ABSTRACT
Methanogens, reductive acetogens and sulfate-reducing bacteria play an important role in disposing of hydrogen in gut ecosystems. However, how they interact with each other remains largely unknown. This in vitro study cocultured Blautia hydrogenotrophica (reductive acetogen), Desulfovibrio piger (sulfate reducer) and Methanobrevibacter smithii (methanogen). Results revealed that these three species coexisted and did not compete for hydrogen in the early phase of incubations. Sulfate reduction was not affected by B. hydrogenotrophica and M. smithii. D. piger inhibited the growth of B. hydrogenotrophica and M. smithii after 10 h incubations, and the inhibition on M. smithii was associated with increased sulfide concentration. Remarkably, M. smithii growth lag phase was shortened by coculturing with B. hydrogenotrophica and D. piger. Formate was rapidly used by M. smithii under high acetate concentration. Overall, these findings indicated that the interactions of the hydrogenotrophic microbes are condition-dependent, suggesting their interactions may vary in gut ecosystems.
Introduction
The gut microbiota consists of a wide variety of microbial species with the ability to ferment dietary fibers and other complex substrates that escape digestion and absorption, resulting in the production of short chain fatty acids as well as carbon dioxide and hydrogen.Citation1–3 The accumulation of hydrogen thermodynamically restricts further microbial fermentation and growth.Citation4–6 Hydrogenotrophic microbes using hydrogen as the electron donor for their anaerobic respiration play an important role in maintaining the hydrogen balance in gut ecosystems.Citation4,Citation7 Moreover, hydrogenotrophic microbes have been suggested to play an important role in human health.Citation5,Citation6 Methanogens are considered beneficial or harmful for human health, and the associations of methanogens with obesity, anorexia, colorectal cancer, inflammatory bowel disease, irritable bowel syndrome, diverticulosis, atherosclerosis and periodontitis were described by Chaudhary and colleagues.Citation8 Hydrogen sulfide has been implicated in the development of colorectal cancer.Citation9,Citation10 Higher sulfate-reducing bacteria (SRB) abundance or hydrogen sulfide concentrations have been reported in ulcerative colitis patients compared to healthy individuals.Citation11,Citation12
Hydrogenotrophic microbes in humans consist of three major functional groups, namely methanogens, reductive acetogens and SRB.Citation6 Methanogens reduce carbon dioxide to methane using hydrogen as electron donor (4 H2 + CO2 → CH4 + 2 H2O).Citation13 Reductive acetogens use hydrogen and carbon dioxide producing acetate via the Wood-Ljungdahl metabolic pathway (4 H2 + 2 CO2 → CH3COOH + 2 H2O).Citation13 SRB reduce sulfate to hydrogen sulfide using hydrogen as electron donor (4 H2 + SO4Citation2− + 2 H+ → H2S + 4 H2O).Citation13 The prevalence of gut methanogens varies between populations with estimates of ~ 30% prevalence in the Western world and ~ 80% in Africa with Methanobrevibacter smithii as the dominant methanogenic archaeal species in the human gut.Citation5,Citation14 SRB that colonize the guts of ∼50% of humans show greater taxonomic diversity than methanogens with Desulfovibrio piger described as the most common species.Citation15 Reductive acetogens are phylogenetically diverse, and Blautia hydrogenotrophica is the most well-known and studied reductive acetogenic species.Citation5 It has been estimated that one-third or one-fourth of acetate in the gut is produced via reductive acetogenesis.Citation16
Competition between the three hydrogenotrophic functional groups has been considered because all of them can use hydrogen as an energy source.Citation4,Citation5 Thermodynamically, sulfate reduction with hydrogen is more favorable with a Gibbs free energy change under standard conditions of −152.2 kJ mol−1, compared to methanogenesis and acetogenesis with Gibbs free energy changes of −131 kJ mol−1 and −95 kJ mol−1, respectively.Citation17 It has been reported that SRB and methanogens are mutually exclusive.Citation18 SRB were rarely detected in the gut microbiota of so-called methane excretors that are subjects with an above average methanogen abundance, while the gut microbiota of non-methane excretors harbors a higher abundance of SRB.Citation18 In addition, lower acetogenesis has been found in the presence of methanogens, and inhibition of methanogens concomitantly led to higher acetate production in fecal cultures.Citation19 However, mutual exclusivity is not always found, and several studies have reported that no significant relationship was observed between methanogens and SRB.Citation20,Citation21 A recent study by Wang et al (2022) found that methanogens, reductive acetogens and SRB abundances did not show a negative correlation with each other indicating their coexistence in adult fecal samples.Citation22 Although these previous studies indicate that the three hydrogenotrophic functional groups may impact each other considering all of them use hydrogen as energy source, detailed insights into the interactions between them remains unknown.
