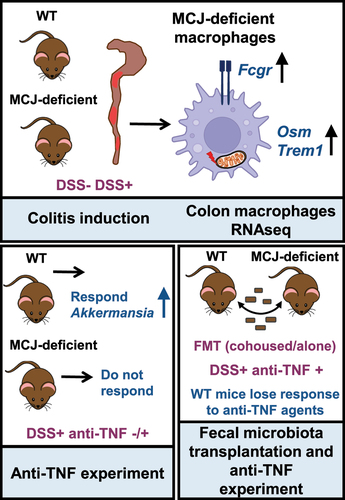
ABSTRACT
Anti-TNF therapy can induce and maintain a remission status during intestinal bowel disease. However, up to 30% of patients do not respond to this therapy by mechanisms that are unknown. Here, we show that the absence of MCJ, a natural inhibitor of the respiratory chain Complex I, induces gut microbiota changes that are critical determinants of the lack of response in a murine model of DSS-induced inflammation. First, we found that MCJ expression is restricted to macrophages in human colonic tissue. Therefore, we demonstrate by transcriptomic analysis of colon macrophages from DSS-induced mice that MCJ-deficiency is linked to the expression of genes belonging to the FcγR signaling pathway and contains an anti-TNF refractory gene signature identified in ulcerative colitis patients. The gut microbial composition changes observed upon DSS treatment in the MCJ-deficient mice revealed the increased presence of specific colitogenic members, including Ruminococcus gnavus and Oscillospira, which could be associated with the non-response to TNF inhibitors. Further, we show that the presence of a microbiota associated resistance to treatment is dominant and transmissible to responsive individuals. Collectively, our findings underscore the critical role played by macrophage mitochondrial function in the gut ecological niche that can substantially affect not only the severity of inflammation but also the ability to successfully respond to current therapies.
Introduction
Ulcerative colitis (UC) is a chronic, heterogeneous and severe inflammatory disease that primarily affects the gastrointestinal tract. Although its etiology remains unknown, evidence suggests a complex interplay between microbiota, the immune system, host genetics and environmental factors. Whilst there is no cure for UC, anti-tumor necrosis factor (TNF) agents are the most effective to induce and maintain a remission status in intestinal bowel disease (IBD) patients as they target the excessive production of TNF, a key proinflammatory cytokine produced mainly by activated immune cells, improving the quality of life of patients. However, there are IBD patients that are refractory to anti-TNF treatment and they can be categorized into two groups; those who never respond to anti-TNF induction therapy (primary failure), and 50% of those who initially respond to anti-TNF treatment but subsequently lose response overtime (secondary failure).Citation1 Furthermore, the treatment poses risks as serious adverse effects including severe infections have been reported.Citation2 Therefore, the identification of key factors that could allow clinicians to identify patients who will respond to anti-TNF therapy seems a priority to promote personalized treatment to IBD patients.
Recently, a study of the UC mucosal transcriptome revealed a decrease in mitochondrial electron transport chain Complex I activity, which was found to correlate with disease severity and treatment response in both adult and pediatric patients.Citation3 In addition, Mottawea et al. stated that host mitochondria-microbiota crosstalk was disturbed in IBD.Citation4 Consequently, mitochondrial dysfunction and microbial composition might play an important role in the response to therapy. Therefore, microbial-host mitochondria interactions seem to be a milestone in the development of effective therapies for UC patients.
Aberrant immune responses against bacteria are extensively described in the pathogenesis of UC. During UC, macrophages are activated by translocated pathogenic bacteria and are associated with the increased proinflammatory status characteristic of the disease.Citation5 In this regard, methylation-controlled J protein (MCJ, Dnajc15), a natural negative regulator of mitochondrial respiration that inhibits complex I activity of the electron transport chain regulating oxidative phosphorylation and ATP production,Citation6,Citation7 has been linked to macrophages. Deficiency of MCJ in bone marrow-derived macrophages leads to the upregulation of the tumor necrosis factor α-converting enzyme (TACE) inhibitor, tissue inhibitor of metalloproteinase 3 (TIMP-3), which inhibits TNF shedding from the plasma membrane.Citation8 As macrophages are pivotal for coordinating processes in the gut and a significant infiltration is produced during colitis, we investigated the expression levels of both genes in colon samples from UC patients reporting MCJ downregulation and TIMP3 upregulation.Citation9 These results led us to study the impact of MCJ deficiency in acuteCitation9 and chronicCitation10 experimental colitis and are in line with parameters disrupted during IBD activity in individuals suffering microbial dysbiosis.Citation11 In addition, we have rencently reported that the influence of MCJ deficiency on microbial composition has a detrimental effect on the severity of UC.Citation12
Because of the intimate relationship that is reported to exist between mitochondrial dysfunction and the efficacy of current therapies, particularly anti-TNF treatments, as well as the role played by macrophages during UC, we have now investigated the effect of mitochondrial dysfunction on the colon response to anti-TNF therapy. Our results show that the mitochondrial electron transport chain Complex I activity is determinant on the response to anti-TNF therapy. Importantly, our results link mitochondrial dysfunction with a microbiota composition leading to a lack of therapeutic efficacy that is dominant and transferable to otherwise responsive individuals. These data highlight the relevance of the physiological response of intestinal macrophages to maintain a homeostatic ecological niche that affects both the severity of gut inflammation and the response to current therapies.
Results
MCJ deficiency prevents a protective response to anti-TNF agents during experimental colitis
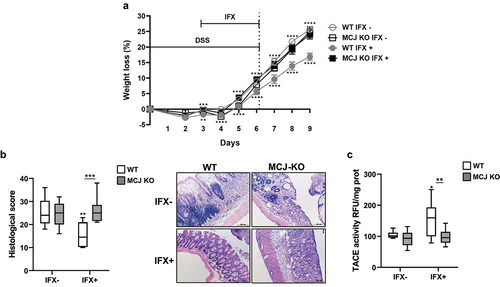
Mitochondria dysfunction, including the reduction of complex I activity in active patients,Citation3 has been related to disease severity and a refractory response to treatment during UC. We have shown that the absence of the complex I negative regulator, MCJ, results in increased DSS-induced pathology.Citation9 In order to evaluate the impact of mitochondrial dysfunction on the responsiveness to anti-TNF therapy during the disease, we evaluated the response of MCJ-deficient mice to Infliximab (IFX, anti-TNF) treatment. MCJ deficiency diminished the therapeutic efficacy of the infliximab biosimilar, as evidenced by the absence of a significant improvement in body weight () and histological scores (), in contrast to WT mice that responded successfully to the treatment. Moreover, TACE levels within the colon only increased in treated WT mice independently of TNF () and infiltrated macrophage numbers that did not vary (Figure S1a,b). TACE activity is responsible for the final release of mature TNF protein from cells. MPO levels were significantly increased (p ≤ .05) in both MCJ KO groups compared to WT groups independently of treatment (Figure S1c).
