 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Supplementation with probiotics has emerged as a promising therapeutic tool to manage metabolic diseases. We investigated the effects of a mix of Bifidobacterium animalis subsp. lactis LA804 and Lactobacillus gasseri LA806 on high-fat (HF) diet -induced metabolic disease in mice. Supplementation with the probiotic mix in HF diet-fed mice (HF-Pr2) reduced weight and fat mass gains, decreased hepatic lipid accumulation, and lowered plasma triglyceride peak during an oral lipid tolerance test. At the molecular level, the probiotic mix protected against HF-induced rise in mRNA levels of genes related to lipid uptake, metabolism, and storage in the liver and white adipose tissues, and strongly decreased mRNA levels of genes related to inflammation in the white adipose tissue and to oxidative stress in the liver. Regarding intestinal homeostasis, the probiotic mix did not prevent HF-induced gut permeability but slightly modified microbiota composition without correcting the dysbiosis induced by the HF diet. Probiotic supplementation also modified the cecal bile acid (BA) profile, leading to an increase in the Farnesoid-X-Receptor (FXR) antagonist/agonist ratio between BA species. In agreement, HF-Pr2 mice exhibited a strong inhibition of FXR signaling pathway in the ileum, which was associated with lipid metabolism protection. This is consistent with recent reports proposing that inhibition of intestinal FXR activity could be a potent mechanism to overcome metabolic disorders. Altogether, our results demonstrate that the probiotic mix evaluated, when administered preventively to HF diet-fed mice could limit obesity and associated lipid metabolism disorders, likely through the inhibition of FXR signaling in the intestinal tract.
Introduction
Energy imbalance due to unhealthy eating habits and low physical activity contributes to the dramatic rise in the prevalence of chronic metabolic diseases and associated cardiovascular risk.Citation1–3 More than one billion people worldwide are presently suffering from obesity and/or metabolic syndrome (MetS), defined as a pathological state characterized by at least three of the following conditions: abdominal obesity, insulin resistance, hypertension, and hyperlipidemia.Citation4 This cluster of conditions increases the risk of heart diseases, stroke, nonalcoholic fatty liver disease (NAFLD), and type 2 diabetes.Citation1,Citation4 Facing the severity of these pathologies as well as their considerable economic burden for public health systems, new therapeutic but also preventive strategies are needed.
Obesity and MetS have been associated with gut microbiota alterations and disruption of intestinal homeostasis with an impact on nutrient absorption, inflammation, intestinal barrier functions, gut hormone secretion, and production of important vitamins and metabolites.Citation5–7 The gut microbiota is generally altered, both in terms of richness and diversity, in situations of obesity and metabolic diseases in rodent models as well as in humans.Citation6,Citation7 Furthermore, the use of germ-free animals and experiments of fecal transplantation from obese donors (mice or humans) to recipient lean mice support a causal role of intestinal bacteria in the development and maintenance of these pathologies.Citation8–10 However, the precise mechanisms linking the changes in gut microbiota and host physiology are still largely unraveled because several processes could occur in synergy or independently. These include the production of short chain fatty acids (SCFAs), the production or modulation of a variety of bioactive molecules (indoles, bile acids, polar lipids, vitamins, amino acids, etc.), regulation of the intestinal mucus layer, intestinal permeability, as well as inflammation and immunity.Citation5–7,Citation11–13 These effects could also vary according to location in the intestinal tract. Consequently, targeting the gut microbiota to fight obesity and MetS is an important avenue of research, and the use of probiotics to restore intestinal homeostasis has appeared over the years as one of the promising strategies for this aim.
A recent systematic review of randomized clinical trials testing lactic acid bacteria has identified 23 studies reporting significant weight loss.Citation14 Some reports have also shown the effects of probiotics on biomarkers associated with MetS. In a randomized trial, administration of a mixture of four probiotic strains to obese children was associated with a decrease in liver markers (ALAT and ASAT), cholesterol, low-density lipoprotein-cholesterol, and triglyceride (TG) levels, as well as a decrease in waist circumference.Citation15 The use of another multi-strain probiotic mix improved levels of ALAT, cytokines, and endotoxins, as well as liver histology in adult patients with NAFLD.Citation16 The consumption of Lactilactobacillus curvatus HY7601 and Lactiplantibacillus plantarum KY1032 by non-diabetic patients with hypertriglyceridemia also reduced lipid catabolism and lipoprotein lipase activity, hence lowering plasma TG levels.Citation17 There is therefore evidence of efficacy that supplementation with probiotics could be useful for the management and potential prevention of MetS and related pathologiesCitation18. However, additional work is clearly needed to better define the mechanisms of action of promising probiotics in pre-clinical models prior to clinical evaluation. Recently, screening of candidates identified several strains with potential for the treatment or prevention of obesity and MetS.Citation19,Citation20 Beneficial effects were generally observed when different strains were administered simultaneously such as Lacticaseibacillus rhamnosus Lb12 and Bifidobacterium animalis subsp. lactis Bf141, which were able to improve the integrity of the intestinal barrier,Citation19 or B. animalis subsp. lactis LA804 and Lactobacillus gasseri LA806, which were able to reduce body weight gain and inflammationCitation20 in mice fed a high-fat (HF) diet.
The objective of this study was to characterize in detail the beneficial effects of the combination of the two probiotic strains B. animalis subsp. lactis LA804 and Lactobacillus gasseri LA806,Citation20 and to gain insight into their mechanisms of action in preventing metabolic disorders in a mouse model of obesity and MetS induced by the HF diet. Markers of lipid metabolism, inflammation, and oxidative stress, as proxies of MetS, were specifically assessed. A thorough evaluation of intestinal homeostasis was also performed to gain a better understanding of the mechanisms of action of the probiotic mix.
Results
Probiotic mix supplementation reduced weight gain, fat accumulation, and hyperinsulinemia in HF diet-fed mice
Mice were fed a HF diet for 12 weeks. The probiotic mix was administered daily from the first day of the HF diet until the end of the experiment. Compared to Chow diet fed mice, HF diet-fed mice showed greater body weight gain () and significantly higher weights of the three white adipose tissue deposits (). Supplementation with the probiotic mix significantly protected against the effects of the HF diet on body weight gain () and adipose tissue accumulation in all fat depots (). Importantly, the probiotic mix did not influence food intake (). The HF diet also increased the amount of TG and the size of lipid droplets accumulated in the liver () whereas probiotic mix intake counteracted these effects () and reduced liver weight (). Hepatic cholesterol levels were not modified in any of the groups ().
Figure 1. Supplementation with the probiotic mix attenuates weight gain and fat mass accumulation and metabolic alterations in HF diet-fed mice. (a) body weight evolution (n = 12–16). (b) body weight gain at 12 weeks. (c) epididymal white adipose tissue (eWAT), mesenteric white adipose tissue (mWAT), inguinal white adipose tissue (iWAT) and liver weights at sacrifice. (d cumulative food intake recorded during 10 weeks. (e) liver triglycerides and total cholesterol. (f) oil-red O liver sections and liver lipid droplet size (n = 6–12); 60× magnification; bar = 25 µm. (g) fasting blood glucose level (n = 15–16). (h) fasting plasma insulin level (n = 5). Data are mean ± SEM. $p <0.05, #p <0.01, ¤p <0.001 versus the Chow diet group and *p <0.05, **p <0.01, ***p <0.001 for the HF-Pr2 versus the HF group.
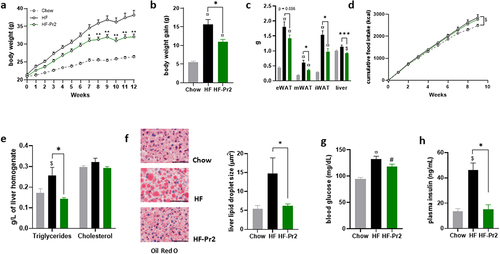
Six-hour fasting plasma TG, cholesterol, glucose, and insulin concentrations were measured at the end of the intervention. TG and total cholesterol levels were significantly increased in the HF diet-fed mice compared to the Chow diet fed group, but the probiotic mix treatment did not affect these values TG: 0.90 ± 0.06 mmol/L (HF), 1.16 ± 0.09 mmol/L (HF-Pr2), and 0.72 ± 0.04 mmol/L (Chow), p = 0.0003 Chow vs HF-Pr2. Total cholesterol: 7.75 ± 0.11 mmol/L (HF), 7.15 ± 0.36 mmol/L (HF-Pr2) and 5.43 ± 0.18 mmol/L (Chow), p = 0.001 HF vs Chow, p = 0.002 HF-Pr2 vs Chow). In the HF-fed group, glycemia and insulinemia were significantly increased compared to the Chow diet fed group (), indicative of a state of insulin resistance and glucose intolerance. Supplementation with the probiotic mix did not fully restore fasting glycemia but maintained fasting insulin concentrations at the level found in Chow diet fed mice ().