Therefore, we cocultured the hydrogenotrophic microbial species B. hydrogenotrophica, D. piger and M. smithii with each other in vitro under hydrogenotrophic conditions, aiming to understand how these three species affect each other’s growth and metabolic activity in vitro. Moreover, the impact of sulfide concentrations on these species was investigated as hydrogen sulfide produced by sulfate reduction is highly reactive and toxic to microbes.Citation23,Citation24 Furthermore, we described the impact of formate and acetate concentrations produced during incubations on the growth of M. smithii as they could serve as substrates for methanogens.Citation25,Citation26
Results
Coculturing with B. hydrogenotrophica or D. piger shortens the lag phase of M. smithii
To study the interactions between the three hydrogenotrophic species, B. hydrogenotrophica DSM 10,507T (B), D. piger DSM 749T (D) and M. smithii DSM 11,975 (M) were cultured in monocultures, in binary cocultures and in triculture (Experiment one, ). Hydrogen was consumed by the three hydrogenotrophic species over time in all the cultures (). As expected, methane was only detected in the presence of M. smithii. Sulfate was consumed and sulfide produced concomitantly, only in the presence of D. piger. Acetate production was only observed in the presence of B. hydrogenotrophica.
Figure 1. Interactions between hydrogenotrophic species. (a) schematic overview of Experiment one. B. hydrogenotrophica, D. piger and M. smithii were cultured in monoculture, or together in binary cocultures and a triculture. (b) hydrogen consumption and metabolite production of B. hydrogenotrophica, D. piger and M. smithii under hydrogenotrophic conditions. (c) log 16S rRNA gene copies per mL of B. hydrogenotrophica, D. piger and M. smithii in monocultures, binary cocultures and a triculture. Data are shown as average ± standard deviation (n = 2). B: B. hydrogenotrophica monoculture; D: D. piger monoculture; M: M. smithii monoculture; BD: B. hydrogenotrophica and D. piger binary coculture; BM: B. hydrogenotrophica and M. smithii binary coculture; DM: D. piger and M. smithii binary coculture; BDM: B. hydrogenotrophica, D. piger and M. smithii triculture.
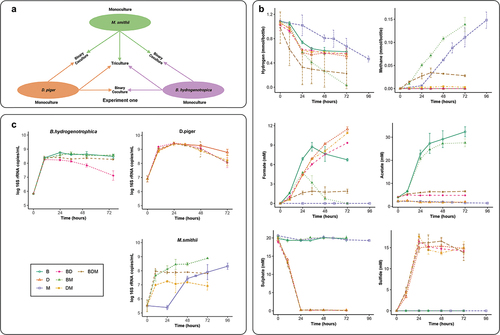
For monocultures, D. piger and B. hydrogenotrophica consumed hydrogen much faster than M. smithii in the early phase of incubation. B. hydrogenotrophica and D. piger grew rapidly and their 16S rRNA gene copy numbers peaked at 24 h. In contrast, M. smithii showed no growth in the first 24 h, but started fast growth afterward and consumed hydrogen throughout the incubation ().
When cocultured with B. hydrogenotrophica or M. smithii, sulfate reduction and the growth of D. piger were not affected. In contrast, B. hydrogenotrophica and M. smithii growth and metabolism were impacted when cocultured with other hydrogenotrophic species. The monoculture of B. hydrogenotrophica produced acetate throughout the 72 h incubations. However, B. hydrogenotrophica stopped producing acetate in the presence of D. piger (BD and BDM) after 10 h incubation. The 16S rRNA gene copy numbers of B. hydrogenotrophica stopped increasing as well. When cocultured with M. smithii (BM), the 16S rRNA gene copy number of B. hydrogenotrophica was not affected compared to its monoculture. However, the BM coculture ended up with a lower acetate concentration at the end of the incubation compared to the monoculture of B. hydrogenotrophica. Remarkably, in BM and DM cocultures, M. smithii had a much shorter lag phase compared to its monoculture: < 10 h versus > 24 h. Afterward, M. smithii stopped growing when cocultured with D. piger (DM and BDM). However, it continued to grow in the coculture with B. hydrogenotrophica (BM) with a similar trend as in its monoculture. Consistently, a small amount of methane (0.0018 ± 0.0007 mmol/bottle) was observed in DM in the first 10 h without a further increase afterward, and in the BDM culture methane was only produced in the first 34 h. In contrast, the BM coculture showed a faster methane production in the first 10 h of incubation compared to the monoculture of M. smithii. After 10 h, methane continued to be rapidly produced in the BM coculture with a similar trend as the monoculture of M. smithii ().
Formate was only detected in cultures with B. hydrogenotrophica or D. piger. For monoculture of B. hydrogenotrophica, formate was produced fast and peaked at 34 h, and started to decrease afterward. Interestingly, in the BM coculture, the highest formate concentration was observed at 24 h, after which it started to decrease until depletion at 72 h. In contrast, the formate in the BD and DM cocultures followed a similar trend as the monoculture of D. piger. The BDM triculture had a fast formate production in the first 24 h, after which its concentration remained stable over time ().