Figure 1. In vivo anti-TNF therapeutic response. WT and MCJ-KO mice were treated the first 6 d with DSS and infliximab (IFX) was administrated orally from day 3 to day 6, followed by 3 d of recovery period. All groups are positive for DSS. (a) Weight loss (%) (n = 8 mice per group at minimum). Statistical differences (P value < .05) were observed between WT IFX- and WT IFX+ from day 3 onward (asterisks below WT IFX+ line) and between MCJ KO IFX+ and WT IFX+ at day 3 and from day 5 onward (asterisks above WT IFX- line). (b) Histological scores and representative images of colon tissue (scale bar size, 100 µm). (c) TACE activity in colonic protein extracts (Rfu/mg protein). For statistical analysis two-way ANOVA was performed. Within boxplots, asterisks above boxes versus control genotype (IFX+ versus IFX-) and asterisks above line versus different genotypes in the same experimental group. (Wt_ifxn and MCJ KO_IFXn: DSS positive and IFX negative; WT_IFXp and MCJ KO_IFXp: DSS and IFX positive).
Transcriptional analysis of colon macrophages from MCJ-deficient mice shows overlapping signatures to a refractory response to treatment.
We have shown that MCJ is expressed weakly in the intestinal tissue both in mice and humans,Citation9 and that macrophages are, together with CD8 T cells, the cells of the immune system with the highest expression levels of the protein.Citation6,Citation8 We, therefore, determined the expression of MCJ in inflamed and healthy colon tissue from IBD patients by immunofluorescence staining. The transmembrane scavenger receptor CD163 was also used to identify the macrophage population within the tissue. MCJ was readily detected in both diseased () and adjacent, healthy colon tissue in colocalization with CD163. These data indicate that the impact of MCJ in IBD patients is primarily associated with macrophages.
Figure 2. Transcriptomic analysis of intestinal tissue macrophages. (a) Immunofluorescence of MCJ (red) and CD163 (green) in inflamed and healthy adjacent colon tissue from IBD patients. Colocalization is represented in yellow. (b) Principal component analysis showing differences between WT and MCJ-deficient DSS positive colon macrophages´ transcriptomes. (c) HOMER identified several transcription factors enriched in DSS-induced MCJ-KO colon macrophages. (d) venn diagram representing 339 differentially expressed genes found in colon macrophages due to MCJ-KO and a human core rectal UC gene expression signature consisting of 5296 genes.Citation3 Out of 305 genes upregulated in colitis induced MCJ-deficient mice colon macrophages, 183 were shared with the human UC gene signature. (e) venn diagram showing shared genes between upregulated genes due to MCJ-KO in DSS-induced mice colon macrophages and patients refractory to anti-TNF and corticosteroid treatments.
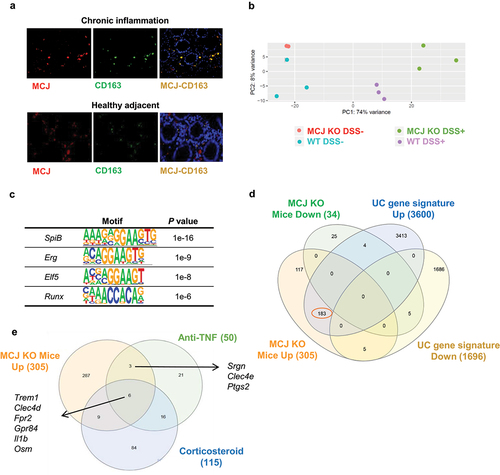
We then examined the role of MCJ in the immune response during the disease. We performed a transcriptome analysis of colon macrophages under DSS-induced colitis. Principal component analysis showed changes according to the disease status of the animals (). The analysis of the 50 most regulated genes showed different patterns of expression associated with the presence/absence of MCJ. MCJ-deficient macrophages expressed 305 upregulated genes compared to WT controls. Among them, there were several confirmed susceptibility-related genes for UC such as Irf5, Tnfrsf9, Fcgr2b Slc11a1, Itgal, Gpr65, Cd40, and Lsp1, suggesting a relevant role of mitochondria dysfunction in immune cells such as macrophages during the course of the disease.
The HOMER package identified a set of transcription factors putatively responsible for the expression changes in macrophages from DSS-induced colitis animals.Citation13 The analysis of the genes upregulated in MCJ-deficient macrophages compared to WT mice identified several transcription factors (, Table S2). Two of them (SpiB and Elf5) are related to Fcgr2b expression, while Runx is involved in Irf5 expression and Erg in the expression of Slc11a1 and Tnfrsf9. Moreover, Elf5 and SpiB possess motifs that regulate Il1b expression, while Erg showed a motif that regulates the expression of Tnf, Il10 and Tgfbi. On the other hand, Erg and Runx may regulate Cxcl3 expression and therefore be involved in leucocyte recruitment.
In order to discern whether transcriptional patterns associated with mitochondrial dysfunction in murine colon macrophages are related to the disease in humans, UC transcriptomes from human rectal samples were compared to experimental colitis macrophage transcriptomes. Out of the 305 upregulated genes in macrophages isolated from the colons of MCJ-deficient mice with induced experimental colitis compared to DSS+ controls (WT), 183 were shared between both cohorts (mouse model and patients) (, Table S3), suggesting a critical role for mitochondrial function during disease progression.
A subset of genes obtained from transcriptional analyses were linked to treatment response and shifts in microbial composition. In this regard, the 305 upregulated genes due to MCJ-deficiency in macrophages from experimental colitis mice were compared to the list of genes proposed to identify patients that are refractory to TNF blockade,Citation14 and corticosteroids.Citation3 Strikingly, 18 genes were shared with these 2 gene signatures (), in which Trem1 and Osm, connected to decreased responsiveness to anti-TNF therapy, were shared between the three gene signatures. Besides, the high expression of activating Fcgr1 and Fcgr3 found in inflammatory MCJ-deficient macrophages, which are cell surface glycoproteins that bind to the Fc portion of IgG antibodies, and the high levels of colonic IgG found (), suggested that the MCJ-deficient genotype might be associated with resistance to TNF blockade.