Probiotic mix supplementation was associated with reduced expression of lipid storage genes and of oxidative stress and inflammatory markers in liver and adipose tissue
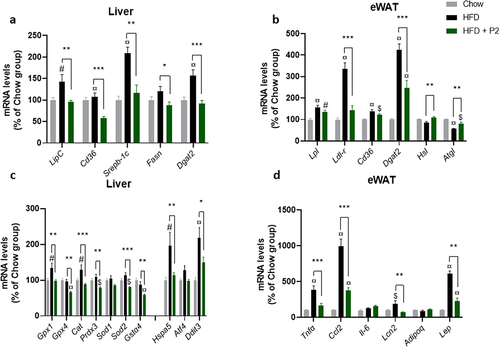
Compared to the Chow diet, 12 weeks on the HF diet induced a concerted increase in the mRNA levels of a set of genes involved in the handling and storage of lipids in the liver and adipose tissue (), such as LipC (encoding hepatic lipase), Lpl (lipoprotein lipase) and Ldl-r (LDL receptor), Cd36 (encoding fatty acid transporter) and Dgat2 (diacylglycerol O-acyltransferase 2, catalyzing the final reaction in the synthesis of triglycerides). These findings are in line with the lipid accumulation observed in hepatic and adipose tissues. Supplementation with the probiotic mix reduced the expression of LipC in the liver (−30%) and of Ldl-r in eWAT (−60%) compared to the HF diet-fed group (). The relative abundance of mRNA for Cd36 fatty acid transporter also decreased (by almost 50%) upon probiotics in the liver. With regard to genes related to hepatic de novo lipogenesis, Srebp-1c, Fasn, and Dgat2 expression was significantly reduced in the HF-Pr2 group compared with the HF group. Furthermore, in the eWAT, supplementation with the probiotic mix was associated with a significant increase in the abundance of Hsl (hormone-sensitive lipase) and Atgl (adipose triglyceride lipase) mRNA, the genes encoding the main lipolysis enzymes ().
Figure 2. Effects of the probiotic mix on gene expression in the liver and eWAT. (a) relative mRNA expression levels of lipid metabolism-related genes in the liver: hepatic lipase (LipC), CD36 antigen (CD36), sterol regulatory element binding transcription factor 1 (Srebp-1c), fatty acid synthase (Fasn), and diacylglycerol O-acyltransferase 2 (Dgat2) (n = 7–10). (b) relative mRNA expression levels of lipid metabolism-related genes in eWAT: lipoprotein lipase (Lpl), low density lipoprotein receptor (Ldl-r), CD36 antigen (CD36), diacylglycerol O-acyltransferase 2 (Dgat2), hormone sensitive lipase (Hsl) and adipose triglyceride lipase (Atgl) (n = 5–8). (c) relative mRNA expression levels of oxidative stress and ER stress-related genes in the liver: glutathione peroxidase 1 (Gpx1) and 4 (Gpx4), Catalase (Cat), peroxiredoxin 3 (Prdx3), superoxide dismutase 1 (Sod1) and 2 (Sod2), glutathione S-transferase alpha 4 (Gsta4), heat shock 70kDa protein 5 (Hspa5), activating transcription factor 4 (Atf4) and DNA-damage inducible transcript 3 (Ddit3). (d) relative mRNA expression levels of inflammation-related genes in eWAT: tumor necrosis factor alpha (Tnfα), chemokine (C-C motif) ligand 2 (Ccl2), interleukin 6 (iIl-6), Lipocalin 2 (Lcn2), Adiponectin (Adipoq) and leptin (Lep). Data are mean ± SEM. $p <0.05, #p <0.01, ¤p <0.001 versus the Chow diet group and *p <0.05, **p <0.01, ***p <0.001 for the HF-Pr2 versus the HF group.
To further explore possible mechanisms explaining the probiotic mix-induced reduction of weight gain and adipose tissue accumulation, the expression of genes related to brown fat activation and white adipose tissue browning was evaluated in fat depots, as well as the expression of genes related to fatty acid oxidation in the liver. These different pathways were not affected by probiotic mix intake, although the HF diet was associated with an increase in the expression of the uncoupling protein 1 (Ucp1) gene in brown fat (Supplementary Figure S2).
Accumulation of lipids in the liver is classically associated with endoplasmic reticulum (ER) stress and oxidative stress. Accordingly, we observed that the HF diet induced a significant increase in the expression of the anti-oxidant Gpx1 and Cat genes and the ER stress markers Hspa5 (encoding the chaperone Bip) and Ddit3 (encoding the C/EBP homologous protein Chop) (). Supplementation with the probiotic mix completely prevented these HF-induced effects. Furthermore, the expression of a set of additional anti-oxidant genes (Gpx4, Prdx3, Sod2, and Gsta4) were significantly reduced in the HF-Pr2 group compared with the HF group (), clearly indicating a protective effect of the probiotic mix on oxidative and ER stresses in the liver of mice fed a HF diet.
The expansion of adipose tissue upon a hypercaloric diet is generally associated with an increase in inflammatory markers, reflecting modifications in the population of macrophages and lymphocytes infiltrating the tissue.Citation5 In agreement, we observed an increase in the expression of TNFα and the chemokine MCP-1 (coded by Ccl2 gene) in eWAT (). The pro-inflammatory adipokine leptin (encoded by Lep gene) was also markedly overexpressed in HF diet-fed mice (). Importantly, supplementation with the probiotic mix strongly prevented HF-induced expression of inflammatory markers in eWAT (), consistent with its protective effect against fat accumulation.
Probiotic mix supplementation was associated with a reduction in postprandial plasma triglycerides and lower expression of genes involved in fatty acid uptake in the small intestine
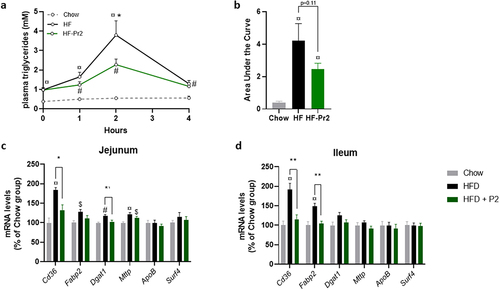
The fact that the probiotic mix prevented weight gain and adipose tissue accumulation without significantly modifying food intake and lipid oxidation pathways the liver in brown adipose tissue (BAT) could suggest an impact of the probiotic mix on lipid absorption. This hypothesis was tested using an oral lipid tolerance test (OLTT) performed after 10 weeks of supplementation. During the 4 h following gavage of mice with oil, plasma concentrations of TG were significantly higher in the HF group than in the Chow diet fed group, peaking 2 h after gavage and returning almost to basal values after 4 h (). Interestingly, supplementation with the probiotic mix was associated with a significantly lower TG peak at 2 h (p = 0.033 between the HF-Pr2 and HF groups, ). The areas under the curves (AUCs) of postprandial plasma TG were significantly higher in the HF and HF-Pr2 groups than in the Chow diet fed group, and there was a trend (p = 0.11) toward a reduction in the group supplemented with the probiotic mix compared with HF diet-fed mice (). These results might suggest a reduction of lipid absorption in the presence of the probiotic mix. However, the postprandial excursion of TG during OLTT results from both intestinal absorption of lipids and their clearance from plasma. The lack of an increase in circulating TG after the oil bolus in Chow diet mice suggests a highly efficient rate of TG clearance in this group and thus potentially impaired clearance in the HF diet groups. To better clarify this point, we evaluated the expression of key genes involved in fatty acid uptake and chylomicron production in the jejunum and the ileum, the two main regions of the small intestine involved in the absorption of dietary lipids. The expression of Cd36 (allowing transport of fatty acids from the lumen to the enterocytes) and Fabp2 (encoding an intracellular transporter of fatty acids) was increased in both the jejunum and ileum of HF diet-fed mice compared to Chow diet-fed mice. Supplementation with the probiotic mix prevented the overexpression of Cd36 in both tissues (). However, other key genes of lipid absorption linked to intracellular TG synthesis (Dgat1) and chylomicron production by enterocytes (Mttp, ApoB, and Surf4) were globally not affected ().
Figure 3. Effects of supplementation with the probiotic mix on intestinal lipid uptake. (a) plasma triglyceride concentrations throughout the oral lipid tolerance test (OLTT) (n = 9–11). (b) area under the curve (AUC) of plasma TGs during OLTT. Relative mRNA expression levels of lipid absorption-related genes in the jejunum (c) and ileum (d) Cd36, fatty acid binding protein 2 (Fabp2), diacylglycerol O-acyltransferase 1 (Dgat1), Microsomal triglyceride transfer protein (Mttp), apolipoprotein B (ApoB) and Surfeit4 (Surf4) (n = 7–10). Data are mean ± SEM. $p <0.05, #p <0.01, ¤p <0.001 versus the Chow diet group and *p <0.05, **p <0.01 for the HF-Pr2 versus the HF group.
Probiotic mix supplementation did not affect intestinal permeability and was associated with some modifications of gut microbiota composition
Since increased intestinal permeability has been associated with inflammation and metabolic disturbances in rodent models fed high-fat diets,Citation12 we evaluated this parameter in vivo using the lactulose-mannitol test. While HF diet-fed mice showed a clear alteration in the integrity of the intestinal barrier, as illustrated by a large increase in the recovery of lactulose in urine, supplementation with the probiotic mix did not correct the HF diet-induced alteration of intestinal permeability (Supplementary Figure S3). In addition, mRNA expressions of Muc2 and Tjp1 (encoding the tight junction protein ZO1) in the ileum were not affected by the probiotic mix consumption (Supplementary Figure S3).