High sulfide concentrations inhibit the growth of M. smithii
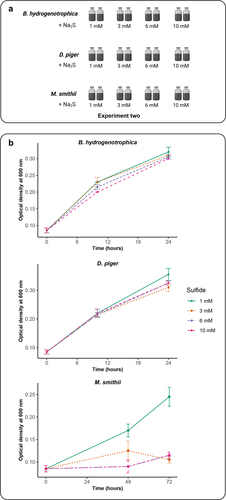
When cocultured with D. piger, methane production by M. smithii and acetate production by B. hydrogenotrophica were inhibited in the late phase of incubations. We speculated this could be caused by the increased sulfide concentration produced by D. piger during incubations as hydrogen sulfide is highly reactive and toxic to microbes.Citation27 To confirm the effect of sulfide on the growth of the hydrogenotrophic species, monocultures of the three species were performed with different sulfide concentrations ranging from 1 to 10 mM (Experiment two, ). We observed that the growth of D. piger and B. hydrogenotrophica was not impacted by sulfide concentrations. Interestingly, low concentration sulfide (1 mM) had no effect on the growth of M. smithii, whereas 3 mM, 6 mM and 10 mM sulfide inhibited the M. smithii growth. These findings suggest that produced sulfide in the DM and BDM cocultures over time inhibited the growth of M. smithii ().
Figure 2. Impact of sulfide concentrations on the growth of B. hydrogenotrophica, D. piger and M. smithii. (a) schematic overview of Experiment two (b) the optimal density of B. hydrogenotrophica, D. piger and M. smithii under different sulfide concentrations during incubations. Data are shown as average ± standard deviation (n = 2).
High acetate concentrations contribute to the usage of formate by M. smithii
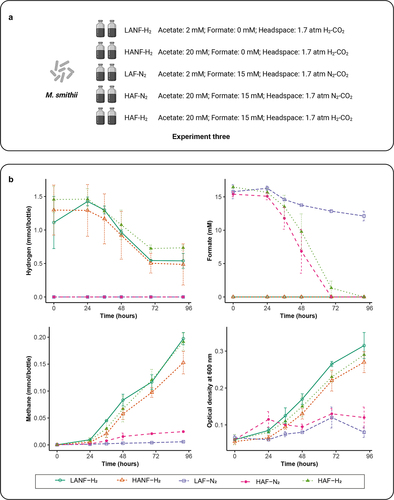
The lag phase of M. smithii was shortened in the presence of B. hydrogenotrophica or D. piger (). We determined whether the production of acetate and formate could explain this phenomenon by growing M. smithii in media supplemented different concentrations of acetate and formate in hydrogenotrophic and non-hydrogenotrophic conditions (Experiment three, ). We observed that both hydrogen and formate were consumed, concomitantly with an increase of methane and culture density (). Compared to HANF-H2 (20 mM acetate, 0 mM formate, 1.7 atm H2-CO2 in the headspace), LANF-H2 (2 mM acetate, 0 mM formate, 1.7 atm H2-CO2 in the headspace) showed higher methane production after 92 h indicating that a lower acetate concentration (2 mM) was more favorable for hydrogenotrophic methanogenesis than the higher one (20 mM). Using formate as an electron donor, HAF-N2 (20 mM acetate, 15 mM formate, 1.7 atm N2-CO2 in the headspace) showed increased formate consumption with increased methane production compared to LAF-N2 (2 mM acetate, 15 mM formate, 1.7 atm N2-CO2 in the headspace) after 92 h incubations, indicating that a higher acetate concentration (20 mM) was more favorable for formate-dependent methanogenesis. Moreover, compared to HANF-H2, HAF-H2 (20 mM acetate, 15 mM formate, 1.7 atm H2-CO2 in the headspace) showed more methane produced with concomitant depletion of formate after 92 h incubation. Although we observed differences in metabolic activities when the concentrations of formate and acetate were modified, a shortened lag phase of M. smithii was not observed, suggesting that a yet unidentified mechanism in the cocultures is responsible for this.
Figure 3. Impact of acetate and formate concentrations on the growth of M. smithii. (a) schematic overview of Experiment three. (b) hydrogen and formate consumption, methane production and microbial density of M. smithii during incubations. Data are shown as average ± standard deviation (n = 2). LANF-H2: 2 mM acetate, 0 mM formate, H2-CO2 headspace; HANF-H2: 20 mM acetate, 0 mM formate, H2-CO2 headspace; LAF-N2: 2 mM acetate, 15 mM formate, N2-CO2 headspace; HAF-N2: 20 mM acetate, 15 mM formate, N2-CO2 headspace; HAF-H2: 20 mM acetate, 15 mM formate, H2-CO2 headspace.
Discussion
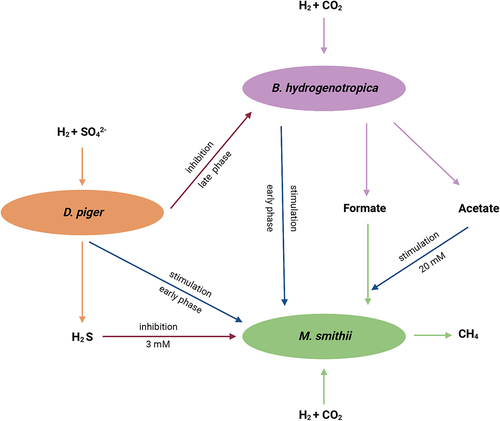
In this study, B. hydrogenotrophica, D. piger and M. smithii were cocultured to investigate their interactions under hydrogenotrophic conditions. The main findings are summarized in and indicated that the three hydrogenotrophic species coexisted and did not compete for hydrogen in the early phase of incubations. Coculturing with B. hydrogenotrophica and D. piger shortened the lag phase of M. smithii, concomitantly resulting in faster methane production. However, the presence of D. piger inhibited the growth of B. hydrogenotrophica and M. smithii and their metabolite production in the late phase of incubations. In addition, we found that high sulfide concentrations inhibited methanogenesis. A higher acetate concentration stimulated the usage of formate by M. smithii.