Figure 3. Functional annotation enrichment analysis of inflamed intestinal macrophages according to MCJ levels. RNA-seq data analysis shows 305 upregulated and 34 downregulated genes in MCJ-KO mice treated with DSS compared to control colitis mice (FDR <0.05 and fold change ≥1.5). ClueGO charts related to immunological function reveals (a) upregulation of FCγR signaling pathway (46,7%) in MCJ-KO colitis mice compared to WT colitis (Log2 > 2, P value < 0.05) and (b) downregulation of chaperone cofactor refolding (50%) and microtubule polymerization (50%). Detailed functional annotation enrichment analyses of (c) 305 upregulated and (c) 34 downregulated genes in MCJ-KO colitis mice using ToppGene, ToppCluster and Cytoscape are represented. (c) Pathways related to upregulated genes; immune system (purple), cytokines (orange), cell adhesion and migration (yellow) and other pathways (green). (d) Biological processes enriched by downregulated genes; mitochondrial respiration (blue), cytokines (orange), NOD-like receptors, the intracellular sensors of PAMPS (purple) and heat acclimation (red). (e) Fecal live bacteria percentage (n = 3) and (f) fecal IgG-coated bacteria percentage obtained by flow cytometer (n = 3). For statistical analysis two-way ANOVA was used.
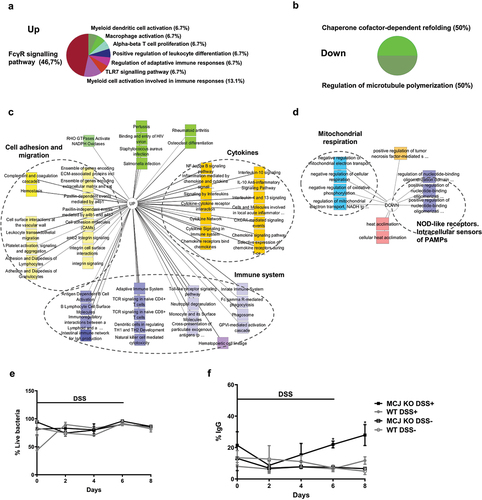
Functional annotation enrichment analysis using ToppGene, ToppCluster, and ClueGO mapped groups of related genes to immunological processes.Citation3 Overview of the ClueGo-derived immune system-related pathways () comparing gene expression based on MCJ deficiency during DSS-induced colitis showed 46% enrichment for the Fc gamma receptor (FCGR) signaling pathway, followed by myeloid cell (13,1%), macrophage (6,7%) and dendritic cell activation (6,7%), regulation of adaptive immune response (6,7%) and leukocyte differentiation (6,7%). The P values for the top specific biological processes and pathways were obtained as an output from ToppGene. A more detailed ToppCluster pathways analysis output showed genes related to immunological and biological processes (). MCJ-deficient mice presented higher expression of genes involved in immune system activation and therefore, higher production of cytokines and chemokines, and higher transendothelial migration of leukocytes. Importantly, IgA production was upregulated in MCJ-deficient colitis mice. Furthermore, the study of microbiota–host interactions along the DSS-induced colitis period showed distinct immune response kinetics. Live bacteria and immunoglobulin G (IgG)-coated fecal bacteria were quantified by flow cytometry. Starting at day 2 after the initiation of DSS treatment, the percentage of live bacteria was similar in all experimental groups (). MCJ-deficient DSS-induced mice exhibited higher IgG-coated fecal bacteria than WT mice increasing gradually until day 8, being significantly higher from day 6 (p = 0.022) to day 8 (p = 0.011) (). These data might indicate earlier microbial-host cross-talk and a faster activation of the immune system. Overall, these data suggest that MCJ modifies the kinetics of the immune response.
We then sought to find correlations between genes expressed in colon macrophages and specific microbial operational taxonomic units (OTUs). We analyzed the microbiota composition in MCJ-deficient and WT mice, both under homeostatic and DSS-induced pathology. As expected,Citation9 MCJ deficiency resulted in significant changes in microbiota composition (Figure S2(a,b)).
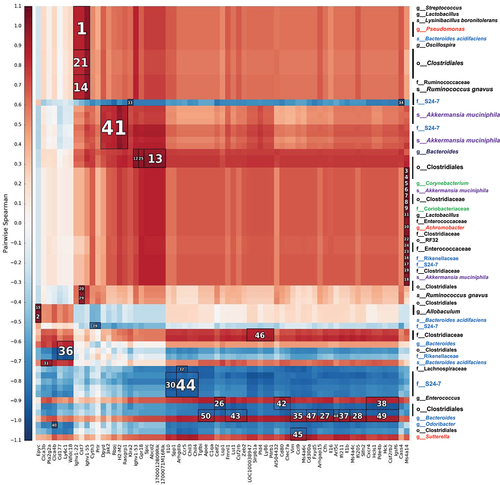
shows the 50 strongest correlations between genes differentially expressed due to MCJ deficiency in macrophages infiltrated during colon inflammation and OTUs present in the 4 experimental groups. Those included associations between Oscillospira and Ruminococcus gnavus, often increased with disease activity, and Cst7 that may play a role in immune regulation through the hematopoietic system. Akkermansia muciniphila presence, often regarded as an immunomodulator, correlated with Jak2 expression. Jak2 is involved in the production of key pro-inflammatory cytokines and consequently, Jak inhibitors are currently being investigated as therapeutic agents for UC. Enterococcus abundance, which is associated with disease severity, correlated with the expression levels of Lsp1, a gene involved in neutrophil transmigration. Different OTUs belonging to the genus Bacteroides, unbalanced in IBD patients, were linked to Ly6c1 (marker of resident macrophages) and Il1b gene expression. On the other hand, the Bacteroides S24–7 family showed a negative correlation with Il10 expression, which is related to gut homeostasis. These correlations suggest that the lack of MCJ provides the optimal environmental conditions for specific bacteria that may act as immunomodulators contributing to UC severity and the response to specific treatments, such as anti-TNF therapy.
Figure 4. Heatmap of Spearman´s rank correlation coefficients. Bacterial abundances from the 4 experimental groups that were not treated with antibiotic and genes modified by the level of MCJ in colon macrophages upon intestinal inflammation (DSS+) were used. To find associations hierarchical all against all association (HAIIA) was performed. A. muciniphila correlates with Jak2 and R. gnavus with Cst7. The numbers indicate the highest correlation between bacteria and gene expression, having number 1 the highest correlation. Red color illustrates positive correlation and blue negative. In the x axis each column shows a different gene and in the y axis each row shows different bacterial species; Firmicutes (black font), Proteobacteria (red), bacteroidetes (blue), actinobacteria (green) and Verrucomicrobia (purple).