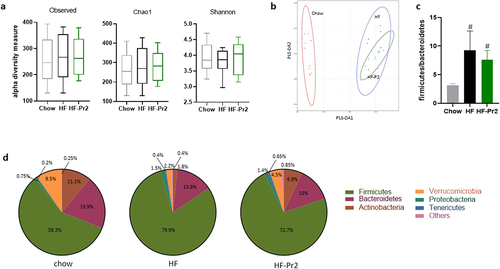
Gut microbiota composition was analyzed by sequencing of the V3-V4 region of 16S rRNA in feces collected at the end of the protocol. A total of 766 different amplicon sequence variants (ASVs) were identified. Alpha diversity estimated using different indexes did not differ between groups (). However, a classical multidimensional scaling (MDS) using Bray-Curtis dissimilarity indicated that mice fed the HF diet had a different microbiota structure and composition than those fed the Chow diet, but this was not corrected by probiotic mix intake (). After taxonomic assignment, comparisons between groups at the phylum level showed that mice fed the HF diet had increased abundance of Firmicutes (now called Bacillota) and reduced levels of Bacteroidetes (Bacteroidota), Actinobacteria (Actinomycetota), and Verrucomicrobia (). The Firmicutes/Bacteroidetes (Bacillota/Bacteroidota) ratio was significantly increased by the HF diet and not corrected by the probiotic mix (). The only significant difference observed at the phylum level between HF diet-fed mice and probiotic mix-supplemented mice was an increase in the abundance of Actinobacteria (Actinomycetota) in the HF-Pr2 group (6.9% vs 1.8%, p = 0.0096, ).
Figure 4. Impacts of supplementation with the probiotic mix on gut microbiota composition. (a) alpha-diversity based on observed OTUs, Chao1 index and Shannon indexes. (b) multidimensional scaling of Bray-Curtis dissimilarity (MDS). (c) Firmicutes/Bacteroidetes ratio. (d) pie charts showing relative abundance of the most abundant phyla. Data are mean ± SEM. #p <0.01 versus the Chow diet group.
At the genus level, the HF diet induced several changes in the composition of gut microbiota when compared with the Chow diet (Supplementary Table S1). The relative abundance of 12 genera were significantly modified by the HF-diet: five were decreased (Akkermansia, Bifidobacterium, Coprococcus, Ruminococcus, and Turicibacter) and seven increased (Adlercreutzia, Allobaculum, Bilophila, Gemmiger, Mucispirillum, Oscillospira, and Streptococcus). However, the relative abundance of only two genera was significantly modified by the probiotic mix when compared to the HF diet-fed group: a decrease in Adlercreutzia (−2.8-fold) and an increase in Bifidobacterium (7.5-fold) (Supplementary Table S1).
Among the 766 ASVs, 301 showed significant differences between groups according to Kruskal–Wallis tests with multiple corrections. When comparisons between groups were performed using pairwise Wilcoxon tests, 269 ASVs had different relative abundances between the Chow diet and HF-diet groups and 277 between the Chow diet and HF-Pr2 groups. Only 17 ASVs displayed significantly different levels between the HF diet and the HF diet plus probiotic mix groups, 7 being increased and 10 being decreased in response to supplementation with the probiotic mix (). We then manually verified the assignments of these 17 ASVs (using BLAST-NCBI), which allowed us to better identify the bacteria affected by supplementation with the probiotic mix. We found that the largest increases in relative abundance were for Bifidobacterium animalis subsp. lactis (ASV_79) and Lactobacillus gasseri (ASV_673) (), in agreement with consumption of LA804 and LA806, the two strains of the probiotic mix. Among the other up-regulated ASVs, we found ASV_418 (Olsenella) and ASV_379 (Lawsonibacter), which were specifically induced in the presence of the probiotic mix and not modified by the HF diet itself (Supplementary Figure S4). The three other ASVs with a significant increase in relative abundance ASV_449 (Christensenella), ASV_87 (Roseburia) and ASV_233 (Flintibacter butyricus) were more than twofold higher in the HF-Pr2 group than in the HF-diet group (), but they were also increased by the HF diet per se (Supplementary Figure S4).
Table 1. Description of the 17 ASVs with significant change in relative abundance in the feces of the probiotic-treated mice compared to HF fed mice.
Of the bacteria whose relative abundance was reduced by supplementation with the probiotic mix, six belong to the Lachnospiraceae family, including bacteria belonging to genera previously associated with obesity or diabetic phenotypes in rodents and humans,Citation21–24 such as Dorea (ASV_554), Fusimonas (ASV_438), or Enterocloster (ASV_736) (). Interestingly, these bacteria were not detectable in the feces of mice supplemented with the probiotic mix (). Furthermore, four ASVs appeared to be of great interest with respect to the benefit of supplementation with the probiotic mix, since their relative abundance was oppositely regulated by the probiotic mix compared to the HF diet alone. ASV_235 (Lachnospiraceae Simiaoa), ASV_152 (Acetivibrio), ASV_554 (Dorea), and ASV_276 (Adlercreutzia muris) were induced by the HF diet, and supplementation with the probiotic mix was able to prevent these inductions (Supplementary Figure S5). The other ASVs whose relative abundance in feces was lower in probiotic mix-supplemented mice than in HF diet-fed mice corresponded to bacteria already strongly down-regulated in HF diet-fed mice compared to Chow diet fed mice, leading to undetectable levels in mice supplemented with the probiotic mix, as seen for ASV_137 (Bifidobacterium pseudolongum), ASV_301 (Muribaculum), ASV_438 (Fusimonas) or ASV-17 (Herbinix or Anaerobium) (Supplementary Figure S5). For this subgroup of bacteria, it is likely that the observed reduction in relative abundance did not contribute to the beneficial action of probiotic supplementation since the changes appeared mainly due to the HF diet.
To gain more insight on how modifications of the composition of the microbiota in the presence of the probiotic mix could have had an impact on host metabolism, we first focused on changes in the concentration of SCFAs in feces because several modified bacteria are potential SCFA producers. However, quantification of the main SCFAs did not reveal any significant effects of probiotic intake. In fact, while the HF diet was associated with a coordinated reduction of the four measurable SCFAs, the intake of the probiotic mix did not modify their concentrations in feces (Supplementary Figure S6).
Probiotic mix supplementation downregulated the FXR-Fgf15 pathway in the ileum
To further investigate whether other intestinal processes could be affected by supplementation with the probiotic mix, we assessed in the ileum, the expression of different sets of genes involved in important metabolic or hormonal pathways previously associated in the literature with beneficial responses to probiotics. A number of lactic bacteria have been shown to stimulate the production of the incretin hormones GIP (glucose-dependent insulinotropic polypeptide) and GLP1 (glucagon-like peptide 1, encoded by the Gcg gene), and of TRG5 receptor (encoded by Gpbar1) in intestinal L cells.Citation25–28 The expression of these genes was not affected in the ileum (). In the jejunum, Gcg and Gip mRNA levels were induced by the HF diet. The probiotic mix only reduced the expression level of Gip (Supplementary Figure S7).
Figure 5. Effects of supplementation with the probiotic mix on regulatory pathways in the ileum. (a) incretin pathway. Relative mRNA expression levels of G protein-coupled bile acid receptor 1 (Gpbar1), glucagon (Gcg) and gastric inhibitory polypeptide (Gip) in the ileum. (b) endocannabinoid pathway. Relative mRNA expression levels of N-acyl phosphatidylethanolamine phospholipase D (Napepld), N-acylethanolamine acid amidase (Naaa) and fatty acid amide hydrolase (Faah) in the ileum. (c) Ahr pathway. Relative mRNA expression levels of Aryl-hydrocarbon receptor repressor (Ahrr), cytochrome P450 family 1 subfamily a polypeptide 1 (Cyp1a1) and cytochrome P450 family 1 subfamily b polypeptide 1 (Cyp1b1) in the ileum. (d) FXR-Fgf15 pathway. Relative mRNA expression levels of nuclear receptor subfamily 1 group H member 4 (Nr1h4), nuclear receptor subfamily 0 group B member 2 (Nr0b2) and fibroblast growth factor 15 (Fgf15) in the ileum. Data are mean ± SEM (n = 7–10). ¤p <0.001 versus the Chow diet group and **p <0.01, ***p <0.001 for the HF-Pr2 versus the HF group.
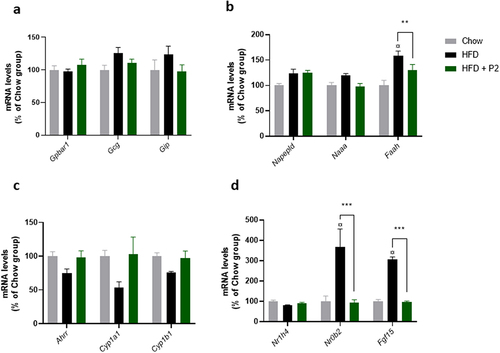
We then measured the expression of genes involved in the synthesis (Napepld) and the degradation (Naaa and Faah) of endocannabinoids and N-acetylamines, important biomolecules involved in several biological processes, including the control of food intake and inflammation.Citation29 The expression of Faah was induced by the HF diet, this overexpression being prevented by the probiotic mix, whereas the other tested genes were not significantly affected (). Because Faah gene is coding for the fatty acid amide hydrolase that catalyzes the degradation of both anandamide (AEA) and other N-acetylamines, and to a lesser extend 2-arachidonoylglycerol (2-AG), we measured their concentrations in the small intestine. The concentrations of AEA and 2-AG were not significantly different between groups (Supplementary Figure S8). Regarding N-acetylamines, stearoylethanolamide (SEA) concentration was increased by the HF diet but not affected by the probiotic mix (Supplementary Figure S8).
