PS, Liu XS, Lu X, Li Z, Guo XE, Mann JJ, Balapure AK, Gershon MD, et al. Pharmacological inhibition of gut-derived serotonin synthesis is a potential bone anabolic treatment for osteoporosis. Nat Med. 2010;16:308–312. doi:10.1038/nm.2098.
- Camilleri M. LX-1031, a tryptophan 5-hydroxylase inhibitor, and its potential in chronic diarrhea associated with increased serotonin. Neurogastroenterol Motil. 2011;23(3):193–200. doi:10.1111/j.1365-2982.2010.01643.x.
- Coleman NS, Foley S, Dunlop SP, Wheatcroft J, Blackshaw E, Perkins AC, Singh G, Marsden CA, Holmes GK, Spiller RC, et al. Abnormalities of serotonin metabolism and their relation to symptoms in untreated celiac disease. Clin Gastroenterol Hepatol. 2006;4:874–881. doi:10.1016/j.cgh.2006.04.017.
- Wan M, Ding L, Wang D, Han J, Gao P. Serotonin: a potent immune cell modulator in autoimmune diseases. Front Immunol. 2020;11:186. doi:10.3389/fimmu.2020.00186.
- Oh CM, Namkung J, Go Y, Shong KE, Kim K, Kim H, Park BY, Lee HW, Jeon YH, Song J, et al. Regulation of systemic energy homeostasis by serotonin in adipose tissues. Nat Commun. 2015;6:6794. doi:10.1038/ncomms7794.
- Zhang X, Beaulieu JM, Sotnikova TD, Gainetdinov RR, Caron MG. Tryptophan hydroxylase-2 controls brain serotonin synthesis. Science. 2004;305:217. doi:10.1126/science.1097540.
- Kulikov AV, Osipova DV, Naumenko VS, Popova NK. Association between Tph2 gene polymorphism, brain tryptophan hydroxylase activity and aggressiveness in mouse strains. Genes Brain Behav. 2005;4:482–485. doi:10.1111/j.1601-183X.2005.00145.x.
- Wang H, Kwon YH, Dewan V, Vahedi F, Syed S, Fontes ME, Ashkar AA, Surette MG, Khan WI. TLR2 Plays a Pivotal Role in Mediating Mucosal Serotonin Production in the Gut. J Immunol. 2019;202:3041–3052. doi:10.4049/jimmunol.1801034.
- Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. doi:10.1038/nature25979.
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi:10.1016/0378-1119(85)90120-9.
- Nakagawa A, Matsumura E, Koyanagi T, Katayama T, Kawano N, Yoshimatsu K, Kumagai H, Sato F, Minami H. Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli. Nat Commun. 2016;7:10390. doi:10.1038/ncomms10390.
- Perez-Casal J, Caparon MG, Scott JR. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi:10.1128/jb.173.8.2617-2624.1991.
- Thurlow LR, Thomas VC, Hancock LE. Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J Bacteriol. 2009;191:6203–6210. doi:10.1128/JB.00592-09.
- Lane DJ. 16S/23S rRNA sequencing. Nucleic Acid Techniques in Bacterial Systematics. 1991;125–175.
- Clifford RJ, Milillo M, Prestwood J, Quintero R, Zurawski DV, Kwak YI, Waterman PE, Lesho EP, Mc Gann P. Detection of bacterial 16S rRNA and identification of four clinically important bacteria by real-time PCR. PLoS One. 2012;7:e48558. doi:10.1371/journal.pone.0048558.
- Sedgley CM, Nagel AC, Shelburne CE, Clewell DB, Appelbe O, Molander A. Quantitative real-time PCR detection of oral Enterococcus faecalis in humans. Arch Oral Biol. 2005;50:575–583. doi:10.1016/j.archoralbio.2004.10.017.
- Perez M, Calles-Enriquez M, Nes I, Martin MC, Fernandez M, Ladero V, Alvarez MA. Tyramine biosynthesis is transcriptionally induced at low pH and improves the fitness of Enterococcus faecalis in acidic environments. Appl Microbiol Biotechnol. 2015;99:3547–3558. doi:10.1007/s00253-014-6301-7.
- Zhang B, Wu X, Liu J, Song L, Song Q, Wang L, Yuan D, Wu Z. beta-Actin: not a Suitable Internal Control of Hepatic Fibrosis Caused by Schistosoma japonicum. Front Microbiol. 2019;10:66. doi:10.3389/fmicb.2019.00066.
- Sakanaka M, Sugiyama Y, Kitakata A, Katayama T, Kurihara S. Carboxyspermidine decarboxylase of the prominent intestinal microbiota species Bacteroides thetaiotaomicron is required for spermidine biosynthesis and contributes to normal growth. Amino Acids. 2016;48:2443–2451. doi:10.1007/s00726-016-2233-0.
- Muller T. Levodopa/carbidopa and entacapone in the treatment of Parkinson’s disease: efficacy, safety and patient preference. Patient Prefer Adherence. 2009;3:51–59. doi:10.2147/PPA.S4084.
- Bender DA, Coulson WF. Aromatic amino acid decarboxylase: pH-dependence of substrates and inhibitors [proceedings]. Biochem Soc Trans. 1977;5:1353–1356. doi:10.1042/bst0051353.
- Jonkers N, Sarre S, Ebinger G, Michotte Y. Benserazide decreases central AADC activity, extracellular dopamine levels and levodopa decarboxylation in striatum of the rat. J Neural Transm (Vienna). 2001;108:559–570. doi:10.1007/s007020170056.
- Miller JH. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1992.
- Sugiyama Y, Nakamura A, Matsumoto M, Kanbe A, Sakanaka M, Higashi K, Igarashi K, Katayama T, Suzuki H, Kurihara S, et al. A Novel Putrescine Exporter SapBCDF of Escherichia coli. J Biol Chem. 2016;291:26343–26351. doi:10.1074/jbc.M116.762450.
- Morita H, Kuwahara T, Ohshima K, Sasamoto H, Itoh K, Hattori M, Hayashi T, Takami H. An Improved DNA Isolation Method for Metagenomic Analysis of the Microbial Flora of the Human Intestine. Microbes and Environments. 2007;22:214–222. doi:10.1264/jsme2.22.214.
- Seishima J, Iida N, Kitamura K, Yutani M, Wang Z, Seki A, Yamashita T, Sakai Y, Honda M, Yamashita T, et al. Gut-derived Enterococcus faecium from ulcerative colitis patients promotes colitis in a genetically susceptible mouse host. Genome Biol. 2019;20:252. doi:10.1186/s13059-019-1879-9.