Anti-TNF treatment alters the microbiota balance
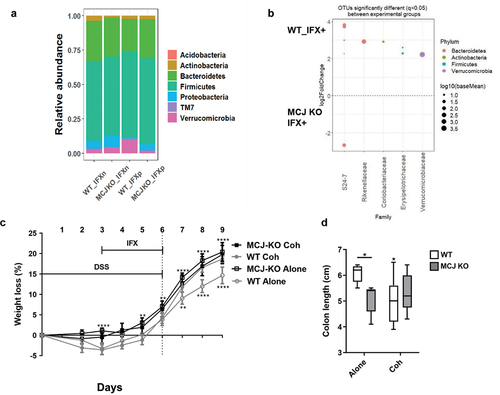
To assess the effect of anti-TNF agents on microbial composition and the impact on the therapeutic response, luminal bacterial communities were studied. They yielded a total of 1.189.812 paired and merged sequences, ranging from 56.923 to 101.477 sequences per sample after quality filtering. The good’s coverage was over 98%. Remarkably, the taxa barplot showed the significant enrichment of the Verrucomicrobia phylum (p = 0.01) and the reduction (p = 0.02) of Proteobacteria in WT mice treated with anti-TNF therapy compared to MCJ-deficient mice (). Although alpha diversity was not significantly altered, DESeq2 and LEfSe identified Akkermansia and OTUs from the S24–7 family augmented in the WT group ( and S1d) pointing to these microorganisms as critical in the primary response to the therapy. Overall, these data show that in contrast to their MCJ-deficient counterparts, WT mice group partially restore their gut microbiota in response to anti-TNF treatment. This microbial restoration may play a critical role in therapy response.
Figure 5. Microbiota composition impact on the anti-TNF therapeutic efficacy. (a) stacked bar plot representation of the relative abundances of colon content taxa at phylum level. (b) differentially abundant OTUs identified through DESeq2 testing (p.Adj <0.05) between WT and MCJ-KO mice treated with anti-TNF agents (WT_P vs MCJ-KO_P). Each point represents a single OTU colored by phylum and grouped by taxonomic family, and point´s size reflect the mean abundance of the sequenced data. To allow microbial transmission between genotypes, WT and MCJ-KO animals were cohoused during 4 weeks prior to colitis induction. (c) weight loss percentage shown as means ± SEM. Asterisks (*) above the line indicate statistical differences between WT and MCJ-KO mice housed alone. Asterisk (*) below the line shows significant differences between WT mice housed alone or cohoused. (d) colonic length (cm). Within boxplots, asterisks above boxes versus control genotype (cohoused versus alone) and asterisks above line versus different genotypes in the same experimental group. Two-way ANOVA (c, e, f) and (d) Mann-Whitney U test were used for statistical analysis. All experimental groups are DSS+ and IFX+.
Cohousing affects IFX treatment efficacy in WT but not MCJ-deficient animals
Then, we studied the link between gut microbiota and the therapeutic response. WT and MCJ-deficient mice were cohoused for 4 weeks prior to DSS and IFX administration. As before, WT mice housed alone effectively responded to IFX since they lost significantly less body weight () and they had longer colons compared to the other experimental groups (). As expected, MCJ-deficient mice housed alone did not respond to IFX treatment. Surprisingly, while MCJ-deficient, cohoused mice did not respond to IFX, cohoused WT mice displayed greater weight loss percentages from day 7 onward () and shorter colons () compared WT housed alone. These results indicate that microbiota changes induced by mitochondrial dysfunction are responsible for the refractory response to anti-TNF treatment. They also show that these changes are transmissible to otherwise responders, independently of their genotype. Furthermore, we have observed that after cohousing of WT and MCJ-deficient and treating them with DSS (not anti-TNF agents), colitis-induced MCJ-deficient mice had lower levels of Prevotella, Oscillospira and R. gnavus and increased Lactobacillus abundance compared to the MCJ-deficient mice housed alone.Citation12
Taken together, our study indicates that the mitochondrial dysfunction in macrophages from DSS-induced colitis mice affects a wide range of signaling pathways critical in the response to anti-TNF therapy. These data confirm a regulation of mitochondria and microbiota, and a pivotal role of the microbial community in macrophage immune responses during colitis. Our results show that around two-thirds of genes impacted by MCJ deficiency are conserved between the murine model and human colitis. They also support the use of animal models in order to avoid the heterogeneity observed in patients and focus on those genes that are relevant and contribute to intestinal inflammation.
Discussion
Previously, other research groups have shown evidence that support the relevance of the reduction in mitochondrial genes and associated energy production pathways during the disease.Citation3,Citation4 However, the individual variability related to therapy response is still unclear. Here, we uncover mitochondrial dysfunction as an important factor to identify UC patients at baseline that will not benefit from the TNF inhibitors’ therapeutic response.
Tissue samples are highly heterogeneous, and the amount of different cell types in a biopsy will have a certain impact on gene expression profiles. Here, we determined that MCJ is expressed in gut macrophages from IBD patients. Therefore, we isolated macrophages from mice colon tissue in order to improve the quality of the results obtained by RNA-seq technology and to characterize the role of MCJ in the early immune response. In agreement with a recent work from Czarnewski et al. that demonstrated a conserved transcriptomic profile between mouse and human colitis, our data show that the DSS mouse model is relevant to study mitochondrial dysfunction in UC patients.Citation15 MCJ deficiency in macrophages had a strong impact on the expression of susceptibility genes identified in UC. Previous studies analyzing mucosal transcriptomes revealed mitochondriopathy in a cohort of UC patients. Among the genes differentially regulated, they observed 13 genes associated with reduced ATP production and complex I activity in addition to the master regulator of mitochondrial biogenesis PPARGC1A (PGC-1α).Citation3 Altogether, this evidence supports the implication of a colonic mitochondriopathy in the course of UC suggesting that approaches targeting the mitochondrial electron transport axis could represent potential therapeutic strategies improving the response to those currently in use.
Lamina propria macrophages are strongly phagocytic for live microorganisms.Citation16 MCJ deficiency in colon macrophages revealed a high impact on Fc-gamma receptor signaling pathways. They recognize the Fc portion of IgG, triggering the engulfment and destruction of opsonized microorganisms. Our results are in agreement with previous reportsCitation17 using a chronic DSS mouse model and mirror observations in human UC patients. As described in the MCJ-deficient murine model, a higher proportion of luminal commensals bound by IgG were observed in UC stool samples, correlating with disease severity and pointing to anti-commensal IgG as a key player in the intestinal inflammatory response. This work shows that MCJ deficiency in macrophages isolated from the colon during colitis significantly increased the expression of Fcgr3 and Fcgr2b in line with Il1b and Cxcl2 levels. The former was found to play a critical role in determining the extent of inflammation in an in vivo model of UC deficient in FCGR2B.Citation5 In addition, Cxcl2 has been shown to control mucosal lymphocytes and neutrophil migration.Citation18 Altogether, these data suggest that anti-commensal IgG might contribute to intestinal inflammation via FCGR-dependent induction of Il1b.
