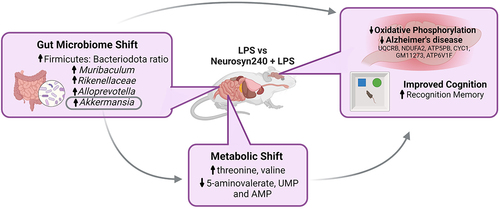
ABSTRACT
The Mediterranean diet (MD) and its bioactive constituents have been advocated for their neuroprotective properties along with their capacity to affect gut microbiota speciation and metabolism. Mediated through the gut brain axis, this modulation of the microbiota may partly contribute to the neuroprotective properties of the MD. To explore this potential interaction, we evaluated the neuroprotective properties of a novel bioactive blend (Neurosyn240) resembling the Mediterranean diet in a rodent model of chronic low-grade inflammation. Behavioral tests of cognition, brain proteomic analysis, 16S rRNA sequencing, and 1H NMR metabolomic analyses were employed to develop an understanding of the gut-brain axis interactions involved. Recognition memory, as assessed by the novel object recognition task (NOR), decreased in response to LPS insult and was restored with Neurosyn240 supplementation. Although the open field task performance did not reach significance, it correlated with NOR performance indicating an element of anxiety related to this cognitive change. Behavioral changes associated with Neurosyn240 were accompanied by a shift in the microbiota composition which included the restoration of the Firmicutes: Bacteroidota ratio and an increase in Muribaculum, Rikenellaceae Alloprevotella, and most notably Akkermansia which significantly correlated with NOR performance. Akkermansia also correlated with the metabolites 5-aminovalerate, threonine, valine, uridine monophosphate, and adenosine monophosphate, which in turn significantly correlated with NOR performance. The proteomic profile within the brain was dramatically influenced by both interventions, with KEGG analysis highlighting oxidative phosphorylation and neurodegenerative disease-related pathways to be modulated. Intriguingly, a subset of these proteomic changes simultaneously correlated with Akkermansia abundance and predominantly related to oxidative phosphorylation, perhaps alluding to a protective gut-brain axis interaction. Collectively, our results suggest that the bioactive blend Neurosyn240 conferred cognitive and microbiota resilience in response to the deleterious effects of low-grade inflammation.
1. Introduction
Mediterranean diet (MD) adherence, which consists of a proportionally higher intake of unprocessed cereals, legumes, extra virgin olive oil, fruits, nuts, and vegetables, along with moderate consumption of fish, dairy, and meat products,Citation1 has been frequently associated with brain health.Citation2–4 The characteristics of MD (as described above) lead to an increased intake of bioactive constituents (e.g., polyphenols, vitamins, and omega-3 polyunsaturated fatty acids) which have been reported to contribute to the neuroprotective effects of MD.Citation5 Inflammation and oxidative stress, which occur in the development and progression of neurodegenerative disorders, have been consistently shown to be mitigated by both MD adherence and intake of bioactive compounds,Citation6–8 providing further insight into the mechanistic underpinnings. More recently, modulation of the composition and metabolism of the gut microbiota has been posited as a potential mediator of these health-related outcomes, although evidence to date has been inconsistent.Citation9
Given the strength of evidence supporting MD in the prevention of neurodegenerative diseases (and other cardiometabolic diseases), MD is widely advocated by dietitians, the NHS, and charities alike. Despite this, the proportion of individuals adhering to MD in England is minimal.Citation10 This is likely owing to the difficulties in achieving behavioral changes, and the prohibitive cost nature of some of these dietary constituents. As such, methods (e.g., dietary supplement) of delivering these bioactive compounds in an easy and convenient manner (if similarly beneficial), would be an advantageous alternative option for individuals unable to or only partially adhering to MD.
In the present manuscript, we explore the impact of a novel blend of bioactive compound supplementation resembling the constituents associated with MD in the context of chronic low-grade inflammation. Microbiota, metabolomic and proteomic analyses were conducted in an attempt to understand the potential microbial contribution with an emphasis on metabolomic changes.
2. Results
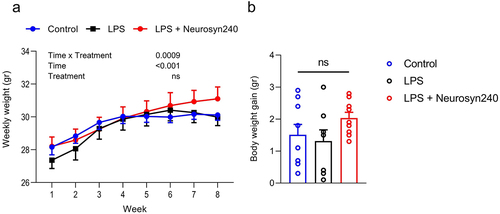
2.1. Chronic LPS treatment and subsequent Neurosyn240 intervention has minimal influence on body weight
Body weight increased over the 8-week period experiments; however, it did not significantly differ across the treatment groups (). Neither LPS nor Neurosyn240 affected the overall body weight gain (). No significant difference in mean food intake was observed over the course of the study, with mice consuming on average 3.19 ± 0.16 g per day per mouse, providing the animals with 183.23 ± 11.07 mg per kg body weight per day of Neurosyn240 (not shown). Using an allometric scaling based on body surface area,Citation11 we calculated a human equivalent dose of 893.80 ± 54.02 mg Neurosyn240 per day for a person of 60 kg.
Figure 1. Body weight in response to LPS and Neurosyn240 treatments. a) There was no significant weight difference across groups throughout the 8-week experimentation, b) Total body weight gain did not significantly differ across groups. Data are represented as the mean ± standard error of the mean (s.e.m.). n = 10 mice per group. ns, not significant. LPS: Lipopolysaccharide.
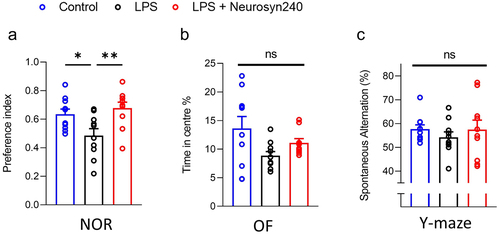
2.2. Neurosyn240 supplementation ameliorates LPS-mediated deficit in recognition memory
NOR performance was significantly different across experimental groups (p = 0.0026) with a significant 27% reduction in NOR performance observed in LPS treated animals compared to control ( p < 0.05). Interestingly, supplementation with Neurosyn240 mitigated LPS-mediated deficit in recognition memory (; p < 0.01). Travel distance and movement speed did not differ during NOR suggesting that this result was not a product of locomotion issues (Figure S2A and B). However, NOR performance significantly correlated with OF performance (Figure S2C; p = 0.0339), indicating that an element of anxiety may contribute to this finding, although both OF performance and exploration time with objects did not significantly differ across groups (Figure S2D). Indeed, OF performance did not significantly differ across experimental groups (Kruskal–Wallis test p = 0.1161; ), although a non-significant reduction was observed in the LPS group. Similarly, Y maze performance was not significantly altered by LPS treatment nor Neurosyn240 co-supplementation (; p = 0.2216).