Figure 4. Summary of the interactions between B. hydrogenotrophica, D. piger and M. smithii under hydrogenotrophic conditions.
It has been considered that the three hydrogenotrophic functional groups may compete because all of them use hydrogen as an energy source in the human gut.Citation4,Citation5 However, our results by coculturing the three hydrogenotrophic species clearly showed that the interactions between the three hydrogenotrophic functional groups are condition dependent. Accordingly, considering the complexity of the human gut ecosystem including its nutrient supply, the variable environmental conditions throughout the gut, as well as the metabolic flexibility of some hydrogenotrophic microbes, the interactions between hydrogenotrophic microbes in the gut are potentially complex and certainly environment-dependent,Citation5,Citation28,Citation29 which might explain the inconsistent findings between studies.
SRB have the greatest affinity for hydrogen and dissimilatory sulfate reduction is thermodynamically more favorable than methanogenesis and reductive acetogenesis.Citation17,Citation30 It has been suggested this advantage is negated when sulfate is depleted in the gut. Consistently, in our study, we observed that 20 mM of sulfate was quickly depleted by D. piger within 24 h, which limits the growth of D. piger under hydrogenotrophic conditions. Hydrogen sulfide is highly reactive and toxic to microbes since it can diffuse across the cell membrane and is involved in protein denaturation and enzyme inhibition.Citation23,Citation24 Moreover, this toxicity has been suggested to be associated with hydrogen sulfide concentrations.Citation24 We indeed confirmed that 3 mM sulfide inhibited the growth of M. smithii. However, the growth of D. piger and B. hydrogenotrophica was not affected by high sulfide concentrations, indicating that the toxicity of sulfide is species dependent. Sulfide concentrations vary between individuals. It has been reported that the mean total sulfide content in wet feces was 0.66 mmol/kg.Citation31 However, a study in which individuals consumed a high-meat diet (600 g/day) rich in sulfur-containing amino acids, showed a much higher fecal sulfide content reaching levels of 3.38 mmol/kg.Citation32 This suggests that interactions between SRB and M. smithii may vary and are partially determined by the daily diet consumed as multiple sources of sulfur are present in the gut, including organic components from consumed plant-based diets as well as host-secreted components, such as mucus. SRB and methanogens have been reported to be coexisted or mutually exclusiveCitation18,Citation20,Citation21 and these inconsistent observations could be associated with the luminal sulfide concentrations, which should be further confirmed to illustrate the importance of hydrogen sulfide in the hydrogen metabolism in gut ecosystems.
Synergistic metabolic relationships have been proposed between M. smithii and hydrogen-producers.Citation29,Citation33 Bacteroides thetaiotaomicron is a well-known saccharolytic bacterium, and its fermentation resulting in production of hydrogen, formate and acetate supports the growth of M.smithii.Citation29,Citation34 In turn, the presence of M. smithii increases the metabolic efficiency of Bacteroides thetaiotaomicron.Citation29 Interestingly, cocolonization of M. smithii and Bacteroides thetaiotaomicron in a humanized gnotobiotic mouse model showed an increased population size of both in cecum and distal colon.Citation35 In addition, coculturing M. smithii and the species of the bacterial family Christensenellaceae indicated a syntrophic relationship via interspecies hydrogen transfer, resulting in higher acetate but lower butyrate production compared to monocultures of Christensenellaceae strains.Citation33 Although B. hydrogenotrophica and D. piger are hydrogenotrophic microbes, they are not obligatorily dependent on hydrogen and could even potentially have similar syntrophic interactions as described above to B. thetaiotaomicron and strains of Christensenellaceae. B. hydrogenotrophica can use glucose and fructose to grow and produce acetate and formate.Citation36,Citation37 Besides hydrogen consumption D. piger is also able to perform fermentation of pyruvate resulting in the production of hydrogen, formate and acetate.Citation38 Both formate and acetate can favor the growth of M. smithii as indicated in a study published recently,Citation26 suggesting possible syntrophic relationships between the M. smithii and the B. hydrogenotrophica and D. piger. Interestingly, we found that the lag phase of M. smithii was consistently shortened by both B. hydrogenotrophica and D. piger. However, this was not due to the increased formate and acetate concentrations. The exact mechanism of this stimulation effect by B. hydrogenotrophica and D. piger remains unknown and needs further studies. Moreover, we found that formate was rapidly converted to methane only under high acetate concentration (20 mM). Therefore using formate as an energy source with high acetate concentration can be an alternative of hydrogen and carbon dioxide for growing M. smithii.Citation26
In this study, we cocultured the three hydrogenotrophic functional groups to give insights into their interactions, which could improve our understanding in their interactions in the human gut. However, our study, like any in vitro study, cannot completely mimic the complexity of the gut environment in vivo and further research is needed to evaluate our findings in vivo. Unfortunately, given the multiple comparative cultivations in our study, we could only obtain duplicate samples that are not sufficient to support sound statistical assessment of potential differences between groups. Nevertheless, although having at least triplicate measurements would be necessary to allow for statistical analyses, our data generally showed that the duplicate measurements followed the expected trend with acceptable hydrogen balances (Supplementary Table S1).