Moreover, it was reported that elevated FCGR2A expression in mucosal UC biopsies taken prior to infliximab intervention was associated with disease refractory to treatment and predictive of subsequent resistance to TNF blockade.Citation17 Our results confirm that elevated FCGR expression in colon macrophages associated with mitochondrial dysfunction and high colonic IgG levels predicted non-responder individuals to anti-TNF therapy. Noteworthy, the overlapping between upregulated genes in UC patients and alterations due to MCJ levels in colon macrophages including tumor necrosis factor receptor superfamily members (Tnfrsf9, Tnfsf14, Cd40), cytokines and chemokines (Jak2, Cxcr4, Ccr5, Ccr3, Ccr2 and Cxcl2, Cxcl3), eosinophil-associated gene Alox15 (linked to corticosteroid response), related to anti-TNF resistance such as oncostatin M (Osm), Trem1 or cyclooxygenase-2 (Cox2 [Ptgs2]), could provide a new research line to understand the basis of disease in order to guide trials of new strategies for treatment-resistant patients. Moreover, the presence of R. gnavus in MCJ-deficient mice might serve as a potential biomarker for predicting the severity and non-response to anti-TNF treatment.Citation11,Citation19,Citation20 On the contrary, the augmented abundance of A. muciniphila and several OTUs of the family S24–7 within the WT genotype indicated induction of a healing gut microbiota. Microbial changes in patients with Crohn’s Disease after IFX therapy displayed a significant gain in short-chain fatty acids (SCFA),Citation21 the main metabolite of A. muciniphila. SCFA may mitigate intestinal inflammation through both intestinal epithelial barrier maintenance and immune regulation.Citation22 Accordingly, previous studies reported that a murine A. muciniphila strain improved chronic colitis disease severity parameters probably due to the release of anti-inflammatory cytokines.Citation22 In this sense, the Bacteroidales S24–7 family was negatively correlated with the expression of several proinflammatory genes in the colon, suggesting its beneficial role in inflammatory conditions.Citation9 Interestingly, there seems to be a negative association between R. gnavus and A. muciniphila abundance, although no molecular mechanisms to explain these correlations have been developed. Meanwhile, A. muciniphila resides in the mucus layer of the large intestine, where it is involved in maintaining intestinal integrity, R. gnavus colonizes the intestinal mucosal surface, where it can use sialic acid from mucin glycans as a carbon source. Therefore, since A. muciniphila utilizes mucin as the unique carbon, nitrogen and energy source, the presence of R. gnavus might affect A. muciniphila growth by limiting the mucus availability in MCJ-deficient mice. Indeed, a reduced abundance of A. municiphila was reported in IBD patients, while the total mucosa-associated bacteria, including R. gnavus and R. torques, increased.Citation23 In this sense, stable colonization of A. muciniphila has been shown to drive anti-inflammatory macrophage polarization in the gut, which correlated with improved therapeutic efficacy.Citation24 Given that it was shown that IL-10 signaling in macrophages is critical to achieve a good response to anti-TNF treatment,Citation25 A. muciniphila supplementation to MCJ deficient individuals might be helpful to improve anti-TNF treatment outcome.
Recent studies have revealed the potential role of gut microbiota to affect and predict the anti-TNF response or relapse.Citation20 For instance, decreased Firmicutes levels, specially, F. prausnitzii was predictive of relapse after IFX treatment in CD patients.Citation26 Therefore, microbial transplant by cohousing was performed in our murine model. Interestingly, we found that WT mice failed to respond properly to IFX treatment after microbial transplant from MCJ-deficient mice. We reported that MCJ-deficiency regulates microbiota composition and promotes specific proinflammatory bacteria during the course of UC.Citation9 The transmission of these pro-inflammatory bacteria to WT mice gut community may enhance pro-inflammatory cytokine production and counteract the effect of anti-TNF antibodies. Nevertheless, MCJ-deficient mice failed to respond to anti-TNF therapy after microbial transplant, suggesting that mitochondria dysfunction is determinant for treatment failure, independently of the gut microbiota composition.
In light of these results, MCJ may emerge as a valuable biomarker for non-response in UC patients, particularly when its expression is reduced. These findings might promote the adoption of personalized medicine, ultimately leading to reductions in treatment costs and time required.
Conclusions
Our results point to a relevant role of MCJ deficiency during colon inflammation. The high impact of MCJ deficiency on immune cell activation, Fc-gamma receptor signaling pathways, and colonic IgG indicate important implications in disease severity. The data on MCJ-deficient mice implicates mitochondria dysfunction in UC pathogenesis mirroring the colonic mitochondriopathy described in UC patients that is also associated with anti-TNF therapy success. We also show that mitochondrial deficiency influences transmissible changes in the gut microbiota ultimately leading to an impaired response to anti-TNF treatments. These findings open up new research lines to predict disease outcome and the response to anti-TNF therapy at baseline in UC patients, and might inform the search for new treatments to improve immune therapies based on rational microbiota manipulation.
Methods
Animals and experimental design
MCJ-deficient mice on a C57BL/6 background and wild-type B6 mice (8–10 wk) were used under the approval of the Animal Research Ethics Board of CIC bioGUNE (Spain; permit number CBBA-0615).Citation27 Mice were maintained under specific pathogen-free conditions, applying standard housing conditions (21–23°C temperature and 12/12-h light/dark cycles) and were fed ad libitum on standard mouse chow diet (Global diet 2914, Harlem, Madison, USA).
Colitis was promoted administering dextran sodium sulfate (DSS) (36–50 kDa; TdB Consultancy) in the drinking water (3%) for 6 d followed by a 2-d recovery period with autoclaved water. Once DSS administration started, DAI (disease activity index) was evaluated daily in all mice by a blind technician. This index is a score based on animal body weight loss percentage, stool consistency and the presence of blood in feces. The criteria proposed by Camuesco et al. were used to assign scores.Citation27 Weight loss percentage alone is an adequate and sufficient method to assess DSS-induced colitis in mice and it was calculated by dividing the total weight loss throughout the experiment by the initial weight and multiplied by 100.
Response to anti-TNF therapy
To evaluate the response to anti-TNF treatment, all experimental mice were administered 2% of DSS ad libitum in drinking water for 6 d, followed by 3 d of recovery period. Mice from each genotype were divided into two groups, with one group receiving an intraperitoneal administration of Infliximab (IFX+) (7.5 mg/kg Inflectra, Pfizer, Brussels, Belgium), an Infliximab biosimilar proven to be effective for UC and able to bind murine TNF,Citation28 while the second group received saline solution (IFX-). The mouse/human chimeric monoclonal IgG1 antibody against TNF was provided from day 3 to 6 during DSS administration.