Figure 2. Neurosyn240 improves LPS mediated reduction in cognitive performance a) nor performance significantly dropped in response to LPS treatment but was subsequently restored through Neurosyn240 supplementation. b) Although not significant, of time in center appeared to drop in response to LPS and was partially recovered by Neurosyn240 supplementation. c) Similar to OF, Y maze performance although not significant appeared to be reduced by LPS treatment with slight recovery observed through Neurosyn240 supplementation. Data are represented as the mean ± standard error of the mean (s.e.m.). n = 10 mice per group. *p < p < 0.05, **p < 0.01. ns: not significant. OF: Open field; NOR: Novel object recognition; LPS: Lipopolysaccharide.
2.3. Neurosyn240 supplementation promotes a more favorable microbial composition that correlates with nor performance
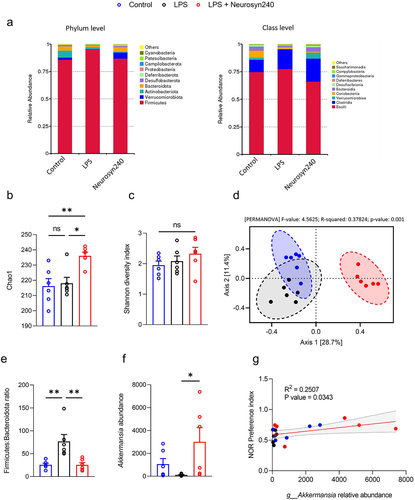
After demonstrating the beneficial effect of Neurosyn240 on cognitive behavior, we next sought to investigate whether the gut microbiota-brain axis was involved in the protective effect against chronic low-grade inflammation. The top 10 relative abundance of key OTUs showed an enrichment of Bacteroidota (class Bacteroidia) and Verrucomicrobiota (class Verrucomicrobia) and a reduction in Firmicutes (class Bacilli (76%) and Clostridia (24%)) in the Neurosyn240 treatment group (). Additionally, consumption of Neurosyn240 increased richness as assessed by Chao1 index (p < 0.05; ) but did not affect diversity (Shannon index >0.05; ). Principle Component analysis (PCA) based on OTUs revealed a significant and distinct clustering of gut microbial communities as observed between groups based on Bray-Curtis distance (PERMANOVA p < 0.05; ). Further analyses at both the phylum and genus levels revealed numerous differences between the experimental groups (Table S1). Notable changes at the phylum level included a significant increase in Verrucomicrobiota in response to Neurosyn240 supplementation (p < 0.05; Table S1) and the Neurosyn240 mediated restoration of the Firmicutes: Bacteroidota ratio which was decreased by LPS treatment (p < 0.01; ). At the genus level, in line with the aforementioned increase in Verrucomicrobiota, the abundance of Akkermansia was similarly increased by Neurosyn240 supplementation (p < 0.05; ) which correlated significantly with NOR performance (p < 0.05; ).
Figure 3. Neurosyn240 alters gut microbial profile which may influence nor performance a) Relative abundance of microbiota at the phylum (left) and class (right) levels in the indicated groups. b) Alpha diversity as analyzed using chao1 was significantly increased through Neurosyn240 treatment, c) but no significant difference was established using Shannon diversity index. d) PCA of beta diversity measured through Bray-Curtis analysis was significantly different across experimental groups. e) the Firmicutes to Bacteroidota ratio was significantly altered in response to LPS insult but was subsequently restored through Neurosyn240 treatment. f) at the genus level the Neurosyn240 intervention resulted in an increased in Akkermansia g) the abundance of Akkermansia correlated with nor performance. Data are represented as the mean ± standard error of the mean (s.e.m.). (n = 6 per group). *p < 0.05, **p < 0.01 ns: not significant.
2.4. Metabolomic profile is associated with Akkermansia abundance and nor performance
To further investigate the mechanisms underlying the protective effect of the Neurosyn240, we carried out metabolomic assessments of cecal content from Control, LPS, and LPS+Neurosyn240 mice. The metabolomic profile was distinctly different across the experimental groups as showcased by the PLS-DA plot, which depicts a clear separation of each group indicating a metabolomic shift in response to both LPS and Neurosyn240 treatments (). This is further emphasized by the volcano plot which depicts the concentration of the 19 significantly modulated metabolites (; FDR q < 0.05) A full heatmap is given in the supplementary data which displays interindividual difference/variability within each group (Figure S3). As with the microbiome analysis, we performed a Spearman correlation analysis between the significantly altered metabolites and NOR performance revealing eight metabolites (Valerate, 5-Aminovalerate, Proline, Threonine, Valine, UMP, Tartrate, and AMP) which were significantly associated with NOR performance (p < 0.05; ). Further correlation analysis this time between the significantly altered metabolites and Akkermansia identified nine significantly associated metabolites (Isovalerate, 5-Aminovalerate, Ornithine, Threonine, Valine, UMP, CMP, AMP, and Guanosine), five of which were identified in the previous analysis to correlate with NOR performance (p < 0.05; ).
Figure 4. The gut metabolomic profile is altered by LPS and premix treatment a) PLS-DA plot showed separation of groups indicative of a metabolomic shift in response to treatment. (n ≥ 5); b) Volcano plot depicting the 19 significantly altered metabolites (FDR q < 0.05). c) Correlation analysis between metabolomics data and microbiome data conducted using M2IA. *p < 0.05, **p < 0.01.
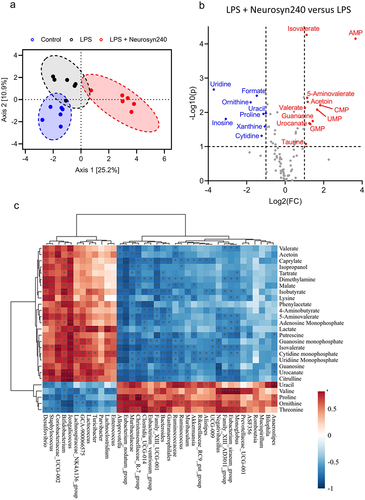
Table 1. The gut metabolomic profile associated with LPS and premix treatment correlates with nor performance and Akkermansia abundance. Metabolites in red highlight metabolites significantly associated with both nor performance and Akkermansia abundance. p-Adjusted values provided using Benjamini-Hochberg FDR correction.