Recently, some Lactobacillus strains have been reported to improve metabolic homeostasis in HF diet-fed mice through the modulation of indole derivatives in the gut, leading to stimulation of the intestinal AhR pathway.Citation26 We measured the expression of three classical target genes of the nuclear receptor AhR in the ileum (Ahrr, Cyp1a1, and Cyp1b1). Despite a trend toward a concerted reduction of their expression in the HF-diet group compared to the Chow diet fed group and an up-regulation with the probiotic mix, the differences observed between groups were not statistically significant ().
Finally, we investigated the regulation of the FXR-FGF15 pathway in the ileum. The intestinal bile acid-activated nuclear receptor FXR (farnesoid X receptor, coded by the Nr1h4 gene) has emerged as a major player in metabolic regulation.Citation30 FXR transcriptional activity was stimulated during the HF diet, as evidenced by a strong induction of its two major target genes Fgf15 (coding for the fibroblast growth factor 15) and Nrob2 (coding for the small heterodimer partner SHP) (). Supplementation with the probiotic mix completely prevented the overexpression of these two genes, thus supporting an inhibition of FXR activity in the ileum. The expression of Nr1h4 (coding FXR) itself was not affected by the diets ().
Probiotic mix supplementation modified bile acid profile favoring FXR antagonists in the caecum
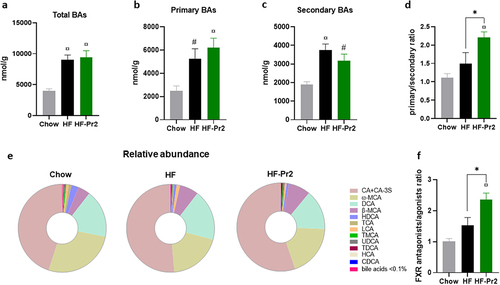
Intestinal FXR activity is directly under the control of BA molecules present in the gut lumen. We therefore determined, by targeted LC-MS/MS, the concentrations of the main BAs in the caecum. The total BA concentration was significantly increased in HF diet-fed mice compared to Chow diet fed mice (). Supplementation with the probiotic mix did not affect the total amount but tended to increase the concentration of primary () and to reduce the amount of secondary () BAs, leading to a significant increase in the primary to secondary BA ratio (). The BA pool composition was moderately modified by the HF diet, with an increase in cholic acid (CA and its related compounds, mainly CA 3-sulfate), deoxycholic acid (DCA), β-muricholic acid (βMCA), and lithocholic acid (LCA) levels in the caecum of HF diet-fed mice ( and Supplementary Figure S9). No major differences in the BA profile were observed between HF diet-fed and HF-Pr2 mice, except for LCA levels that were significantly reduced with the probiotic mix (Supplementary Figure S9). Among the different BAs, several can stimulate the transcriptional activity of FXR (such as CA, DCA, CDCA, LCA, and MCA), while others are well-described antagonists of intestinal FXR (especially β-MCA, TMCA, UDCA, and TUDCA). It is then important to note that the ratio of FXR antagonist/agonist BAs was significantly higher in HF-Pr2 mice than in HF diet-fed mice (), in line with the observed reduction of FXR activity in the ileum of mice supplemented with the probiotic mix.
Figure 6. Effects of the probiotic mix on bile acid (BA) profile in the caecum. (a) concentration of total BAs in the caecum. (b) amount of primary BAs (CA, CA-3S, CDCA, TCDCA, TCA, HCA, β-MCA and TMCA). (c) amount of secondary BAs (DCA, TDCA, LCA, ω-MCA, HDCA and THDCA). (d) primary and secondary BA ratio. (e) BA profile in the caecum. (f) FXR antagonists/FXR agonists ratio. Data are mean ± SEM (n = 6–10). #p <0.01, ¤p <0.001 versus the Chow diet group and *p <0.05 for the HF-Pr2 versus the HF group.
Modulations of the BA profile in the gut could result from changes in the rate of their production and secretion by the liver and/or from enzymatic modifications by bacterial enzymes in the intestinal lumen. The mRNA levels of key genes of BA synthesis, modification, and export from hepatocytes were not different in the liver of HF diet-fed mice with or without probiotic supplementation (Supplementary Figure S10). The hepatic expression of Cyp7a1, coding the rate-limiting enzyme of the classical pathway, was significantly increased in the two groups fed with the HF diet (Supplementary Figure S10), in agreement with the higher levels of CA and total BAs observed in the gut of the HF diet-fed mice.
Inhibition of intestinal FXR pathway was associated with improvement of host phenotype markers during supplementation with the probiotic mix
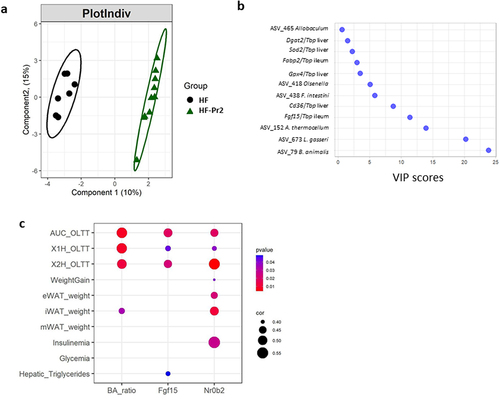
When the whole dataset recorded and/or generated in the present study was taken into account (n = 1474 variables), a Partial Least-Squares Discriminant Analysis (PLS-DA) clearly separated the HF and the HF-Pr2 groups (). The Variable Importance in Projection (VIP) score identified the changes in the abundance of the two probiotic strains (ASV_79 and ASV_673), of ASV_152 (Acetivibrio), and that of Fgf15 expression in the ileum, as the four most discriminant parameters between HF and HF-Pr2 mice (). This analysis thus highlighted the reduction of Fgf15 expression, and therefore the inhibition of FXR activity in the ileum, as a major contributor to the differences between the two groups. To verify whether intestinal inhibition of the FXR pathway could be associated with improved anthropometric and metabolic parameters, we looked for correlations between indicators of intestinal FXR activity (i.e. the FXR antagonist to agonists bile acid ratio and the gene expression of Fgf15 and of Nr0b2 in the ileum) and the different variables related to obesity and MetS (weight gain, adipose tissue weights, liver triglycerides, results of OLTT, insulinemia, and glycemia) in the three groups of mice. The intestinal FXR activity indicators were significantly correlated with OLTT data (AUC and peaks of TG after 1 and 2 h) (). The expression level of Nr0b2 was correlated with eWAT and iWAT weights. When Benjamini–Hochberg (BH) multiple correction was applied, only the correlation with the AUC of OLLT remained significant (Supplementary Figure S11). We also identified significant correlations between Fgf15 and Nr0b2 gene expression in the ileum and the expression of Cd36 and Fabp2, key genes of fatty acid uptake in ileal and jejunal enterocytes (Supplementary Figure S12).
Figure 7. A multivariate approach highlights significant separation between HF and HF-Pr2 groups and the potential role played by the inhibition of intestinal FXR pathway in the beneficial effects of the probiotic mix. (a) partial least squares-discriminant analysis (PLS-DA) plot of individual mice from HF and HF-Pr2 groups and (b) Variable Importance in the project (VIP) representation of the most discriminant features identified by PLS-DA. (c) correlation plot of indexes of intestinal FXR inhibition (Ba_ratio = ratio of FXR antagonist/agonist bile acid species), Fgf15 (Fgf15/Tbp mRNA ratio measured in the ileum) and Nr0b2 (Nr0b2/Tbp mRNA ratio measured in the ileum) and MetS related parameters (X1H_OLTT and X2H_OLTT correspond to the plasma TG values measured during the OLTT 1 and 2 hours after administration of the oil bolus). Spearman correlation coefficients are indicated by the size of the dots, p-values are indicated by the color.
Discussion
The use of probiotics to help ameliorate obesity and associated metabolic disorders is now considered a valid and promising strategy.Citation14,Citation15,Citation18 Lactobacillus and Bifidobacterium strains are commonly used for this purpose, and a number of studies have clearly evidenced the beneficial effects of such strains on obesityCitation14 and, for some of them, on the characteristics of Mets.Citation15–17 However, the multiple possible mechanisms of action involved in the improvement of metabolic health are complex and generally remain to be clarified.Citation31 Furthermore, the beneficial effects often appear to be related to the individual properties of specific strains rather than to a general probiotic effect.Citation32 The validation and characterization of the mechanisms of action of effective strains are therefore of the utmost importance. Using a multi-criteria in vitro screening strategy followed by in vivo tests in pre-clinical models, Alard and colleagues identified a high-potential mixture of B. animalis subsp. lactis LA804 and L. gasseri LA806 strains, having a significant impact on obesity in HF diet-fed miceCitation20. In the present study, we further characterized the potential of the combination of these two strains to prevent MetS, obesity onset, and lipid metabolism dysfunction in mice. We identify the contribution of an inhibition of the intestinal FXR pathway as a possible mechanism to explain the beneficial effects of these probiotics.