To study the effect of microbiota composition on the response to anti-TNF response in mice, a previous microbiota transplantation was made. For this purpose, WT and MCJ-deficient female mice between 3 and 5 weeks of age were cohoused for 4 weeks before inducing colitis. Then, colitis was induced by administrating 3% of DSS in the drinking water for 6 d, followed by 3 d of recovery period. IFX was provided intraperitoneally from day 3 to 6 to all mice during the DSS administration period.
Myeloperoxidase activity assay
One centimeter of the distal colon was used to measure the enzymatic activity as described previously by Pascual-Itoiz et al.Citation9
FACS analysis to quantify IgG coated faecal bacteria
Faeces of each mouse were collected and homogenized in sterile PBS at a concentration of 100 mg/ml. Faeces debris was removed after several centrifugations and the pellet was resuspended in sterile PBS and stained with anti-mouse IgG-FITC (Sigma-Aldrich) or LIVE/DEAD™ BacLight™ Bacterial Viability Kit (ThermoFisher scientific) to check bacterial viability. BD FACSCanto II flow cytometer and BD FACSDiva software were used to obtain data that were analyzed with FlowJo software (TreeStar).
Histology, immunohistochemistry, and immunofluorescence
Colon tissue from mice was fixed in 10% formalin fixative, dehydrated, embedded in paraffin, and cut into 5 μm-thick sections. For histopathology, sections were deparaffined, hydrated, and stained with hematoxylin and eosin according to standard protocols. Stained sections were analyzed and given a score by a blinded pathologist. For F4/80 staining, tissue sections were deparaffined, hydrated and subjected to antigen retrieval using proteinase K for 15 min. Endogenous enzymes were first blocked with 3% hydrogen peroxide and then with goat serum. Sections were incubated with rat anti-mouse F4/80 primary antibody (1:50) for 2 h at 37°C (Biolegend), followed by 30 min incubation with ImmPRESS HRP Goat Anti-Rat IgG secondary antibody (Vector). Detection was performed with DAB substrate (Vector). Images were captured with a Zeiss Axioimager A1 microscope and analyzed with ImageJ software.
Colon samples from inflamed and healthy areas were obtained after surgery from IBD patients under the approval of the Clinical Research Ethics Board of Euskadi (CEIC-E; code 16–12). Colocalization of MCJ with CD163 in inflamed and adjacent healthy colon tissues was determined by immunofluorescence. Colon tissues were embedded in paraffin, cut, rehydrated, and unmasked with proteinase K for 15 min. Then, tissues were incubated with anti-human MCJ antibody at 4°C overnight and a Cy3-conjugated secondary antibody was added for 30 min. After the first staining, tissues were incubated with anti-human CD163 antibody for 1 hr at 37°C followed by the addition of FITC-conjugated secondary antibody (30 min). Photographs were taken with a fluorescence microscope (Axioimager.D1 Zeiss). The area of each color was quantified using the CellProfiler tool and Frida software was used to analyze the colocalization of MCJ with CD163.
TACE activity
Colon protein extracts were subjected to fluorimetric SensoLyte 520 TACE (α-Secretase) activity assay kit (AnaSpec). Enzyme activity was monitored using a BioTek Synergy HT microplate fluorescence reader (BioTek, Winooski, VT) at an excitation wavelength of 355 nm and an emission wavelength of 500 nm. Results are expressed as RFUs per mg of protein.
Colon macrophages RNAseq
Intestinal cells from murine colon tissues were extracted by several washing steps in 25 ml of Hank’s Balanced Salt Solution (HBSS) and by collagenase type IV-mediated cleavage (Gibco) in 2% FBS-RPMI media, a matrix-degrading enzyme. For macrophage isolation, extracted cells were labeled with 10 µl of anti-F4/80-Biotin antibody (Miltenyi Biotec) for 30 min followed by the addition of 10 µl of anti-biotin microbeads (Miltenyi Biotec). A multistand magnet and LS columns from Miltenyi Biotec were used to isolate macrophages by positive selection. RNA from eluted macrophages was purified using DEAD™ Reagent (ThermoFisher), and the pellet was resuspended in 10 µl of HyClone water (ThermoFisher). Finally, an off column DNAse treatment was performed with RNase-Free DNase Set kit from Qiagen and resulting RNA was stored at −80°C. The quantity and quality of the RNAs were evaluated using Qubit RNA HS Assay Kit (Thermo Fisher Scientific), and Agilent RNA 6000 Nano and Pico Chips (Agilent Technologies), respectively.
Sequencing libraries were prepared using the NuGEN Universal Plus mRNA-Seq kit (PART NO. 0508) following the user’s guide (M01442 v2). Libraries were visualized on an Agilent 2100 Bioanalyzer using Agilent High Sensitivity DNA kit (Agilent Technologies), quantified using Qubit dsDNA HS DNA Kit (Thermo Fisher Scientific) and sequenced in a HiSeq2500 with single-reads of 50 nucleotides.
Quality control of sequenced samples was performed with FASTQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were mapped against the mouse (mm10) reference genome with the STAR program to account for spliced junctions.Citation29 The resulting BAM alignment files were then used to generate a table of raw counts using Rsubread.Citation30 The raw counts table was the input for the Differential Expression (DE) analysis, carried out by DESeq2 to detect differentially expressed genes among the different conditions.Citation31 To analyze the functional GO annotation enrichment of immunological and biological functions, and the pathways involved in colon macrophages, the Database for Annotation, Visualization and Integrated Discovery (DAVID),Citation32 ToppGeneCitation33 and ToppClusterCitation34 were used. Visualization of networks was obtained using Cytoscape.v3.0.2 software.Citation35 Related genes to immunological processes were mapped with the ClueGO tool.Citation36 For enriched motif discovery in colon macrophages genomic regions, the HOMER software was used using the findMotifs.pl script. Motifs of lengths between 10 and 12 nucleotides were considered in this analysis. RNAseq was validated by Real Time-PCR with Il10, Il1b, Lcn2 and Reg3b genes, which were chosen according to their log2 fold change. For that purpose, we synthesized cDNA with M-MLV reverse transcriptase (ThermoFisher Scientific). RT-PCR was performed on 384 well plates with the QuantStudio 6 Flex Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA, USA) with PerfeCTa SYBR Green SuperMix Low ROX (Quantabio, Beverly, MA, USA) and amplification was analyzed by QuantStudio RT PCR software v1.3. Primers for Il10, Il1b, Lcn2, Reg3b and Rpl19 genes were optimized (see annealing temperature in Supplementary Table S1) and to normalize colonic mRNA expression, the murine Rpl19 gene was used. mRNA relative quantification was calculated using the ΔΔCt method,Citation37 and PCR efficiency was always between 90 and 110%.