2.5. Proteomic changes in the brain relate to oxidative phosphorylation, retrograde endocannabinoid signalling, and neurodegenerative disease
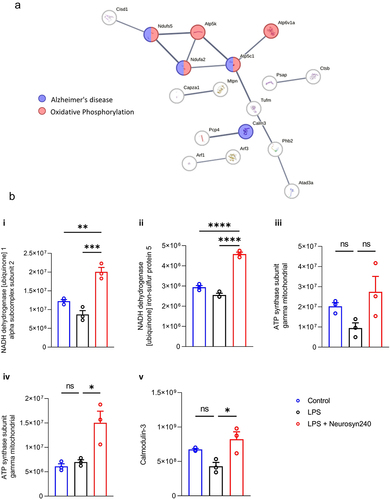
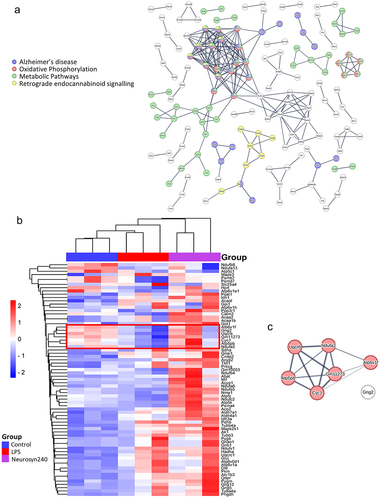
Shotgun and 2DE proteomic analyses identified a large shift in the proteomic profile of the brain in response to LPS treatment and Neurosyn240 supplementation, with shotgun identifying 314 proteins significantly altered across experimental conditions (Figure S4 and ). 2DE proteomic analysis returned 46 significant proteins which were highly aligned with the shotgun analysis, thus validating the shotgun proteomic analysis (Figure S5 and ). Analysis of protein–protein interactions was conducted using STRING with the minimum required interaction score set to 0.9 (highest confidence) (). Subsequent KEGG pathway analysis highlighted 76 pathways significantly altered in response to LPS treatment and Neurosyn240 supplementation according to shotgun proteomic analysis (p < 0.05; Table S2). Of these pathways, Parkinson’s disease and Alzheimer’s disease (along with other neurodegenerative conditions), oxidative phosphorylation, metabolic pathways, and retrograde endocannabinoid signaling emerged as highly significant pathways (p < 0.0001; and Table S2). The abundance of these proteins appeared to be predominantly upregulated in response to Neurosyn240 intervention when compared to control (). A cluster of proteins (highlighted on the heatmap) namely V-type proton ATPase subunit F (ATP6V1F), guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 (GNG2), cytochrome b-c1 complex subunit 7 (UQCRB), cytochrome c oxidase subunit 5B (GM11273), cytochrome c1 (CYC1), ATP synthase F(0) complex subunit B1 (ATP5PB), and NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 (NDUFA2) were reduced by LPS treatment but were subsequently recovered by Neurosyn240 supplementation. Six of these seven proteins were linked (STRING high confidence 0.7) and highly associated with oxidative phosphorylation (; p < 0.0001).
Figure 5. Brain proteomic profile is altered by LPS and Neurosyn240 treatment a) Protein–protein interaction analysis using STRING highlight Alzheimer’s disease, Oxidative phosphorylation, metabolic pathways and retrograde endocannabinoid signaling as highly altered pathways; b) Subsequent heatmap depicting the highlighted proteins associate with these pathways; c) Cluster of proteins that are reduced by LPS and recovered by Neurosyn240 are highly related to oxidative phosphorylation.
2.6. Akkermansia abundance correlates with proteomic changes in the brain relating to oxidative phosphorylation and Alzheimer’s disease
Having established a correlation between Akkermansia and both NOR performance and key metabolomic changes, we further explored whether Akkermansia similarly correlated with proteomic changes in the brain. Akkermansia significantly correlated with 61 proteins identified by shotgun proteomics analysis (p < 0.05; Table S3). These proteins were analyzed utilizing STRING with an interaction score set to 0.7 (high confidence) and subsequent KEGG pathway analysis, which highlighted pathways related to Alzheimer’s disease and oxidative phosphorylation (). These proteins were largely increased by Neurosyn240 supplementation ().
3. Discussion
Low-grade chronic inflammation, also known as “inflammageing,” accompanies ageing and is likely a significant factor in the progressive decline that is observed throughout the ageing process. It is driven by cell senescence, immune system dysfunction, metabolic disturbances, and microbiome dysbiosis.Citation12 Crucially, of these factors, the microbiome represents a modifiable and therefore more targetable component of the underlying process. Here, using an LPS model of chronic low-grade inflammation, we evaluated the efficacy of a novel bioactive blend resembling the Mediterranean diet (Neurosyn240) in counteracting the deleterious impacts of chronic inflammation. Utilizing behavioral tests of cognition, 16S rRNA sequencing, 1H NMR metabolomics, and shotgun proteomics analyses, we were able to determine the efficacy of Neurosyn240 supplementation and simultaneously explore potential interactions across the gut – brain axis, with an emphasis on metabolomic changes. Collectively, our results suggest that the bioactive blend conferred cognitive resilience, mitigating the impact of LPS induced chronic inflammation. These behavioral differences were associated with changes in the microbiome, with restoration of the Firmicutes: Bacteroidota ratio and some strains including Muribaculum, Rikenellaceae, Alloprevotella, and notably Akkermansia, which closely correlated with metabolic changes. The proteomic profile within the brain was dramatically influenced by both interventions, with oxidative phosphorylation emerging as an influential pathway. Intriguingly, a subset of these proteomic changes simultaneously correlated with Akkermansia abundance, perhaps alluding to a protective gut-brain axis interaction.
The reduction in NOR performance resulting from chronic low-grade inflammation was significantly ameliorated by the Neurosyn240 diet. We and others have shown recognition memory to be sensitive to both LPS administrationCitation13–17 and a pro-inflammatory profile.Citation18,Citation19 Indeed, recognition memory appears to be particularly sensitive and has been forwarded as a method of distinguishing prodromal AD.Citation20 As such, considering all the behavioral analysis, it appears that the animals were in a relatively early state of cognitive decline. Similarly, it has been reported that the Mediterranean diet,Citation21 as well as its bioactive constituents,Citation22 can mitigate inflammation, leading to improvements in cognition and recognition memory alike.Citation15,Citation23–25 It is likely that the anti-inflammatory actions of Neurosyn240 contribute to this effect, however this is difficult to establish as the intervention is a low-grade inflammation and thus sub-clinical inflammatory response as described by our group previously.Citation13 However, analysis at the gene level provides insight into this process (Supplementary Figure S6), with neurosyn240 displaying anti-inflammatory effects in both the colon and the brain. This was particularly true for Tnfa (Supplementary Figure S6C) in the brain and Il10 (Supplementary Figure S6F) in the colon. However, other genes only showed nominal changes, reiterating that this is a sub-clinical inflammatory response. Recognition memory can be to some extent influenced by anxiety, and it is therefore important to consider anxiety when determining the results of NOR. Extensively reviewed,Citation26 it is evident that the Mediterranean diet, and likely the associated bioactives, have considerable impact upon mitochondrial function. Most notably in this experimentation, an LPS mediated reduction in a cluster of proteins associated with oxidative phosphorylation (UQCRB, NDUFA2, ATP5PB, CYC1, GM11273, ATP6V1F), was prevented by Neurosyn240 supplementation. Reduced expression/abundance of these mitochondrial proteins is observed in the development of Alzheimer’s disease,Citation27 and other neurodegenerative conditions.Citation28 Indeed, UQCRB and CYC1C which encode proteins part of mitochondria complex III have been reported to have key functions in both metabolic syndrome and AD,Citation29 perhaps relating to their association with reactive oxygen species (ROS) production and/or hypoxic signaling and/or angiogenesis.Citation30–33 NDUFA2 is a subunit of NADH dehydrogenase (complex 1), which is located in the mitochondrial inner membrane, and has also been associated with AD development.Citation27,Citation34 ATP5BP encodes a subunit of mitochondrial ATP synthase, and this protein has been found to be altered in diabetic vascular dementia,Citation35 again highlighting the link between metabolic disturbances, the mitochondria and the development of neurodegenerative conditions. GM11273 or COX5B, a subunit of Complex IV, the terminal complex of the electron transfer chain, has also been linked to neurodegenerative diseaseCitation36 and mental health conditions.Citation37,Citation38 Together, these mitochondrial disturbances and subsequent recovery are likely key to the actions of the Neurosyn240 intervention.