We first confirmed that the supplementation with the probiotic mix limited body weight gain and adipose tissue accumulation, without modifying food intake, in HF diet-fed mice. These effects were accompanied by protection against inflammation in the adipose tissue and against lipid accumulation, oxidative stress, and ER stress in the liver. These beneficial effects on various hallmarks of MetS and obesity were not associated with a reorientation of the fate of excessive lipid intake during the HF diet toward catabolic pathways such as brown fat stimulation or hepatic mitochondrial and peroxisomal oxidations. The browning of WAT was recently proposed to contribute to the effects of L. amylovorus KU4 (Amylolactobacillus amylophilus),Citation33 but here we did not find any evidence of modification of browning markers after intake of the probiotic mix. To better characterize the effects on lipid metabolism, we evaluated the impact of the probiotic mix on postprandial lipemia in vivo using an oral lipid tolerance test. We found that the postprandial plasma TG peak was lowered during OLTT in mice treated with the probiotic mix compared to the HF diet-fed mice. At the molecular level in the jejunum and ileum, the probiotic mix protected against HF-induced rise in mRNA levels of Cd36 and Fabp2, two transporters involved in fatty acid uptake by enterocytes. These results suggest that the pathways involved in intestinal lipid uptake are globally modulated by the consumption of the probiotic mixture. Nevertheless, since OLTT results are the balance between lipid absorption and TG clearance from the plasma, the impacts of the probiotic mix on these mechanisms, and to which extent they contribute to lowering the accumulation of fat in tissues and improving metabolic parameters, remains to be elucidated.
Various mechanisms have been proposed to explain the beneficial effects of probiotics on metabolic health, including improvement of the gut barrier function.Citation34 Increased intestinal permeability is considered a major defect in obesity and MetS, participating in endotoxin translocation from gut lumen to circulation and induction of a pro-inflammatory state in the adipose tissue, liver, and other peripheral organs.Citation35 Some probiotics have been reported to restore intestinal permeability in in vitro models, animal models, and, in some cases, in humans.Citation34,Citation36 Here, in agreement with numerous studies in rodents,Citation37 we found that the HF diet was associated with a significant increase in gut permeability assessed in vivo by the lactulose-mannitol test. However, the probiotic mix did not protect or improve the HF diet-induced leaky gut. This important result demonstrated that, under our experimental conditions, improvement of intestinal permeability was not required for the beneficial action of the tested probiotic mix on obesity, metabolic health, and lipid metabolism.
Another classical mechanism proposed to explain the beneficial actions of probiotics is the production of SCFAs,Citation38 either directly by the probiotic strains able to synthesize them or by modulation of the gut microbiota composition favoring SCFA-producing bacteria. Although Lactobacillus and Bifidobacteria are known producers of SCFAs and their consumption in the present study was accompanied by an increase in the relative abundance of other species able to produce SCFAs, including Olsenella,Citation39 Lawsonibacter,Citation40 Christensenella ,Citation41 Roseburia,Citation42 and Flintibacter butyricus,Citation43 we were surprised to find that the supplementation with the probiotic mix was not associated with a change in the levels of the main SCFAs (acetate, propionate, butyrate, and valerate) in fecal samples. Indeed, the HF diet induced, as expected, a strong reduction in the levels of these four molecules, but this effect was not prevented or restored by the probiotic mix. However, we cannot exclude that changes in SCFAs have occurred in other compartments such as in plasma or caecum. It has recently been demonstrated that circulating rather than fecal SCFAs are more directly linked to metabolic health in a cross-sectional study in humans.Citation44 One of the described beneficial effects of SCFAs on metabolic dysfunctions is the stimulation of the production of incretin hormone GLP1 by intestinal endocrine cells.Citation45 Consistent with the absence of SCFA modifications in this study, no changes in the expression of Gcg gene (coding for GLP1) were found in the ileum and jejunum of supplemented mice.
Improved metabolic health in HF-fed mice was reported after administration of Limosilactobacillus reuteri, which is able to produce indoles and their derivatives that can activate the aryl hydrocarbon receptor (AhR).Citation26 These AhR agonists are microbiota metabolites derived from tryptophan and are strongly reduced in the gut lumen and feces in obesity and MetS.Citation26 Activation of AhR by indoles in the small intestine is associated with the restoration of gut barrier defects and the production of GLP1.Citation26 Although these two parameters were not modified in the study, we assessed the possible involvement of the AhR pathway by evaluating the expression of classical target genes of Ahr in the ileum (Ahrr, Cyp1a1, and Cyp1b1). The lack of difference between groups further supported that the AhR pathway did not play a major role in this study.
Another potential mechanism involved in the positive effects of the probiotic mix may be the modulation of the endocannabinoid system in the gut. An interplay between metabolic health, intestinal barrier integrity, and endocannabinoid system balance has already been demonstrated,Citation13 and some studies have reported effects of probiotics on endocannabinoid concentration in the ileum, such as an increase in 2-AG, after administration of Akkermansia muciniphila in HF-diet fed mice, which was associated with improved metabolic dysfunctions.Citation46 While we observed a modification of Faah gene expression, the enzyme involved in the degradation of endocannabinoids, in the ileum of the supplemented mice, we did not find any modifications in the concentration of anandamide and the main N-acetylamines in small intestinal tissues.
FXR plays a pivotal role in the metabolism of BA, lipids, and glucose, but also in inflammation and intestinal barrier function.Citation30,Citation47 In the ileum, FXR controls the expression of Fgf15 (fibroblast growth factor 15 in rodent and FGF19 in humans), an important regulator of glucose and lipid homeostasisCitation48 in addition to its role in the control of BA synthesis in the liver.Citation49 BAs are natural ligands of FXR and can be either activators or inhibitors, with CDCA (chenodeoxycholic acid) and DCA (deoxycholic acid) being the more potent agonists, whereas UDCA (ursodeoxycholic acid) and β-MCA (muricholic acid) are antagonists of FXR. Supplementation of HF diet-fed mice with the probiotic mix did not affect the total amount of BAs but led to a significant increase in the primary to secondary BA ratio in the caecum and a reduction in LCA abundance. Furthermore, we found a significant increase in the FXR antagonists/agonists ratio, in line with a marked inhibition of FXR activity in the ileum of the supplemented mice, as evidenced by a drastic reduction in the expression of the FXR target genes Fgf15 and Nr0b2. Interestingly, it was recently reported that inhibition of intestinal FXR activity by caffeic acid phenethyl ester specifically in the gut exerts potent anti-diabetic effects.Citation50 Similarly, increased levels of intestinal T-β-MCA (the tauro-conjugated β-muricholic acid) which is a strong antagonist of FXR has been shown to improve HFD-induced obesity, hepatic steatosis, and glucose intolerance via the inhibition of intestinal FXR activity.Citation51 In addition, the interplay between BA pool composition, intestinal FXR activity, and lipid absorption has recently been highlighted.Citation52 Altogether, these data indicate that an inhibition of intestinal FXR signaling could be involved in the mechanisms of action of the probiotic mix tested in this study.
Using a PLS-DA with all the parameters and data measured in the study (1474 variables), we found that the decrease in the expression of Fgf15 in the ileum, reflecting the inhibition of intestinal FXR activity, was among the most discriminant variables between the HF and the HF-Pr2 groups. Furthermore, the changes in the expression of Fgf15 and Nrob2 and of the FXR antagonist/agonist bile acid ratio were correlated with the evolution of the postprandial lipemia assessed by OLTT and, to a lower extent, to the size of adipose tissue depots. Fgf15 and Nr0b2 expressions were also correlated with small intestinal expression of Cd36 and Fabp2, key actors of fatty acid uptake by enterocytes. Altogether, these data are thus indicative of the contribution of the inhibition of intestinal FXR signaling in the mechanisms of action of the tested probiotic mix in preventing obesity and metabolic deteriorations upon HF diet, potentially through a limitation of intestinal fatty acid uptake.
Recent works have demonstrated the potential of the inhibition of FXR activity specifically in the small intestine to counteract obesity and to improve metabolic disturbances associated with diabetes and nonalcoholic fatty diseases.Citation47,Citation51,Citation53 This original concept derived from the observation that intestine-specific Fxr gene invalidation in the mouse protects against diet-induced obesity.Citation51 Contradictory results have been reported in this genetically modified model showing, for example, exacerbation of alcoholic fatty liver disease in intestinal FXR deficient mice.Citation54 However, the use of inhibitors or antagonists acting selectively on intestinal FXR, such as caffeic acid phenethyl ester, T-β-MCA, or glycine-β-MCA, clearly supports a beneficial role of the inhibition of FXR.Citation50,Citation51,Citation55 Clinical trials with UDCA, a strong FXR antagonist, also showed an increase in insulin sensitivity in patients with nonalcoholic steatohepatitis or obesity.Citation47 Furthermore, it was demonstrated that the beneficial effect of metformin in type 2 diabetic patients is associated with a change in the intestinal BA profile, favoring FXR antagonists (especially GUDCA) and the inhibition of FXR signaling.Citation56 We recently found that a similar mechanism may occur in mice treated with metformin.Citation57
Little is known regarding the possibility that probiotics could specifically target intestinal FXR signaling to improve metabolic health. It was shown that the probiotic mix VSL#3 was able to down-regulate the FXR-Fgf15 pathway via modulation of BA metabolism, enhancing in turn BA deconjugation and their fecal excretion.Citation58 VSL#3 is currently sold as a mixture of eight different strains of Lactobacillus, Bifidobacterium, and Streptococcus thermophilus BT01, with effects on intestinal inflammation.Citation59 Interestingly, studies in mice have also reported beneficial metabolic effects of this mix,Citation60 and in humans a protective effect of VSL#3 against increased fat mass has been evidenced in healthy young adults consuming a HF diet.Citation61 The mechanism of action was, however, not reported in these studies. Very recently, two studies reported that improvement of Mets parameter in HF-diet fed mice by probiotic supplementation (a mix of Limosilactobacillus vaginalis FN3 and Bifidobacterium animalis subsp. lactis F1–7Citation62 and Lactobacillus plantarum LP104)Citation63 was possibly associated with regulation of BA metabolism and reduce FXR-Fgf15 pathway in the ileum.