In conclusion, this in vitro study gives a detailed overview of interactions between the three hydrogenotrophic species under hydrogenotrophic conditions. Results revealed that the interactions and metabolisms of these hydrogenotrophic functional groups are condition-dependent in vitro. Their relationships are complex and may vary throughout the gut considering the variable environmental conditions in vivo, and thus not easy to extrapolate to the in vivo situations in the gut. Our study thus may explain why inconsistent observations are reported about the coexistence of hydrogenotrophs in the gut.
Materials and methods
Study set up
To study the interactions between the three hydrogenotrophic species, Blautia hydrogenotrophica DSM 10,507T (B), Desulfovibrio piger DSM 749T (D) and Methanobrevibacter smithii DSM 11,975 (M) were cultured in duplicate, in monoculture, in binary cocultures: B. hydrogenotrophica and D. piger (BD), B. hydrogenotrophica and M. smithii (BM), D. piger and M. smithii (DM), and all of them together (BDM) (, Experiment one) using a basal medium (Supplementary Table S2) as previously describedCitation39 with some modifications that included the addition of 2 mM sodium acetate, 20 mM sodium sulfate, 1 g/L yeast extract (OXOID) and 1 g/L tryptone (OXOID). Incubation was performed in 30 mL serum bottles containing 10 mL medium. The bottle headspace consisted of a mixture of H2 and CO2 (80:20, v/v; 1.7 atm). Individual precultures of the three species were prepared prior to the inoculations with the same medium. After inoculations of each species, the optical density (OD) at 600 nm with a spectrophotometer (600 DiluPhotometer, Implen GmbH, Munich, Germany) was measured when their growth was in the mid-log phase. Subsequently, precultures were diluted with the same medium and 0.5 mL precultures (OD = 0.2) were used to inoculate monocultures, binary cocultures and tricultures for each species. Cultures were incubated at 37°C with 150 rpm. All samples were taken at 0 h, 10 h, 24 h, 34 h, 48 h and 72 h except for the monoculture of M. smithii for which samples were taken at 0 h, 24 h, 48 h, 58 h, 72 h and 96 h due to its slower growth compared to other cultures.
To study the effect of sulfide concentrations on the growth of B. hydrogenotrophica, D. piger and M. smithii, the same medium as described above with different sulfide concentrations (1 mM, 3 mM, 6 mM and 10 mM) modified by adding sodium sulfide were used to incubate these three species (, Experiment two). The incubation condition and inoculation were the same as described above. For B. hydrogenotrophica and D. piger, samples were taken at 0 h, 10 h, 24 h. For M. smithii, samples were taken at 0 h, 48 h, 72 h due to its slower growth compared to other cultures.
To study the effect of formate and acetate concentrations on the growth of M. smithii, the same medium as mentioned in Experiment one was used with the following modifications (, Experiment three): no modification (2 mM acetate, 0 mM formate, 1.7 atm H2-CO2 in the headspace; LANF-H2); addition of 20 mM acetate without modification of the headspace (HANF-H2); addition of 15 mM formate with the headspace flushed with a mixture of N2-CO2 (80:20, v/v; 1.7 atm; LAF-N2,); addition of 20 mM acetate and 15 mM formate with the headspace flushed with a mixture of N2-CO2 (80:20, v/v; 1.7 atm; HAF-N2); addition of 20 mM acetate and 15 mM formate without modification of the headspace (HAF-H2). For Experiment three, samples were taken at 0 h, 24 h, 36 h, 48 h, 69 h and 92 h for all incubations.
Sampling and analytical methods
For sampling, 0.2 mL gas samples were taken using a sterile 1 mL syringe from the headspace of the serum bottle and were analyzed immediately by gas chromatography (GC). 1 mL culture medium was taken at each sampling timepoint and subsequently centrifuged at 4°C at maximum speed (21130 × g) for 10 min to separate the microbial biomass and supernatant. Afterwards, 200 µL supernatant was mixed with 50 μL of ZnCl2 solution (ZnCl2: 50 g/L; 0.2 mL/L acetic acid) to react and precipitate the sulfide for the subsequent analysis. The remaining supernatant and pellet were stored at −20 °C for further analysis.