Neurosyn240 supplementation influenced the microbiota composition, restoring the Firmicutes: Bacteroidota ratio and increasing abundance of strains such as Muribaculum, Rikenellaceae Alloprevotella, and notably Akkermansia. Intriguingly, Akkermansia abundance significantly correlated with 61 of the 314 significantly altered proteins identified by proteomic analysis, providing evidence that Akkermansia may in part contribute to the neuroprotective effects conferred by the Neurosyn240. These 61 proteins were predominantly related to oxidative phosphorylation and/or mitochondrial dysfunction (and included NDUFA2) suggesting that Akkermansia may contribute to the regulation of this pathway (mitochondrial integrity). This is in line with others who have similarly attributed the effects of bioactives such as tea polyphenols to the upregulation of Akkermansia and subsequent regulation of mitochondrial function.Citation39 It should, however, be noted that the benefits of Akkermansia in the context of cognitive health and neurodegenerative disease are inconsistent across neurological conditions (e.g., Akkermansia abundance is positively associated with Parkinson’s disease and negatively associated with AD).Citation40 Indeed, we found strong associations between pathways associated with both Alzheimer’s disease and Parkinson’s disease. However, upon evaluation of these proteins it is clear that there is considerable overlap between these two pathways, which likely explains the reason for both appearing on the KEGG analysis (and not due to alternative interactions). These results emphasize the potential implications of Akkermansia in neurological diseases and perhaps indicate the importance of balance (e.g., not too much, not too little) which should be considered in the future, particularly when developing therapeutics. As such, it may be wise to use a targeted approach in such situations restricting to those with low Akkermansia abundance.
These results also highlight a relationship that may exist between the gut microbiota and metabolomic profiles, which may in turn contribute to proteomic changes within the brain. Akkermansia was significantly increased by Neurosyn240 supplementation, suggesting that the bioactive compounds contained in the blend promote/protect Akkermansia abundance. This is in line with others who have reported similar results in response to bioactive compounds such as polyphenols, and our previous results in which comparable levels of saffron extract elicited a similar effect.Citation14 Akkermansia abundance positively correlated with NOR performance. The protective/enhancing effects of Akkermansia upon cognition have been widely reported,Citation41,Citation42 and encompass that of recognition memory.Citation43 Intriguingly, five of the significantly altered metabolites that correlated with NOR performance similarly correlated with Akkermansia abundance, offering a route (i.e., metabolic change) by which Akkermansia may confer its protective effects. Surprisingly, despite numerous reports of Akkermansia influencing metabolism in which it has been described as the “holy grail for metabolic disease”Citation44 the mechanisms underpinning these effects have not been well defined and should form the basis for future research endeavors.
Limitations and future directions: We would like to take this opportunity to discuss potential limitations and areas of future research endeavors. Firstly, we were only able to establish cognitive decline and subsequent improvement in the NOR task (recognition memory). A more extensive test battery may be warranted to tease out some of the subtle differences detected. Intriguingly, both Y maze (Spatial memory) and OF (anxiety) task performance mirrored NOR performance with 6% and 37% increases in the Neurosyn240 group compared to LPS group respectively. Although not reaching significance, collectively these results support the notion that Neurosyn240 has beneficial impacts upon cognition. Recognition memory appears to be particularly sensitive to neurological insult, considering the results in their entirety it is possible that the animals at experimental endpoint were in the early stages of cognitive decline, and a few more weeks would have resulted in significant cognitive decline in all cognitive domains tested, due to improved signal to noise ratio. Indeed, weekly dosage of 0.25 mg/kg administered for 12 weeks, 4 weeks longer than in the present study, resulted in a reduction in performance in multiple behavioral tests including NOR, Morris water maze and Y maze.Citation45 Such findings clearly indicate that the LPS dosing regimen along with the experimental set up can strongly influence behavioral test performance. The present study utilized a dose of 0.5 mg/kg body weight administered on a weekly basis for 8 weeks (selected to mimic chronic low-grade inflammation). We have previously utilized this model and described similar defects in recognition memory.Citation13 In comparison to other studies, a onetime 0.33 mg/kg administration did not detect spatial memory impairment in young mice,Citation46 and 5 days of 0.25 mg/kg treatment resulted in reduced NOR performance,Citation15 reporting on no other behavioral changes (no others measured). Administration of 0.25–0.75 mg/kg for 7 sequential days appears to consistently induce cognitive decline across a broad range of cognitive domains,Citation47–50 with reduced spatial and recognition memory reported (Morris water maze, y maze NOR) however OF performance was only quantified (and found to be influenced by LPS) by Oz et al.Citation50 Similar results were also reported in a study in which the seven injections were administered on alternative days (Water maze and Y maze),Citation51 again OF was not reported. Higher doses e.g., 2 mg/kg − 5 mg/kg have been shown to drastically impact cognition across a wide variety of behavioral tests with only one dose necessary,Citation52–55 however this does not model chronic low-grade inflammation. Extending the intervention period in models of chronic low-grade inflammation would therefore appear advantageous from a cognitive perspective to achieve significance across greater cognitive domains and should be trialed in future experiments. In addition to this, the omics analysis (given the nature of this type of analysis) was only conducted on a subset (n = 6 or n = 3 respectively) of samples. A larger N number would have greatly strengthened the validity of our results, however the level of significance achieved utilising these randomly chosen subsets is a compelling indication of the strength of this interaction. Furthermore, there are several routes of transmission by which the gut brain axis can influence the brain and subsequent cognition. In the present study we were particularly interested in the route relating to metabolite production,Citation56 and therefore remained focused upon this aspect of the gut brain axis. As a result, we did not cover other aspects of the communication system, which would have given a more comprehensive picture of the process. This should certainly be followed up in future studies to determine a more complete understanding of the gut brain axis in this process. For example, histology of the colon/intestine, e.g., morphometric analysis, would provide greater evidence relating to the immunomodulatory effect of Neurosyn240, supporting the qPCR evidence provided. Further to this, although we allude to the potential importance of Akkermansia in the neuroprotective actions of Neurosyn240 more sophisticated approaches will be necessary in the future experimentation to validate this connection. Finally, the Mediterranean dietary pattern is characterized by a high consumption of plant-based food such as fruits, vegetables, herbs, and spices, a moderate consumption of olive oil as main fat source, fish, red wine, and low consumption of red/processed meat and refined carbohydrate products.Citation57 Given that Neurosyn240 is predominantly polyphenols and vitamins it only mimics this aspect of the Mediterranean dietary pattern, we envisage future interventions to build upon this given that multifaceted approaches are likely to elicit greater neuroprotective effects.