Our work presents some limitations despite the fact that we tried to explore most of the pathways and mechanisms classically proposed in the literature to explain the beneficial action of probiotics in rodent models of MetS or obesity. First of all, the amplitudes of the gene expression regulation in the different tissues were quite low and the real involvement of such changes in the metabolic outcomes remains to be demonstrated. Secondly, the bile acid composition was studied in the caecum and, even if the caecum is closely located after the ileum, one could not exclude that additional modifications of bile acid species occurred at the level of the ileum, contributing to the inhibition of the FXR activity. In addition, modifications of the microbiota composition in the small intestine might have contributed more directly to the beneficial effects of the probiotic mix rather than the changes detected in the feces. Finally, although the role of colonization of the gut by probiotics in explaining their health effects is still a matter of debate in the scientific community, the possible mucosal adherence and intestinal colonization of our strains were not studied in the present work.
In conclusion, although there have already been several studies, both in rodents and humans, investigating the potential of Lactobacillus and Bifidobacterium strains in improving lipid metabolism in MetS and obesity, here we evaluate the efficacy and the mechanisms of action of B. animalis subsp. lactis LA804 and L. gasseri LA806, two strains carefully selected after a multi-criteria screeningCitation20. We discovered that the inhibition of FXR signaling in the small intestine could contribute to the beneficial action of these strains, independently from the more classically described protective effects of lactic bacteria on gut permeability, SCFA production, or GLP1 induction. Inhibition of the intestinal FXR/Fgf15 pathway was associated with modulation of the cecal BA profile with increased FXR antagonist/agonist ratio among BA metabolites and subtle changes in gut microbiota composition that may have contributed to this process. Although the translation from animals to humans of the beneficial effects and involved mechanisms remains to be established, the fact that several trials with Lactobacillus and Bifidobacterium strains have already demonstrated weight and metabolic improvementsCitation14,Citation15,Citation18,Citation61 positively support B. animalis subsp. lactis LA804 and L. gasseri LA806 strains as a promising probiotic mix for the management of weight and metabolism in MetS and obesity.
Materials and methods
Mouse model, experimental design, and tissue sampling
The study protocol was approved by the local ethics committee with the agreement number CECAPP_LS_2021_006. Forty-eight C57BL/6J male mice were purchased from Envigo, France, at the age of 5 weeks. Animals were housed in an air-conditioned room with a controlled environment of 21 ± 0.5°C and 60–70% humidity with a 12-h light/dark cycle and with free access to food and water. After acclimatization for 1 week, mice were randomly divided into three groups: the Chow diet group, the high-fat diet group (HF), and the high-fat diet + probiotic mix group (HF-Pr2). The Chow diet group received the control Chow diet (LASQC diet® Rod-16 R, LASvendi). Both high-fat diet groups received a diet containing 61% of the energy from fat (292HF-FEDSAFE, composition in Supplementary Table S2) and the HF-Pr2 group was supplemented daily with a mixture of two probiotic strains (detailed below). The nutritional intervention lasted for 12 consecutive weeks. Body weight was measured once a week and food intake monitored three times a week throughout the experiment. At the end of the study, animals were euthanized after a 12-h fasting (Supplementary Figure S1). Blood and tissues were immediately collected, weighed, and treated accordingly to subsequent analysis requirements and then stored at 80°C. Fasting blood glucose levels were measured using a OneTouch glucometer (Johnson&Johnson, New Brunswick, United States).
Probiotic mix
The probiotic mix was provided by PiLeJe Laboratoire (Paris, France) in the form of a freeze-dried mixture of two probiotic strains (B. animalis subsp. lactis LA804 and L. gasseri LA806) with a ratio of 1:1. PBS was added to the mix in order to administer 1.109 CFU/day to each mouse. HF-Pr2 mice were force-fed daily, every morning, with the probiotic mix from the first day of a high-fat diet until the end of the experiment. Mice from the Chow diet and HF groups received PBS only. To conform to the animal ethics committee’s instructions, probiotics and PBS were administered by intra-oral depot during the first 6 weeks of treatment followed by intragastric gavage until the end of the experiment.
Measurements of metabolic markers
Liver triglyceride (TG) and total cholesterol concentrations were measured with commercial kits (BIOLABO, ref #80019 and #K1106 respectively). Plasma insulin level was measured by ELISA (Crystal Chem, ref #90080). All assays were performed according to the instructions of the manufacturers.
Oral lipid tolerance test (OLTT)
Animals fasted for 12 h before OLTT. The test was performed after 10 weeks of treatment and consisted in the administration of 250 µL of Isio4® vegetable oil, a mixture of rapeseed, sunflower, oleic sunflower, and linseed oils with balanced polyunsaturated fatty acid profile (Lesieur). The oil was administered by intragastric gavage. Blood samples were collected from the tail to measure fasting (before the test) and postprandial lipemia (1 h, 2 h, and 4 h post-gavage). Blood was centrifuged to measure plasma triglyceride concentrations using a commercial kit in accordance with the manufacturer’s instructions (BIOLABO ref #80019).
RNA extraction and real-time quantitative PCR (RT-qPCR)
Total RNA from several tissues (liver, white/brown adipose tissues, and intestinal segments) were extracted using TRI-Reagent (Sigma Aldrich, ref #T9424), in accordance with the manufacturer’s instructions. First-strand cDNAs were synthesized from 1 µg of total RNAs using TAKARA Prime Script™ RT Reagent (TAKARA Bio, ref #RR037A). Purity and concentration of RNAs were determined using NanodropOne (Ozyme, Saint-Cyr-L’Ecole, France), and the quality was checked by using a Bioanalyser (Agilent Technologies, Santa Clara, United-States). Real-time PCR assays were performed with Rotor-Gene Q (Qiagen, Hilden, Germany) using SYBR® Premix Ex-Taq™ (TAKARA Bio, ref #RR420L). TATA-box binding protein (TBP) was used as a reference gene to normalize the results, as previously reported.Citation64 Primers are listed in Supplementary Table S3.
Hepatic lipid droplet analysis
After dissection, the liver was immediately fixed in 4% paraformaldehyde and then paraffin-embedded for the analysis of lipid content. The prepared series of 10-µm sections were stained with Oil Red O (Biognost, ref #ORO-k-250) to determine the accumulation of neutral lipids in the liver. The surface area of the lipid droplets (µmCitation2) was quantified per acquisition field using a custom-written Fiji macro.
16s RNA gene sequencing and data processing
Analysis of the intestinal microbiota at the end of the experiment was carried out by sequencing bacterial 16S rRNA in feces by the ProfilExpert platform (Lyon, France). Purified DNA were extracted from 15 mg of feces with the Zymobiomics DNA microprep kit (Zymo Research, ref #D4301). 40 ng of DNA were used for the generation of libraries targeting the V3-V4 regions (Quick-16S NGS Library Prep Kit, Ozyme) and then sequenced on Illumina MiSeq standard v3. The sequences were “demultiplexed” using Bcl2fastq software (v2.17.1.14) then cleaned with cutadapt (v1.9.1). Amplicon sequence variants (ASVs) were identified using Qiime2 DADA2 with a Zero noise OTU approach. Finally, a taxonomic assignment was made with Greengenes-13.8-nr99 base. Alpha and beta diversities were calculated under RStudio (v2021.09.1 build 372) using the phyloseq (v1.34.0) and metagenomeseq (1.32.0) libraries.
Quantification of caecal bile acids
The quantification of bile acids was carried out from 100 mg of frozen caecum by High-Performance Liquid Chromatography coupled with tandem Mass Spectrometry (HPLC-MS/MS) according to the method described by Humbert et al.Citation65 and as previously reported.Citation66 The results are expressed in nmol/g of caecum (wet weight).
Statistical analysis
Statistical analyses and graphical representations were performed using RStudio (V2021.09.1 Build 372) and GraphPad Prism 9. For each parameter, normality (Shapiro test), homogeneity of variances (Levene test), and independence of residuals (Durbin Watson test) were tested. Depending on the results, either a Kruskal–Wallis test followed by Benjamini–Hochberg (BH) multiple correction and a Dunn posthoc test or a one-way Anova with BH multiple correction followed by a posthoc Tukey test were used to compare groups. Nonparametric Spearman rank correlations between markers of metabolic syndrome, intestinal FXR pathway, and intestinal lipid uptake were conducted using nonparametric Spearman’s test and visualized using a correlation matrix and scatter plots. PLS-DA of the data were performed on microbial data and on the whole set of data generated in the study. Variable Importance of Projection (VIP) scores were assessed to rank parameters for their degree of discrimination within the model. Data are presented as the mean ± SEM. The results were considered significant when p < 0.05. Symbols are used to illustrate significant differences with the Chow diet-fed group ($p < 0.05; #p < 0.01 and ¤p < 0.001) and stars are used to illustrate significant differences between HF-Pr2 and HF groups (*p < 0.05; **p < 0.01 and ***p < 0.001).