Hydrogen and methane amount were detected using a Compact GC 4.0 (Global Analyser Solutions, Breda, the Netherlands) equipped with a molsieve 5A column, operated at 100°C coupled to a Carboxen 1010 pre-column. Detection was done via a Thermal Conductivity Detector. Argon was used as carrier gas with a flow rate of 5 mL/min and pressure of 325 kPa. The software Chromeleon (Version 7.2, Thermo Fisher Scientific, Waltham, Massachusetts, USA) was used for data processing with a standard curve to quantify hydrogen and methane concentrations.
Formate and acetate in the supernatant were detected via High-Performance Liquid Chromatography equipped with a Shodex SH1821 column (Showa Denko K.K., Tokyo, Japan) operated at 45°C. Four hundred microliters of supernatant were added to 600 μL of a DMSO solution at 10 mM in 0.1 N H2SO4. DMSO was used as an internal standard. Elution was performed with 0.01 N H2SO4 at a flow rate of 1 mL/min. Detection was done via a refractive index detector. The software Chromeleon (Version 7.2, Thermo Fisher Scientific) was used for data processing with a standard curve to quantify formate and acetate concentrations.
Sulfate was detected via Ion Chromatography (IC, ICS-2100, Thermo Fisher Scientific) using a Dionex IonPac AS16 column, operated at 30°C. Thirty microliters of supernatant were added to 970 µL of an internal standard solution (0.5 mM sodium iodide prepared in ultra-pure water). Ultra-pure water was used as the eluent with a flow rate of 0.1 mL/min. Detection was done via an electrochemical IC detector. The software Chromeleon (Version 7.2, Thermo Fisher Scientific) was used for data processing with a standard curve to quantify sulfate concentrations.
Fifty microliters of the sample and ZnCl2 mixture were used to determine hydrogen sulfide concentrations. Hydrogen sulfide was quantified via the methylene-blue method as described previously.Citation40
Quantitative polymerase chain reaction (qPCR) analysis
Microbial count for each species was determined using qPCR for Experiment one. DNA extraction was performed via the repeated beat-beating method as described previouslyCitation41 with a small modification. Briefly, 300 µL of Stool Transport and Recovery (STAR) buffer (Roche Diagnostics, United States) was mixed with each pellet for the first beat-beating, and 200 µL of the STAR buffer was added for the second bead-beating. DNA concentrations were determined with a Qubit Fluorometer (Life Technologies, Darmstadt, Germany) in combination with the dsDNA BR Assay kit (Invitrogen, Carlsbad, CA, USA), and subsequently was adjusted to 1 ng/µL with nuclease-free water for qPCR.
The three microbial species were quantified using their 16S rRNA gene fragments. A standard template for each hydrogenotrophic species was generated using purified PCR products. The 16S rRNA gene PCR products for B. hydrogenotrophica and D. piger were obtained using universal primers 27F (5’-AGAGTTTGATCMTGGCTCAG-3’) and 1492 R (5’-GGTTACCTTGTTACGACTT-3’) with the DNA extracted from B. hydrogenotrophica DSM 10,507 and D. piger DSM 749 as template, respectively.Citation42 The 16S rRNA gene PCR product for M. smithii was obtained using the primers A109F (5’- ACKGCTCAGTAACACGT −3’) and Arch1492R (5’- GGCTACCTTGTTACGACTT −3’) with the DNA extracted from M. smithii DSM 11,975 as template.Citation43,Citation44 All PCR products were purified using the GeneJET PCR Purification Kit (Thermo Fisher Scientific) and quantified using a Qubit Fluorometer (Life Technologies) in combination with the dsDNA BR Assay kit (Invitrogen). A dilution corresponding to 1010 copies/µL was prepared for each standard. Then all standards were serially ten-fold diluted with nuclease-free water and dilution from 109 copies/µL to 102 copies/µL was used for the standard curve. All qPCRs were carried out in triplicate with an iCycler iQ real-time detection system (Bio-Rad Laboratories BV). Each reaction mixture with a total volume of 10 μL contained 5 μL 2× iQ SYBR green (Bio-Rad Laboratories B.V.), 2 µL of DNA template (samples at 1 ng/μL or standards), 300 nM forward (1 µL) and reverse (1 µL) primers, and nuclease-free water (1 µL). Primers and qPCR conditions for each species are listed in Supplementary Table S3.
Author contributions
TW: study concept and design, laboratory work, statistical analysis and interpretation of data, drafting this manuscript. NL: study design and laboratory work, interpretation of data and critically revised the manuscript. CP: assisted in study design and critically revised the manuscript. HS: contributed to study supervision, assisted in study design and critically revised this manuscript. EZ: study concept and design, interpretation of data, lead the study supervision and critically revised this manuscript.
Supplemental Material
Download MS Word (69.9 KB)Acknowledgments
Dr. H. R. Gaskins from the University of Illinois at Urbana-Champaign is acknowledged for critically reading the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data generated or analyzed during this study are included in this report and supplementary tables .
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2023.2261784
Additional information
Funding
References
- Zoetendal EG, de Vos WM. Effect of diet on the intestinal microbiota and its activity. Curr Opin Gastroenterol. 2014;30(2):189–12. doi:10.1097/MOG.0000000000000048.
- Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, Lawley TD, Finn RD. A new genomic blueprint of the human gut microbiota. Nature. 2019;568(7753):499–504. doi:10.1038/s41586-019-0965-1.
- Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661. doi:10.1038/nrmicro3344.
- Nakamura N, Lin HC, McSweeney CS, Mackie RI, Gaskins HR. Mechanisms of microbial hydrogen disposal in the human colon and implications for health and disease. Annu Rev Food Sci Technol. 2010;1(1):363–395. doi:10.1146/annurev.food.102308.124101.
- Smith NW, Shorten PR, Altermann EH, Roy NC, McNabb WC. Hydrogen cross-feeders of the human gastrointestinal tract. Gut Microbes. 2019;10(3):270–288. doi:10.1080/19490976.2018.1546522.
- Carbonero F, Benefiel AC, Gaskins HR. Contributions of the microbial hydrogen economy to colonic homeostasis. Nat Rev Gastro Hepat. 2012;9(9):504–518. doi:10.1038/nrgastro.2012.85.
- Strocchi A, Levitt MD. Factors affecting hydrogen production and consumption by human fecal flora. The critical roles of hydrogen tension and methanogenesis. J Clin Invest. 1992;89(4):1304–1311. doi:10.1172/JCI115716.
- Chaudhary PP, Conway PL, Schlundt J. Methanogens in humans: potentially beneficial or harmful for health. Appl Microbiol Biotechnol. 2018;102(7):3095–3104. doi:10.1007/s00253-018-8871-2.
- Kanazawa K, Konishi F, Mitsuoka T, Terada A, Itoh K, Narushima S, Kumemura M, Kimura H. Factors influencing the development of sigmoid colon cancer - bacteriologic and biochemical studies. Cancer. 1996;77(8):1701–1706.
- Cai WJ, Wang MJ, Ju LH, Wang C, Zhu YC. Hydrogen sulfide induces human colon cancer cell proliferation: role of akt, ERK and p21. Cell Biol Int. 2010;34(6):565–572. doi:10.1042/CBI20090368.
- Gibson G, Cummings J, Macfarlane G. Growth and activities of sulphate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol Lett. 1991;86(2):103–112. doi:10.1111/j.1574-6968.1991.tb04799.x.
- Rowan F, Docherty NG, Murphy M, Murphy B, Coffey JC, O‘connell PR. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis Colon Rectum. 2010;53(11):1530–1536. doi:10.1007/DCR.0b013e3181f1e620.
- Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41(1):100–180. doi:10.1128/br.41.1.100-180.1977.
- Levitt MD, Furne JK, Kuskowski M, Ruddy J. Stability of human methanogenic flora over 35 years and a review of insights obtained from breath methane measurements. Clin Gastroenterol Hepatol. 2006;4(2):123–129. doi:10.1016/j.cgh.2005.11.006.
- Rey FE, Gonzalez MD, Cheng J, Wu M, Ahern PP, Gordon JI. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc Natl Acad Sci USA. 2013;110(33):13582–13587. doi:10.1073/pnas.1312524110.
- Miller TL, Wolin MJ. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl Environ Microb. 1996;62(5):1589–1592. doi:10.1128/aem.62.5.1589-1592.1996.
- Gibson GR, Macfarlane G, Cummings J. Sulphate reducing bacteria and hydrogen metabolism in the human large intestine. Gut. 1993;34(4):437. doi:10.1136/gut.34.4.437.
- Gibson GR, Macfarlane GT, Cummings JH. Occurrence of sulphate-reducing bacteria in human faeces and the relationship of dissimilatory sulphate reduction to methanogenesis in the large gut. J Appl Bacteriol. 1988;65(2):103–111. doi:10.1111/j.1365-2672.1988.tb01498.x.
- Bernalier A, Lelait M, Rochet V, Grivet J-P, Gibson GR, Durand M. Acetogenesis from H2 and CO2 by methane-and non-methane-producing human colonic bacterial communities. FEMS Microbiol Ecol. 1996;19(3):193–202. doi:10.1111/j.1574-6941.1996.tb00212.x.
- Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, et al. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci USA. 2011;108(supplement_1):4599–4606. doi:10.1073/pnas.1000071108.
- Doré J, Pochart P, Bernalier A, Goderel I, Morvan B, Rambaud J. Enumeration of H2-utilizing methanogenic archaea, acetogenic and sulfate-reducing bacteria from human feces. FEMS Microbiol Ecol. 1995;17(4):279–284. doi:10.1111/j.1574-6941.1995.tb00152.x.
- Wang T, van Dijk L, Rijnaarts I, Hermes GD, de Roos NM, Witteman BJ, de Wit NJW, Govers C, Smidt H, Zoetendal EG, et al. Methanogen levels are significantly associated with fecal microbiota composition and alpha diversity in healthy adults and irritable bowel syndrome patients. Microbiol Spectr. 2022;10(6):e01653–22. doi:10.1128/spectrum.01653-22.
- Paulo LM, Stams AJ, Sousa DZ. Methanogens, sulphate and heavy metals: a complex system. Rev Environ Sci Biotechnol. 2015;14(4):537–553. doi:10.1007/s11157-015-9387-1.