4. Materials and methods
4.1. Study approval
All experimental procedures and protocols performed were reviewed and approved by the Animal Welfare and Ethical Review Body (AWERB) and were conducted in accordance with the specification of the United Kingdom Animal Scientific Procedures Act, 1986 (Amendment Regulations 2012). Reporting of the study outcomes complies with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelinesCitation58
4.2. Overview of experimental procedure
Thirty male C57BL/6J mice sourced from Charles River (Margate, UK) were maintained in individually ventilated cages (n = 5 per cage), within a controlled environment (21 ± 2°C; 12-h light/dark cycle; light from 7:00 AM) and fed ad libitum on a standard chow diet (RM3-P; Special Diet Services, Horley UK) up to the age of 10 weeks, ensuring normal development and stabilization of the microbiota.Citation59 Then, the mice were transferred onto one of two diets, namely control diet (e.g., AIN93-M) or AIN-93 M supplemented with Neurosyn240 (Activ’Inside, Beychac-et-Caillau, France). Neurosyn240 is a proprietary (patent pending) standardized blend of MemophenolTM, a unique formula of French Grape (Vitis vinifera L.) and North-American Wild Blueberry (Vaccinium angustifolium A.) (patent WO/2017/072219), saffron extract (patent WO/2018/020013), green tea extract, olive leaf extract, trans-resveratrol, zinc, vitamins B5, B9, B12, C, D3, E, polyphenols (mainly flavan-3-ol monomers ≥10%, resveratrol ≥1%, and others more specific such as oleuropein), crocins carotenoids (mainly trans-4-GG, trans-3-Gg; cis-4-GG, trans-2-G) ≥500 ppm and at least 15% of vitamins and mineral reference value at the concentration of 1,794 mg/kg of diet. This represents roughly 90.2% polyphenols and carotenoids rich extracts, 9.3% vitamins, and 0.4% minerals. Neurosyn240 is therefore a high polyphenol blend. Dietary fiber content of this mix is at trace amounts and therefore is unlikely to exert a physiological response. Diets were prepared by Research Diet Inc. (New Brunswick, USA) to comply with animal nutrition requirements. Chronic low-grade inflammation was induced through weekly intraperitoneal injections (i.p.) of 0.5 mg/kg body weight lipopolysaccharide (LPS; from Escherichia coli O55:B5; Sigma Aldrich, UK) for 8 weeks as described previouslyCitation60 or a SHAM injection consisting of saline (Figure S1). At the end of the experiments and following the completion of behavioral testing (see below for detail), 5-month-old animals were sedated with a mixture of isoflurane (1.5%) in nitrous oxide (70%) and oxygen (30%) and transcardially perfused with ice-cold saline containing 10 UI heparin (Sigma-Aldrich, UK). Blood samples were kept on ice for 30 min and sera were isolated via centrifugation at 10,000 × g for 5 min at room temperature. Brains were rapidly removed, halved, snap frozen, and stored at −80°C until biochemical analysis. Additionally, caeca were removed, weighed, and contents were gently extracted. Samples were then snap-frozen in liquid nitrogen and stored at −80°C until further analysis.
4.3. Behavioral assessment
All behavioral tests were performed at the experimental endpoint after the 8-week intervention as described.Citation14 Briefly, Open Field (OF) task was used as a measure of anxiety-like behavior. Animals were individually placed within the (50 cm × 50 cm × 50 cm) square arena illuminated with low 100 lux lighting and were allowed to move freely for a 10-min period. Mice were tracked using Ethovision software which determined travel distance, velocity, and time spent in the center/periphery of the maze, respectively.
The novel object recognition (NOR) task, a measure of recognition memory was performed in low 100 lux lighting. On day 1 (habituation), the animal was placed into an empty maze for 10 min. On day 2, animals were conditioned to a single object for a 10-min period. On day 3, mice were exposed to two identical objects for 15 min. Following an inter-trial interval of 1 h, mice were placed back within the testing arena now containing one familiar object and one novel object. Videos were analyzed for a 5-min period, after which if an accumulative object exploration of 8 s failed to be reached, the analysis continued for the full 10 min or until 8 s was achieved. Those not achieving 8 s of exploration were excluded from the analysis.Citation61 The discrimination index was calculated as DI = (TN − TF)/(TN + TF), where TN is the time spent exploring the novel object, and TF is the time spent exploring the familiar object.
Y maze spontaneous alternation test, a measure of spatial working memory, was performed with each animal for 7 min recording zone transitioning and locomotor activity. Spontaneous Alternation was calculated using the following formula: (Number of alternations/max number of alternations × 100).
4.4. Microbial 16S rRNA extraction and sequencing
Microbial DNA was isolated from approximately 50 mg of cecal content using the QIAamp PowerFecal Pro DNA Kit (Qiagen, Manchester, UK) from n = 6 randomly chosen animals as per the manufacturer’s instructions. DNA quantity was assessed using a Nanodrop 2000 Spectrophotometer (Fisher Scientific, UK). Quality assessment was performed using agarose gel electrophoresis to detect DNA integrity, purity, fragment size, and concentration. The 16S rRNA amplicon sequencing of the V3-V4 hypervariable region was performed with an Illumina NovaSeq 6000 PE250. Sequence analysis was performed using the Uparse software (Uparse v7.0.1001),Citation62 using all the effective tags. Sequences with ≥97% similarity were assigned to the same OTUs. A representative sequence for each OTU was screened for further annotation. For each representative sequence, Mothur software was performed against the SSUrRNA database of SILVA Database 138.Citation63 OTUs abundance information was normalized using a standard of sequence number corresponding to the sample with the least sequences. Alpha-diversity was assessed using both Chao1 and Shannon H diversity indices whilst beta diversity was assessed using Bray-Curtis. Statistical significance was determined by Kruskal–Wallis or Permutational Multivariate Analysis of Variance (PERMANOVA). Comparisons at the Phylum, Family, and Genus level were made using classical univariate analysis using Kruskal–Wallis combined with a false discovery rate (FDR) approach used to correct for multiple testing.