Authors’ contributions
MLB, EJ, MCM, and HV conceived and designed the study. AB, BB, MLB, and EM performed the experiments and data analysis. AP, CC, CP, EL, SC, AVM, MG, and FL contributed to study design, samples collection, or biological or data analyses. VS performed the analysis of short chain fatty acids. AB, BB, MLB, EM, EJ, MCM, and HV performed the interpretation. AB, BB, and HV wrote the first version of the paper. CC, EJ, SH, and MCM critically revised the manuscript. All authors approved the final version before submission.
Supplemental Material
Download MS Word (571.1 KB)Supplemental Material
Download MS Word (16 KB)Acknowledgments
We thank Dr. Dominique Rainteau and his colleagues (at Centre de Recherche Saint-Antoine, Paris) for the analysis of cecal bile acids and Prof. G.G. Muccioli and his colleagues (at Université Catholique de Louvain, Belgium) for the analysis of endocannabinoids. Fecal microbiota sequencing was performed by ProfileXpert (at the Faculté de Médecine et de Pharmacie Rockefeller, Lyon, France). Mrs Claude Blondeau (PiLeJe Laboratoire) was acknowledged for draft review.
Disclosure statement
Sophie Holowacz and Elsa Jacouton are employees of PiLeJe Laboratoire. No other conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The data of 16S sequencing have been deposited in the French repository at “Recherche.data.gouv.fr” and can be accessible with the following link: https://doi.org/10.57745/MXOF51.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2023.2281015
Additional information
Funding
References
- Myers J, Kokkinos P, Nyelin E. Physical activity, cardiorespiratory fitness, and the metabolic syndrome. Nutrients. 2019;11(7):1652. doi:10.3390/nu11071652.
- Saklayen MG. The global epidemic of the metabolic syndrome. Curr Hypertens Rep. 2018;20(2):12. doi:10.1007/s11906-018-0812-z.
- Kopp W. How western diet and lifestyle drive the pandemic of obesity and civilization diseases. Diabetes, Metab Syndr Obes Targets Ther. 2019;12:2221–21. doi:10.2147/DMSO.S216791.
- Després JP, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444(7121):881–887. doi:10.1038/nature05488.
- Malesza IJ, Malesza M, Walkowiak J, Mussin N, Walkowiak D, Aringazina R, Bartkowiak-Wieczorek J, Mądry E. High-fat, Western-style diet, systemic inflammation, and gut microbiota: a narrative review. Cells. 2021;10(11):3164. doi:10.3390/cells10113164.
- Agus A, Clément K, Sokol H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. 2021;70(6):1174–1182. doi:10.1136/gutjnl-2020-323071.
- Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8(1):42. doi:10.1186/s13073-016-0303-2.
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi:10.1038/nature05414.
- de Groot P, Scheithauer T, Bakker GJ, Prodan A, Levin E, Tanweer Khan M, Herrema H, Ackermans M, Serlie MJM, de Brauw M, et al. Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut. 2020;69(3):502. doi:10.1136/gutjnl-2019-318320.
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Sci. 2013;341(6150):1241214. doi:10.1126/science.1241214.
- Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–335. doi:10.1038/nri3430.
- Serino M, Luche E, Gres S, Baylac A, Bergé M, Cenac C, Waget A, Klopp P, Iacovoni J, Klopp C, et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut. 2012;61(4):543–553. doi:10.1136/gutjnl-2011-301012.
- Jansma J, Brinkman F, Van Hemert S, El Aidy S. Targeting the endocannabinoid system with microbial interventions to improve gut integrity. Prog Neuropsychopharmacol Biol Psychiatry. 2021;106:110169. doi:10.1016/j.pnpbp.2020.110169.
- Álvarez-Arraño V, Martín-Peláez S. Effects of probiotics and synbiotics on weight loss in subjects with overweight or obesity: a systematic review. Nutr. 2021;13(10):3627. doi:10.3390/nu13103627.
- Famouri F, Shariat Z, Hashemipour M, Keikha M, Kelishadi R. Effects of probiotics on nonalcoholic fatty liver disease in obese children and adolescents. J Pediatr Gastroenterol Nutr. 2017;64(3):413–417. doi:10.1097/MPG.0000000000001422.
- Duseja A, Acharya SK, Mehta M, Chhabra S, Shalimar R, Das S, Dattagupta A, Dhiman S, Chawla YK. High potency multistrain probiotic improves liver histology in non-alcoholic fatty liver disease (NAFLD): a randomised, double-blind, proof of concept study. BMJ Open Gastroenterol. 2019;6(1):e000315. doi:10.1136/bmjgast-2019-000315.
- Ahn HY, Kim M, Chae JS, Ahn YT, Sim JH, Choi ID, Lee SH, Lee JH. Supplementation with two probiotic strains, lactobacillus curvatus HY7601 and lactobacillus plantarum KY1032, reduces fasting triglycerides and enhances apolipoprotein A-V levels in non-diabetic subjects with hypertriglyceridemia. Atheroscleros. 2015;241(2):649–656. doi:10.1016/j.atherosclerosis.2015.06.030.
- Xavier-Santos D, Bedani R, Lima ED, Saad SMI. Impact of probiotics and prebiotics targeting metabolic syndrome. J Funct Foods. 2020;64:103666. doi:10.1016/j.jff.2019.103666.
- Le Barz M, Daniel N, Varin TV, Naimi S, Demers-Mathieu V, Pilon G, Audy J, Laurin É, Roy D, Urdaci MC, et al. In vivo screening of multiple bacterial strains identifies lactobacillus rhamnosus Lb102 and bifidobacterium animalis ssp. lactis Bf141 as probiotics that improve metabolic disorders in a mouse model of obesity. FASEB J Off Publ Fed Am Soc Exp Biol. 2019;33(4):4921–4935. doi:10.1096/fj.201801672R.
- Alard J, Cudennec B, Boutillier D, Peucelle V, Descat A, Decoin R, Kuylle S, Jablaoui A, Rhimi M, Wolowczuk I, et al. Multiple selection criteria for probiotic strains with high potential for obesity management. Nutr. 2021;13(3):713. doi:10.3390/nu13030713.
- Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, De Angelis M. The controversial role of human gut Lachnospiraceae. Microorgan. 2020;8(4):573. doi:10.3390/microorganisms8040573.
- Li Q, Chang Y, Zhang K, Chen H, Tao S, Zhang Z. Implication of the gut microbiome composition of type 2 diabetic patients from northern China. Sci Rep. 2020;10(1):5450. doi:10.1038/s41598-020-62224-3.
- Kusada H, Kameyama K, Meng XY, Kamagata Y, Tamaki H. Fusimonas intestini gen. Nov., sp. Nov., a novel intestinal bacterium of the family lachnospiraceae associated with diabetes in mice. Sci Rep. 2017;7(1):18087. doi:10.1038/s41598-017-18122-2.
- Park S, Zhang T, Kang S. Fecal microbiota composition, their interactions, and metagenome function in US adults with type 2 diabetes according to enterotypes. Int J Mol Sci. 2023;24(11):9533. doi:10.3390/ijms24119533.
- Simon MC, Strassburger K, Nowotny B, Kolb H, Nowotny P, Burkart V, Zivehe F, Hwang JH, Stehle P, Pacini G, et al. Intake of Lactobacillus reuteri improves incretin and insulin secretion in glucose-tolerant humans: a proof of concept. Diab Care. 2015;38(10):1827–1834. 10.2337/dc14–2690. doi:10.2337/dc14-2690.
- Natividad JM, Agus A, Planchais J, Lamas B, Jarry AC, Martin R, Michel ML, Chong-Nguyen C, Roussel R, Straube M, et al. Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome. Cell Metab. 2018;28(5):737–749.e4. doi:10.1016/j.cmet.2018.07.001.
- Zhang C, Fang R, Lu X, Zhang Y, Yang M, Su Y, Jiang Y, Man C. Lactobacillus reuteri J1 prevents obesity by altering the gut microbiota and regulating bile acid metabolism in obese mice. Food Funct. 2022;13(12):6688–6701. doi:10.1039/D1FO04387K.
- Lopez-Escalera S, Lund ML, Hermes GDA, Choi BSY, Sakamoto K, Wellejus A. In vitro screening for probiotic properties of lactobacillus and bifidobacterium strains in assays relevant for non-alcoholic fatty liver disease prevention. Nutr. 2023;15(10):2361. doi:10.3390/nu15102361.
- Dörnyei G, Vass Z, Juhász CB, Nádasy GL, Hunyady L, Szekeres M. Role of the endocannabinoid system in metabolic control processes and in the pathogenesis of metabolic syndrome: an update. Biomed. 2023;11(2):306. doi:10.3390/biomedicines11020306.
- Sun L, Cai J, Gonzalez FJ. The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer. Nat Rev Gastroenterol Hepatol. 2021;18(5):335–347. doi:10.1038/s41575-020-00404-2.
- Gérard C, Vidal H. Impact of gut microbiota on host glycemic control. Front Endocrinol. 2019;10:29. doi:10.3389/fendo.2019.00029.
- McFarland LV, Evans CT, Goldstein EJC. Strain-specificity and disease-specificity of probiotic efficacy: a systematic review and meta-analysis. Front Med. 2018;5:124. doi:10.3389/fmed.2018.00124.