DNA extraction and microbiome analysis
Colon contents from dysbiosis and anti-TNF experiments were collected at sacrifice, stored at −80°C, and freeze-dried. DNA was isolated using the FavorPrep Stool DNA Isolation Mini kit (Vienna, Austria). Eluted DNA concentration and quality was determined spectrophotometrically with a NanoDrop ND-100 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Microbial community composition of samples taken from colon content was determined using amplicon sequencing of the 16S rRNA gene as described in Pascual-Itoiz et al.Citation9 The V3-V4 hypervariable region of the 16S rRNA gene of colonic bacteria was amplified using Illumina Miseq on a standard protocol from the manufacturer. Bacteria communities were grouped as OTUs (Operational Taxonomic Units) based on 16S rRNA similarities. Data processing was performed using QIIME (v.1.9.0),Citation38 and sequences were clustered as OTUs of 97% similarity with UCLUST.Citation39 As a final step, OTUs were checked for chimeras using RDP gold database, and taxonomy was assigned with the Greengenes database (version 4feb2011).Citation40 Differential abundance of OTUs was obtained using the DESeq2 toolCitation31, and Rbase was used to create graphs.
Human RNAseq samples
Rectal gene expression signature composed of 5296 genes differentially expressed (FDR <0.001 and fold change ≥1.5) between UC and control individuals, were obtained from Supplementary Dataset 1 from Haberman et al.Citation3 Corticosteroid response associated genes were presented by Haberman et al.Citation3, and anti-TNF response data by West et al.Citation14
Statistical analysis
Statistical analysis was performed using GraphPad software (GraphPad Software, San Diego, CA, USA). Results were graphed as box and whisker plots with median, quartiles, and range. The significance was assessed by two-way ANOVA statistical test determined as *P value < 0.05, **P value < 0.01, ***P value < 0.001, and ****P value < 0.0001. Significance is represented by an asterisk or asterisks upside the box to indicate differences within the same genotype in different experimental conditions, infliximab negative (IFX-) versus infliximab positive (IFX+), and cohoused infliximab positive (Coh) versus alone infliximab positive (Alone). Asterisks above the lines indicate differences between the different genotypes in the same experimental group.
Abbreviations
| DSS | = | Dextran sulfate sodium |
| FcγR | = | Fc gamma receptor |
| HRP | = | Horseradish Peroxidase |
| IBD | = | Inflammatory bowel disease |
| IFX | = | Infliximab |
| MCJ | = | Methylation-controlled J protein |
| OTU | = | Operational taxonomic unit |
| SCFA | = | Short-chain fatty acid |
| TACE | = | Tumor necrosis factor α-converting enzyme |
| TIMP3 | = | Tissue inhibitor of metalloproteinase 3 |
| TNF | = | Tumour necrosis factor |
| UC | = | Ulcerative colitis |
Authors’ contributions
Conception and design of the study (LA), data collection (APC, JC, MAPI, VGJ, AP, DB, APa and LA), data analysis (APC, JLL, MGL, MAPI, MF, IS, AMA, JLL and LA), drafting the manuscript (APC, JA and LA), manuscript revision (HR and MLMC), statistical analysis (APC, JLL and LA), obtained funding (AF, IRL, JA, and LA), and technical support (LB and IMR). All authors approved the final version for publication.
Ethics approval
Animal protocols were approved by the Animal Research Ethics Board of CIC bioGUNE (Spain; permit number CBBA-0615). Collection of colon samples from IBD patients were approved by the Clinical Research Ethics Board of Euskadi (CEIC-E; code 16-12).
Supplemental Material
Download TIFF Image (118.8 KB)Supplemental Material
Download TIFF Image (96 KB)Supplemental Material
Download MS Word (42.3 KB)Acknowledgments
We thank Estibaliz Atondo for technical support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Raw sequences used for metagenomics analysis were made available at European Nucleotide Archive (ENA www.ebi.ac.uk/ena) under the project number PRJEB33422 for dysbiosis and PRJEB41595 for infliximab experiment. Raw sequences used to perform the transcriptomic analysis were uploaded to GEO (Gene Expression Omnibus) database under project accession code GSE135033 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE135033).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2023.2266626.
Additional information
Funding
References
- Gisbert JP, Chaparro M. Primary failure to an anti-TNF agent in inflammatory bowel disease: switch (to a second anti-TNF agent) or swap (for another mechanism of action)? J Clin Med. 2021;10(22):5318. doi:10.3390/jcm10225318.
- Gerriets V, Goyal A, Khaddour K. Tumor necrosis factor inhibitors. Florida: StatPearls. Treasure Island (FL); 2022.
- Haberman Y, Karns R, Dexheimer PJ, Schirmer M, Somekh J, Jurickova I, Braun T, Novak E, Bauman L, Collins MH, et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat Commun. 2019;10(1):38. doi:10.1038/s41467-018-07841-3.
- Mottawea W, Chiang CK, Muhlbauer M, Starr AE, Butcher J, Abujamel T, Deeke SA, Brandel A, Zhou H, Shokralla S, et al. Altered intestinal microbiota–host mitochondria crosstalk in new onset Crohn’s disease. Nat Commun. 2016;7(1):13419. doi:10.1038/ncomms13419.
- Peloquin JM, Goel G, Villablanca EJ, Xavier RJ. Mechanisms of pediatric inflammatory bowel disease. Annu Rev Immunol. 2016;34(1):31–16. doi:10.1146/annurev-immunol-032414-112151.
- Hatle KM, Gummadidala P, Navasa N, Bernardo E, Dodge J, Silverstrim B, Fortner K, Burg E, Suratt BT, Hammer J, et al. MCJ/DnaJC15, an endogenous mitochondrial repressor of the respiratory chain that controls metabolic alterations. Mol Cell Biol. 2013;33:2302–2314. doi:10.1128/MCB.00189-13.
- Schusdziarra C, Blamowska M, Azem A, Hell K. Methylation-controlled J-protein MCJ acts in the import of proteins into human mitochondria. Hum Mol Genet. 2013;22:1348–1357. doi:10.1093/hmg/dds541.