4.5. Citation1H NMR metabolomics
Cecal metabolites were analyzed and quantified by1H NMR analysis from the same n = 6 randomly chosen animals utilized in the 16S analysis. The preparation method was as previously described.Citation64 Briefly, frozen cecal contents were thoroughly mixed at 5,000 rpm in a Precellys 24 (Bertin Technologies, France) and diluted to a feces-to-buffer ratio of 13 (e.g., 50 mg in 750 µL) by adding a deuterated phosphate buffer (1.9 mM Na2HPO4, 8.1 mM NaH2PO4, and 1 mM sodium 3-(trimethylsilyl)-propionate-d4 in deuterated water (Goss Scientifics, Crewe, United Kingdom)). After mixing and centrifugation, 500 µl was transferred into a 5-mm NMR tube for spectral acquisition. High resolution1H NMR spectra were recorded on a 600-MHz Bruker Avance spectrometer fitted with a 5-mm TCI proton-optimized triple resonance NMR inverse cryoprobe and a 24-slot autosampler (Bruker, Rheinstetten, Germany). Sample temperature was controlled at 300 K. Each spectrum consisted of 128 scans of 65,536 complex data points with a spectral width of 20 ppm (acquisition time 2.6 s). The noesypr1d presaturation sequence was used to suppress the residual water signal with low power selective irradiation at the water frequency during the recycle delay (D1 = 2 s) and mixing time (D8 = 0.01 s). A 90° pulse length of 11.4 µs was set for all samples. Spectra were transformed with a 0.1-Hz line broadening and zero filling, manually phased, baseline corrected, and referenced by setting the trimethylsilylpropanoic acid methyl signal to 0 ppm. Metabolites were identified using information found in the literature or on the web (Human Metabolome Database, http://www.hmdb.ca/) and quantified using the software Chenomx® NMR Suite 8.6™.
4.6. Proteomic analysis
For proteomic analysis, left brain samples (about 0.2 g of tissue) from a subset n = 3 of the same utilized in the 16S and metabolomic analysis were transferred into a pre-cooled potter, resuspended in 6 volumes (w/v) of rehydration solution (7 M urea, 2 M thiourea, 4% CHAPS, 60 mM dithiothreitol, and 0.002% bromophenol blue), and added with protease and phosphatase inhibitors as reported previously.Citation65
The 2DE was carried out as previously described,Citation66 using Immobiline IPG BlueStrip (SERVA Electrophoresis, GmbH, Heidelberg, Germany), 18 cm, linear-gradient (pH 3–10) for isoelectric focusing and a 12% SDS-PAGE for the second dimension, respectively. The analysis of the images was performed using Same Spot (v4.1, TotalLab; Newcastle Upon Tyne, UK) software. The spot volume ratios among the different conditions (Control, LPS, and Neurosyn240 supplementation) were calculated using the average spot normalized volume of three biological replicates. The software included statistical analysis calculations. For proteins identification, the gel pieces were trypsin digested as previously described.Citation67 FlexAnalysis v. 3.3 was used to process the raw data and generate the peak list to be submitted to the database search using BioTools 3.2 exploiting the free version of MASCOT search engine (version 2.8.0 at http://www.matrixscience.com) against UniProt/Swiss-Pro non-redundant database version 2021–04 restricted to the Mus musculus taxonomy.
Shotgun proteomics analysis was carried out as follows. After protein quantification, a volume corresponding to 50 μg of proteins was loaded onto a Nanosep 10-kDa-cutoff filter (Pall Corporation, Michigan, USA) and digested according to our routine protocol.Citation68 Each digested protein sample was analyzed in technical triplicate by LC-MS/MS using a UltiMate3000 RSLCnano (Thermo Fisher Scientific, Waltham, MA, USA) chromatographic system coupled to an Orbitrap Fusion Tribrid mass spectrometer, operating in positive ionization mode, equipped with a nanoESI source (EASY-Spray NG) (Thermo Fisher Scientific, Waltham, MA, USA). Peptides were loaded on a PepMap100 C18 pre-column cartridge (5 µm particle size, 100 Å pore size, 300 µm i.d. ×5 mm length, Thermo Fisher Scientific, Waltham, MA, USA) and subsequently separated on an EASY-Spray PepMap RSLC C18 column (2 µm particle size, 100 Å pore size, 75 µm i.d. ×15 cm length, Thermo Fisher Scientific, Waltham, MA, USA) at a flow rate of 300 nL/min and a temperature of 38°C, by a one-step linear gradient from 95% eluent A (0.1% FA in water) to 25% eluent B (99.9% ACN, 0.1% FA) in 113 min and total LC run of 120 min. Precursor (MS1) survey scans were recorded in the Orbitrap, at resolving powers of 120 K (at m/z 200). Data-dependent MS/MS (MS2) analysis was performed in top speed mode with a 3 s cycle time, during which the most abundant multiple-charged (2 ± 7+) precursor ions detected within the range of 375–1500 m/z were selected for activation in the order of abundance and detected in an ion trap at a rapid scan rate. Quadrupole isolation with a 1.6 m/z isolation window was used, and dynamic exclusion was enabled for 60 s after a single scan. Automatic gain control targets were 4.0 × 105 for MS1 and 2.0 × 103 for MS2, with 50 and 300 ms maximum injection times, respectively. For MS2, the signal intensity threshold was 5.0 × 103, and the option “Injection Ions for All Available Parallelizable Time” was set. High-energy collisional dissociation (HCD) was performed using 30% normalized collision energy.
Raw data were processed using PEAKS studio XproCitation69 (Bioinformatics Solutions Inc., Waterloo, Ontario, Canada) using the ‘correct precursor only’ option and the filter charge set 2 to 8. Spectra were matched against the UniProt SwissProt database restricted to Mammalia taxonomy, to which a list of common contaminants was appended (67,666 entries). False discovery rate (FDR) was set to 0.5% at the peptide-spectrum matches (PSM) level. The post-translational modification (PTM) profile was set as follows: fixed cysteine carbamidomethylation (ΔMass: 57.02), variable methionine oxidation (ΔMass: 15.99). Non-specific cleavage was allowed to one end of the peptides, with a maximum of two missed cleavages and Trypsin enzyme specificity. The highest error mass tolerances for precursors and fragments were set at 10 ppm and 0.5 Da, respectively. After processing every single raw data, the label-free quantification (LFQ) tool of PEAKS Studio was used to detect differentially expressed proteins. Parameters for LFQ were set as follows. Quantification type: Label-free quantification; Mass Error Tolerance: 10.0 ppm; Retention Time Shift Tolerance: Auto; FDR Threshold: 0.5%. For quantitative analysis, the significance threshold at the protein level was set to ≥20–10 logP with a fold change ≥2.0.