- Park SS, Lee YJ, Kang H, Yang G, Hong EJ, Lim JY, Oh S, Kim E. Lactobacillus amylovorus KU4 ameliorates diet-induced obesity in mice by promoting adipose browning through PPARγ signaling. Sci Rep. 2019;9(1):20152. doi:10.1038/s41598-019-56817-w.
- Camilleri M. Human intestinal barrier: effects of stressors, diet, prebiotics, and probiotics. Clin Transl Gastroenterol. 2021;12(1):e00308. doi:10.14309/ctg.0000000000000308.
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–1772. doi:10.2337/db06-1491.
- Chaiyasut C, Sivamaruthi BS, Lailerd N, Sirilun S, Khongtan S, Fukngoen P, Peerajan S, Saelee M, Chaiyasut K, Kesika P, et al. Probiotics supplementation improves intestinal permeability, obesity index and metabolic biomarkers in elderly Thai subjects: a randomized controlled trial. Foods. 2022;11(3):268. doi:10.3390/foods11030268.
- Teixeira TFS, Collado MC, Ferreira CLLF, Bressan J, Peluzio MDCG. Potential mechanisms for the emerging link between obesity and increased intestinal permeability. Nutr Res. 2012;32(9):637–647. doi:10.1016/j.nutres.2012.07.003.
- Markowiak-Kopeć P, Śliżewska K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutr. 2020;12(4):1107. doi:10.3390/nu12041107.
- Kraatz M, Wallace RJ, Svensson L. Olsenella umbonata sp. Nov., a microaerotolerant anaerobic lactic acid bacterium from the sheep rumen and pig jejunum, and emended descriptions of olsenella, olsenella uli and olsenella profusa. Int J Syst Evol Microbiol. 2011;61:795–803. doi:10.1099/ijs.0.022954-0.
- Sakamoto M, Iino T, Yuki M, Ohkuma M. Lawsonibacter asaccharolyticus gen. Nov., sp. Nov., a butyrate-producing bacterium isolated from human faeces. Int J Syst Evol Microbiol. 2018;68:2074–2081. doi:10.1099/ijsem.0.002800.
- Morotomi M, Nagai F, Watanabe Y. Description of christensenella minuta gen. Nov., sp. Nov., isolated from human faeces, which forms a distinct branch in the order clostridiales, and proposal of christensenellaceae fam. nov. Int J Syst Evol Microbiol. 2012;62:144–149. doi:10.1099/ijs.0.026989-0.
- Nie K, Ma K, Luo W, Shen Z, Yang Z, Xiao M, Tong T, Yang Y, Wang X. Roseburia intestinalis: a beneficial gut organism from the discoveries in genus and species. Front Cell Infect Microbiol. 2021;11:757718. doi:10.3389/fcimb.2021.757718.
- Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, Kumar N, Bresciani A, Martínez I, Just S, Ziegler C, et al. The mouse intestinal bacterial collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nature Microbiol. 2016;1(10):16131. doi:10.1038/nmicrobiol.2016.131.
- Müller M, Hernández MAG, Goossens GH, Reijnders D, Holst JJ, Jocken JWE, Van Eijk H, Canfora EE, Blaak EE. Circulating but not faecal short-chain fatty acids are related to insulin sensitivity, lipolysis and GLP-1 concentrations in humans. Sci Rep. 2019;9(1):12515. doi:10.1038/s41598-019-48775-0.
- Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein–coupled receptor FFAR2. Diabetes. 2012;61(2):364–371. doi:10.2337/db11-1019.
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110(22):9066–9071. doi:10.1073/pnas.1219451110.
- Sonne DP. MECHANISMS in ENDOCRINOLOGY: FXR signalling: a novel target in metabolic diseases. Eur J Endocrinol. 2021;184:R193–R205. doi:10.1530/EJE-20-1410.
- Jahn D, Rau M, Hermanns HM, Geier A. Mechanisms of enterohepatic fibroblast growth factor 15/19 signaling in health and disease. Cytokine Growth Factor Rev. 2015;26:625–635. doi:10.1016/j.cytogfr.2015.07.016.
- Kir S, Beddow SA, Samuel VT, Miller P, Previs SF, Suino-Powell K, Xu HE, Shulman GI, Kliewer SA, Mangelsdorf DJ. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Sci. 2011;331:1621–1624. doi:10.1126/science.1198363.
- Xie C, Jiang C, Shi J, Gao X, Sun D, Sun L, Wang T, Takahashi S, Anitha M, Krausz KW, et al. An intestinal farnesoid X receptor–ceramide signaling axis modulates hepatic gluconeogenesis in mice. Diabetes. 2017;66(3):613–626. doi:10.2337/db16-0663.
- Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, Fabre KM, Mitchell JB, Patterson AD, Gonzalez FJ. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun. 2013;4(1):2384. doi:10.1038/ncomms3384.
- Clifford BL, Sedgeman LR, Williams KJ, Morand P, Cheng A, Jarrett KE, Chan AP, Brearley-Sholto MC, Wahlström A, Ashby JW, et al. FXR activation protects against NAFLD via bile-acid-dependent reductions in lipid absorption. Cell Metab. 2021;33(8):1671–1684.e4. doi:10.1016/j.cmet.2021.06.012.
- Jiang C, Xie C, Li F, Zhang L, Nichols RG, Krausz KW, Cai J, Qi Y, Fang Z-Z, Takahashi S, Tanaka, N., et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest. 2015;125(1):386–402. doi:10.1172/JCI76738.
- Huang M, Kong B, Zhang M, Rizzolo D, Armstrong LE, Schumacher JD, Chow MD, Lee YH, Joseph LB, Stofan M, et al. Enhanced alcoholic liver disease in mice with intestine-specific farnesoid X receptor deficiency. Lab Invest. 2020;100:1158–1168. doi:10.1038/s41374-020-0439-y.
- Gonzalez FJ, Jiang C, Patterson AD. An intestinal microbiota–farnesoid X receptor axis modulates metabolic disease. Gastroenterol. 2016;151(5):845–859. doi:10.1053/j.gastro.2016.08.057.
- Sun L, Xie C, Wang G, Wu Y, Wu Q, Wang X, Liu J, Deng Y, Xia J, Chen B, et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med. 2018;24(12):1919–1929. doi:10.1038/s41591-018-0222-4.
- Bravard A, Gérard C, Defois C, Benoit B, Makki K, Meugnier E, Rainteau D, Rieusset J, Godet M, Vidal H. Metformin treatment for 8 days impacts multiple intestinal parameters in high-fat high-sucrose fed mice. Sci Rep. 2021;11(1):16684. doi:10.1038/s41598-021-95117-0.
- Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, Moschetta A. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the fxr-Fgf15 axis in mice. Cell Rep. 2014;7:12–18. doi:10.1016/j.celrep.2014.02.032.
- Ganji-Arjenaki M, Rafieian-Kopaei M. Probiotics are a good choice in remission of inflammatory bowel diseases: a meta analysis and systematic review. J Cell Physiol. 2018;233:2091–2103. doi:10.1002/jcp.25911.
- Cheng FS, Pan D, Chang B, Jiang M, Sang LX. Probiotic mixture VSL#3: an overview of basic and clinical studies in chronic diseases. World J Clin Cases. 2020;8:1361–1384. doi:10.12998/wjcc.v8.i8.1361.
- Osterberg KL, Boutagy NE, McMillan RP, Stevens JR, Frisard MI, Kavanaugh JW, Davy BM, Davy KP, Hulver MW. Probiotic supplementation attenuates increases in body mass and fat mass during high-fat diet in healthy young adults: probiotic supplementation during high-fat diet. Obesity. 2015;23(12):2364–2370. doi:10.1002/oby.21230.
- Liang X, Zhang Z, Zhou X, Lu Y, Li R, Yu Z, Tong L, Gong P, Yi H, Liu T, Zhang, L., et al. Probiotics improved hyperlipidemia in mice induced by a high cholesterol diet via downregulating FXR. Food Funct. 2020;11(11):9903–9911. doi:10.1039/D0FO02255A.
- Wang Y, Xing X, Ma Y, Fan Y, Zhang Y, Nan B, Li X, Wang Y, Liu J. Prevention of high-fat-diet-induced dyslipidemia by lactobacillus plantarum LP104 through mediating bile acid enterohepatic axis circulation and intestinal flora. J Agric Food Chem. 2023;71(19):7334–7347. doi:10.1021/acs.jafc.2c09151.
- Naville D, Gaillard G, Julien B, Vega N, Pinteur C, Chanon S, Vidal H, Le Magueresse-Battistoni B. Chronic exposure to a pollutant mixture at low doses led to tissue-specific metabolic alterations in male mice fed standard and high-fat high-sucrose diet. Chemosphere. 2019;220:1187–1199. doi:10.1016/j.chemosphere.2018.12.177.
- Humbert L, Maubert MA, Wolf C, Duboc H, Mahé M, Farabos D, Seksik P, Mallet JM, Trugnan G, Masliah J, et al. Bile acid profiling in human biological samples: comparison of extraction procedures and application to normal and cholestatic patients. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;899:135–145. doi:10.1016/j.jchromb.2012.05.015.
- Robert C, Buisson C, Laugerette F, Abrous H, Rainteau D, Humbert L, Vande Weghe J, Meugnier E, Loizon E, Caillet F, et al. Impact of rapeseed and soy lecithin on postprandial lipid metabolism, bile acid profile, and gut bacteria in mice. Mol Nutr Food Res. 2021;65(9):2001068. doi:10.1002/mnfr.202001068.