- Navasa N, Martin I, Iglesias-Pedraz JM, Beraza N, Atondo E, Izadi H, Ayaz F, Fernandez-Alvarez S, Hatle K, Som A, Dienz, O., et al. Regulation of oxidative stress by methylation-controlled J protein controls macrophage responses to inflammatory insults. J Infect Dis. 2015;211(1):135–145. doi:10.1093/infdis/jiu389.
- Pascual-Itoiz MA, Pena-Cearra A, Martin-Ruiz I, Lavin JL, Simo C, Rodriguez H, Atondo E, Flores JM, Carreras-Gonzalez A, Tomas-Cortazar J, et al. The mitochondrial negative regulator MCJ modulates the interplay between microbiota and the host during ulcerative colitis. Sci Rep. 2020;10(1):572. doi:10.1038/s41598-019-57348-0.
- Pena-Cearra A, Pascual-Itoiz MA, Lavin JL, Fuertes M, Martin-Ruiz I, Castelo J, Palacios A, Barriales D, Fullaondo A, Aransay AM, et al. Mitochondrial complex I dysfunction alters the balance of soluble and membrane-bound TNF during chronic experimental colitis. Sci Rep. 2022;12(1):9977. doi:10.1038/s41598-022-13480-y.
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi:10.1038/s41586-019-1237-9.
- Peña-Cearra A, Song, D, Castelo, J, Palacios, A, Lavin, JL, Azkargorta, M, Elortza, F, Fuertes, M, Pascual-Itoiz, MA, Barriales, D, Martin-Ruiz, I, Fullaondo, A, Aransay, AM, Rodriguez, H, Palm, NW, Anguita, J, Abecia, L. Mitochondrial dysfunction promotes microbial composition that negatively impacts on ulcerative colitis development and progression. npj Biofilms and Microbes. 2023;9(74):0709–2023. doi:10.1038/s41522-023-00443-y37805634.
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi:10.1016/j.molcel.2010.05.004.
- West NR, Hegazy AN, Owens BMJ, Bullers SJ, Linggi B, Buonocore S, Coccia M, Gortz D, This S, Stockenhuber K, et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor–neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23(5):579–589. doi:10.1038/nm.4307.
- Czarnewski P, Parigi SM, Sorini C, Diaz OE, Das S, Gagliani N, Villablanca EJ. Conserved transcriptomic profile between mouse and human colitis allows unsupervised patient stratification. Nat Commun. 2019;10(1):2892. doi:10.1038/s41467-019-10769-x.
- Smith PD, Janoff EN, Mosteller-Barnum M, Merger M, Orenstein JM, Kearney JF, Graham MF. Isolation and purification of CD14-negative mucosal macrophages from normal human small intestine. J Immunol Methods. 1997;202(1):1–11. doi:10.1016/s0022-1759(96)00204-9.
- Castro-Dopico T, Dennison TW, Ferdinand JR, Mathews RJ, Fleming A, Clift D, Stewart BJ, Jing C, Strongili K, Labzin LI, et al. Anti-commensal IgG drives intestinal inflammation and type 17 immunity in ulcerative colitis. Immunity. 2019;50(4):1099–1114 e1010. doi:10.1016/j.immuni.2019.02.006.
- Ohtsuka Y, Lee J, Stamm DS, Sanderson IR. MIP-2 secreted by epithelial cells increases neutrophil and lymphocyte recruitment in the mouse intestine. Gut. 2001;49(4):526–533. doi:10.1136/gut.49.4.526.
- Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9(1):103. doi:10.1186/s13073-017-0490-5.
- Dovrolis N, Michalopoulos G, Theodoropoulos GE, Arvanitidis K, Kolios G, Sechi LA, Eliopoulos AG, Gazouli M. The interplay between mucosal microbiota composition and host gene-expression is linked with infliximab response in inflammatory bowel diseases. Microorganisms. 2020;8(3):438. doi:10.3390/microorganisms8030438.
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi:10.1038/nature08530.
- Zhai R, Xue X, Zhang L, Yang X, Zhao L, Zhang C. Strain-specific anti-inflammatory properties of two Akkermansia muciniphila strains on chronic colitis in mice. Front Cell Infect Microbiol. 2019;9:239. doi:10.3389/fcimb.2019.00239.
- Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105(11):2420–2428. doi:10.1038/ajg.2010.281.
- Wang B, Chen X, Chen Z, Xiao H, Dong J, Li Y, Zeng X, Liu J, Wan G, Fan S, et al. Stable colonization of Akkermansia muciniphila educates host intestinal microecology and immunity to battle against inflammatory intestinal diseases. Experimental & Molecular Medicine. 2023;55(1):55–68. doi:10.1038/s12276-022-00911-z.
- Koelink PJ, Bloemendaal FM, Li B, Westera L, Vogels EWM, van Roest M, Gloudemans AK, van ’t Wout AB, Korf H, Vermeire S, et al. Anti-TNF therapy in IBD exerts its therapeutic effect through macrophage IL-10 signalling. Gut. 2020;69(6):1053–1063. doi:10.1136/gutjnl-2019-318264.
- Rajca S, Grondin V, Louis E, Vernier-Massouille G, Grimaud JC, Bouhnik Y, Laharie D, Dupas JL, Pillant H, Picon L, et al. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn’s disease. Inflamm Bowel Dis. 2014;20:978–986. doi:10.1097/MIB.0000000000000036.
- Camuesco D, Comalada M, Rodriguez-Cabezas ME, Nieto A, Lorente MD, Concha A, Zarzuelo A, Galvez J. The intestinal anti-inflammatory effect of quercitrin is associated with an inhibition in iNOS expression. Br J Pharmacol. 2004;143(7):908–918. doi:10.1038/sj.bjp.0705941.
- Lopetuso LR, Petito V, Cufino V, Arena V, Stigliano E, Gerardi V, Gaetani E, Poscia A, Amato A, Cammarota G, et al. Locally injected infliximab ameliorates murine DSS colitis: differences in serum and intestinal levels of drug between healthy and colitic mice. Dig Liver Dis. 2013;45:1017–1021. doi:10.1016/j.dld.2013.06.007.
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi:10.1093/bioinformatics/bts635.
- Liao Y, Smyth GK, Shi W. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi:10.1093/nar/gkt214.
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014-0550-8.
- Huang da W, BT S, RA L. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211.
- Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server):W305–311. doi:10.1093/nar/gkp427.
- Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 2010;38:W96–102. doi:10.1093/nar/gkq418.
- Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Methods. 2012;9(11):1069–1076. doi:10.1038/nmeth.2212.
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi:10.1093/bioinformatics/btp101.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using Real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262.
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi:10.1038/nmeth.f.303.
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi:10.1093/bioinformatics/btq461.
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–618. doi:10.1038/ismej.2011.139.