Protein – protein interactions and pathway analysis were conducted by uploading the names of significantly altered proteins (as determined by proteomic analysis) into String (https://string-db.org/) for PPI network construction.Citation70 The species was set as “Mus musculus,” and other parameters were set as default with the exception of meaning of network edges: which was changed to depict the strength of the interaction (reflected by the thickness of the line), network display options: non-interacting proteins were removed from the figures and Confidence threshold: Confidence threshold used for each string analysis is given in text. Node color depicts pathways of interest (i.e., those with the highest significance).
4.7. RNA isolation and qRT-PCR
RNA isolation, cDNA synthesis, and qRT-PCR were carried out as previously described.Citation14 Briefly, total RNA was isolated from the brain samples using the Qiazol reagent (Qiagen, UK). One μg of total RNA was treated with DNase I (Invitrogen, UK) and used for cDNA synthesis using Invitrogen Oligo (dT) primers and M-MMLV reverse transcriptase. Quantitative real-time PCR (qRT-PCR) reactions were performed using SYBR green detection technology on the Roche light cycler 480 (Roche Life Science, UK). Results are expressed as relative quantity scaled to the average across all samples per target gene and normalized to the reference gene glyceraldehyde 3-phosphate dehydrogenase (Gapdh), which was identified as the optimal housekeeping selection using the software RefFinder.Citation71 Primer sequences of the inflammatory markers and housekeeping are given in the supplementary data (Table S4).
4.8. Statistical analysis
Data analysis was performed in GraphPad Prism version 8 (GraphPad Software, CA, USA). All data are presented as the mean ± standard error of the mean (s.e.m.) unless otherwise stated. After identifying outliers using the ROUT method (q = 1%), data were checked for normality/equal variances using Shapiro–Wilk test. For the analysis of dietary intervention, an ANOVA, or Kruskal Wallis test was used, followed by Tukey or Dunns’s multiple comparison depending on the normality of data. For correlation analysis, Pearson correlation coefficient was utilized (unless stated otherwise). p values of less than 0.05 were considered statistically significant.
Statistical analysis of metabolomics data was carried out using Metaboanalyst 5.0.Citation72 Data were normalized by sum, scaled by autoscaling, and square root-transformed. Partial Least-Squares Discriminant Analysis (PLS-DA) was employed to illustrate the clustering of different metabolites across groups. Univariate analysis was carried out using one-way ANOVA, followed by Tukey HSD. Dendrogram and heatmaps were created with Spearman and Ward. Heatmap shows the significant metabolites based upon ANOVA results.
Correlation analysis between metabolomics data and microbiome data was conducted using M2IA.Citation73 Missing values were filtered if present in more than 80% of samples or the relative standard deviation was smaller than 30%.Citation74 The remaining missing data values were handled using Random Forest. Data were normalized using total sum scaling. All other correlation analyses were conducted using Spearman’s rank-order correlation analysis.Citation75 Exploratory correlation analyses were not corrected for multiple testing unless otherwise stated. When corrected, Benjamini-Hochberg FDR correction was utilized.
Authors contribution
D.V. conceptualized and designed the experiments and analytical approaches; D.V. provided the Home Office Animal License; M.G.P. performed the animal experiments, the cognitive testing, the microbiome analysis, and analyzed the data; E.C. and G.L.G. performed the metabolomic analyses. L.G., L.Z., and M.R. performed the brain proteomic analyses; L.L. performed the RT-qPCR; L.P. and D.G. provided the Neurosyn240 extract. M.G.P. and D.V. wrote the manuscript with contributions from all the coauthors; M.M. and C.A. critically revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Availability of data and materials
The 16S rRNA gene sequence data have been deposited in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA1062272. Other original data will be made available upon request.
Supplemental Material
Download Zip (742.6 KB)Disclosure statement
DV received funding from Activ’Inside. L.P, D.G. work for Activ’Inside and provided the Neurosyn240 extract. Activ’Inside was not involved in the design, implementation, analysis, and interpretation of the data. All the other authors have no conflict of interest to declare.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2024.2363011
Additional information
Funding
References
- Sikalidis AK, Kelleher AH, Kristo AS. Mediterranean Diet. Encyclopedia. 2021;1(2):371–19. doi:10.3390/encyclopedia1020031.
- Gardener H, Caunca MR. Mediterranean Diet in Preventing Neurodegenerative Diseases. Curr Nutr Rep. 2018;7(1):10–20. doi:10.1007/s13668-018-0222-5.
- Radd-Vagenas S, Duffy SL, Naismith SL, Brew BJ, Flood VM, Fiatarone Singh MA. Effect of the Mediterranean diet on cognition and brain morphology and function: a systematic review of randomized controlled trials. Am J Clin Nutr. 2018;107(3):389–404. doi:10.1093/ajcn/nqx070.
- Sánchez-Villegas A, Galbete C, Martinez-González MA, Martinez JA, Razquin C, Salas-Salvadó J, Estruch R, Buil-Cosiales P, Martí A. The effect of the Mediterranean diet on plasma brain-derived neurotrophic factor (BDNF) levels: the PREDIMED-NAVARRA randomized trial. Nutr Neurosci. 2011;14(5):195–201. doi:10.1179/1476830511Y.0000000011.
- Franco GA, Interdonato L, Cordaro M, Cuzzocrea S, Di Paola R. Bioactive compounds of the Mediterranean diet as nutritional support to fight neurodegenerative disease. Int J Mol Sci. 2023;24(8):24. doi:10.3390/ijms24087318.
- De Marchi F, Vignaroli F, Mazzini L, Comi C, Tondo G. New insights into the relationship between nutrition and neuroinflammation in Alzheimer’s disease: Preventive and therapeutic perspectives. CNS Neurol Disord Drug Targets. 2023;23(5):614–627. doi:10.2174/1871527322666230608110201.
- Mathur R, Ahmid Z, Ashor AW, Shannon O, Stephan BCM, Siervo M. Effects of dietary-based weight loss interventions on biomarkers of endothelial function: a systematic review and meta-analysis. Eur J Clin Nutr. 2023;77(10):927–940. doi:10.1038/s41430-023-01307-6.