- Kushkevych I, Dordević D, Vítězová M. Toxicity of hydrogen sulfide toward sulfate-reducing bacteria desulfovibrio piger vib-7. Arch Microbiol. 2019;201(3):389–397. doi:10.1007/s00203-019-01625-z.
- Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, Latreille P, Kim K, Wilson RK, Gordon JI. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci USA. 2007;104(25):10643–10648. doi:10.1073/pnas.0704189104.
- Pilliol V, Guindo CO, Terrer E, Aboudharam G, Drancourt M, Grine G. Culturing clinical Methanobrevibacter smithii using GG medium in a minimal anaerobe atmosphere. J Microbiol Methods. 2023;207:106704. doi:10.1016/j.mimet.2023.106704.
- Karhadkar P, Audic J-M, Faup G, Khanna P. Sulfide and sulfate inhibition of methanogenesis. Water Res. 1987;21(9):1061–1066. doi:10.1016/0043-1354(87)90027-3.
- Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J. 2012;6(1):57–70. doi:10.1038/ismej.2011.90.
- Catlett JL, Carr S, Cashman M, Smith MD, Walter M, Sakkaff Z, Kelley C, Pierobon M, Cohen, MB, Buan, NR. Metabolic synergy between human symbionts Bacteroides and Methanobrevibacter. Microbiol Spectr. 2022;10(3):e01067–22.
- Kristjansson J, Schönheit P. Why do sulfate-reducing bacteria outcompete methanogenic bacteria for substrates? Oecologia. 1983;60(2):264–266. doi:10.1007/BF00379530.
- Florin T. Hydrogen sulphide and total acid-volatile sulphide in faeces, determined with a direct spectrophotometric method. Clin Chim Acta. 1991;196(2–3):127–134. doi:10.1016/0009-8981(91)90065-K.
- Magee EA, Richardson CJ, Hughes R, Cummings JH. Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am J Clin Nutr. 2000;72(6):1488–1494. doi:10.1093/ajcn/72.6.1488.
- Ruaud A, Esquivel-Elizondo S, de la Cuesta-Zuluaga J, Waters JL, Angenent LT, Youngblut ND, Ley RE. Syntrophy via interspecies H2 transfer between Christensenella and Methanobrevibacter underlies their global cooccurrence in the human gut. Mbio. 2020;11(1):11. doi:10.1128/mBio.03235-19.
- Traore SI, Khelaifia S, Armstrong N, Lagier J, Raoult D. Isolation and culture of Methanobrevibacter smithii by co-culture with hydrogen-producing bacteria on agar plates. Clin Microbiol Infect. 2019;25(12):e1561.1–e1561.5. doi:10.1016/j.cmi.2019.04.008.
- Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host–archaeal–bacterial mutualism. Proc Natl Acad Sci USA. 2006;103(26):10011–10016. doi:10.1073/pnas.0602187103.
- Bernalier A, Willems A, Leclerc M, Rochet V, Collins MD. Ruminococcus hydrogenotrophicus sp nov, a new H2/CO2-utilizing acetogenic bacterium isolated from human feces. Arch Microbiol. 1996;166(3):176–183. doi:10.1007/s002030050373.
- Bernalier A, Rochet V, Leclerc M, Dore J, Pochart P. Diversity of H2/CO2-utilizing acetogenic bacteria from feces of non-methane-producing humans. Curr Microbiol. 1996;33(2):94–99. doi:10.1007/s002849900081.
- Moore WC, Johnson J, Holdeman L. Emendation of Bacteroidaceae and Butyrivibrio and descriptions of Desulfomonas gen. nov. and ten new species in the genera Desulfomonas, Butyrivibrio, Eubacterium, Clostridium, and Ruminococcus. Int J Syst Evol Microbiol. 1976;26(2):238–252. doi:10.1099/00207713-26-2-238.
- Plugge CM. Anoxic media design, preparation, and considerations. Methods Enzymol. 2005;397:3–16.
- Cline JD. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14(3):454–458. doi:10.4319/lo.1969.14.3.0454.
- An R, Wilms E, Smolinska A, Hermes GD, Masclee AA, de Vos P, Schols HA, van Schooten FJ, Smidt H, Jonkers DMAE, et al. Sugar beet pectin supplementation did not alter profiles of fecal microbiota and exhaled breath in healthy young adults and healthy elderly. Nutrients. 2019;11(9):2193. doi:10.3390/nu11092193.
- Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microb. 1996;62(2):625–630. doi:10.1128/aem.62.2.625-630.1996.
- Whitehead T, Cotta M. Phylogenetic diversity of methanogenic archaea in swine waste storage pits. FEMS Microbiol Lett. 1999;179(2):223–226. doi:10.1111/j.1574-6968.1999.tb08731.x.
- Teske A, Hinrichs K-U, Edgcomb V, de Vera Gomez A, Kysela D, Sylva SP, Sogin ML, Jannasch HW. Microbial diversity of hydrothermal sediments in the Guaymas basin: evidence for anaerobic methanotrophic communities. Appl Environ Microb. 2002;68(4):1994–2007. doi:10.1128/AEM.68.4.1994-2007.2002.