- Shannon OM, Ranson JM, Gregory S, Macpherson H, Milte C, Lentjes M, Mulligan A, McEvoy C, Griffiths A, Matu J. et al. Mediterranean diet adherence is associated with lower dementia risk, independent of genetic predisposition: findings from the UK biobank prospective cohort study. BMC Med. 2023;21(1):81. doi:10.1186/s12916-023-02772-3.
- Kimble R, Gouinguenet P, Ashor A, Stewart C, Deighton K, Matu J, Griffiths A, Malcomson FC, Joel A, Houghton D. et al. Effects of a Mediterranean diet on the gut microbiota and microbial metabolites: A systematic review of randomized controlled trials and observational studies. Crit Rev Food Sci Nutr. 2022;63(27):8698–8719. doi:10.1080/10408398.2022.2057416.
- Papadaki A, Wood L, Sebire SJ, Jago R. Adherence to the Mediterranean diet among employees in South West England: Formative research to inform a web-based, work-place nutrition intervention. Prev Med Rep. 2015;2:223–228. doi:10.1016/j.pmedr.2015.03.009.
- Nair A, Jacob S. A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm. 2016;7(2):27. doi:10.4103/0976-0105.177703.
- Saavedra D, Añé-Kourí AL, Barzilai N, Caruso C, Cho KH, Fontana L, Franceschi C, Frasca D, Ledón N, Niedernhofer LJ. et al. Aging and chronic inflammation: highlights from a multidisciplinary workshop. Immun Ageing. 2023;20(1):25. doi:10.1186/s12979-023-00352-w.
- Hoyles L, Pontifex MG, Rodriguez-Ramiro I, Anis-Alavi MA, Jelane KS, Snelling T, Solito E, Fonseca S, Carvalho AL, Carding SR. et al. Regulation of blood–brain barrier integrity by microbiome-associated methylamines and cognition by trimethylamine N-oxide. Microbiome. 2021;9(1):235. doi:10.1186/s40168-021-01181-z.
- Pontifex MG, Connell E, Le Gall G, Pourtau L, Gaudout D, Angeloni C, Zallocco L, Ronci M, Giusti L, Müller M. et al. Saffron extract (Safr’inside™) improves anxiety related behaviour in a mouse model of low-grade inflammation through the modulation of the microbiota and gut derived metabolites. Food Funct. 2022;13(23):12219–12233. doi:10.1039/D2FO02739A.
- Di Paolo M, Corsi F, Cerri C, Bisti S, Piano I, Gargini C. A window to the brain: The Retina to monitor the progression and efficacy of saffron repron® Pre-Treatment in an LPS model of neuroinflammation and memory impairment. Pharmaceuticals (Basel). 2023;16(9):1307. doi:10.3390/ph16091307.
- Zheng YR, Tufvesson-Alm M, Trepci A, Imbeault S, Li XQ, Schwieler L, Engberg G, Erhardt S. Dual administration of lipopolysaccharide induces behavioural changes in rats relevant to psychotic disorders. Acta Neuropsychiatr. 2023; 1–13. doi:10.1017/neu.2023.40.
- Khodaei S, Wang DS, Ariza A, Syed RM, Orser BA. The impact of inflammation and general anesthesia on memory and executive function in mice. Anesth Analg. 2023;136(5):999–1011. doi:10.1213/ANE.0000000000006221.
- Schaeffer JD, Newell C, Spann C, Siemens G, Liegey Dougall A. Inflammation, depression, and anxiety related to recognition memory in young adults. J Gen Psychol. 2023;150(1):1–25. doi:10.1080/00221309.2021.1893638.
- Xie W, Kumar S, Kakon SH, Haque R, Petri WA, Nelson CA. Chronic inflammation is associated with neural responses to faces in bangladeshi children. Neuroimage. 2019;202:116110. doi:10.1016/j.neuroimage.2019.116110.
- Goldstein FC, Loring DW, Thomas T, Saleh S, Hajjar I. Recognition memory performance as a cognitive marker of prodromal alzheimer’s disease. J Alzheimer’s Dis. 2019;72(2):507–514. doi:10.3233/JAD-190468.
- Divella R, Marino G, Infusino S, Lanotte L, Gadaleta-Caldarola G, Gadaleta-Caldarola G. The Mediterranean Lifestyle to Contrast Low-Grade Inflammation Behavior in Cancer. Nutrients. 2023;15(7):15. doi:10.3390/nu15071667.
- Rivera Rodríguez R, Jj J. Terpenes: Modulating anti-inflammatory signaling in inflammatory bowel disease. Pharmacol. Ther. 2023;248:108456. doi:10.1016/j.pharmthera.2023.108456.
- Devranis P, Vassilopoulou Ε, Tsironis V, Sotiriadis PM, Chourdakis M, Aivaliotis M, Tsolaki M. Mediterranean diet, ketogenic diet or MIND diet for aging populations with cognitive decline: a systematic review. Life (Basel). 2023;13(1):173. doi:10.3390/life13010173.
- Arjmand G, Abbas-Zadeh M, Eftekhari MH. Effect of MIND diet intervention on cognitive performance and brain structure in healthy obese women: a randomized controlled trial. Sci Rep. 2022;12(1):2871. doi:10.1038/s41598-021-04258-9.
- Mattei J, Bigornia SJ, Sotos-Prieto M, Scott T, Gao X, Tucker KL. The Mediterranean diet and 2-year change in cognitive function by status of type 2 diabetes and glycemic control. Diabetes Care. 2019;42(8):1372–1379. doi:10.2337/dc19-0130.
- Pollicino F, Veronese N, Dominguez LJ, Barbagallo M. Mediterranean diet and mitochondria: new findings. Exp Gerontol. 2023;176:112165. doi:10.1016/j.exger.2023.112165.
- Armand-Ugon M, Ansoleaga B, Berjaoui S, Ferrer I. Reduced mitochondrial activity is early and steady in the entorhinal cortex but it is mainly unmodified in the frontal cortex in alzheimer’s disease. Curr Alzheimer Res. 2017;14(12):1327–1334. doi:10.2174/1567205014666170505095921.
- Garcia-Esparcia P, Koneti A, Rodríguez-Oroz MC, Gago B, Del Rio JA, Ferrer I. Mitochondrial activity in the frontal cortex area 8 and angular gyrus in Parkinsons disease and Parkinsons disease with dementia. Brain Pathol. 2018;28(1):43–57. doi:10.1111/bpa.12474.
- Li J, Zhang Y, Lu T, Liang R, Wu Z, Liu M, Qin L, Chen H, Yan X, Deng S. et al. Identification of diagnostic genes for both Alzheimer’s disease and metabolic syndrome by the machine learning algorithm. Front Immunol. 2022;13:1037318. doi:10.3389/fimmu.2022.1037318.
- Monti E, Mancini A, Marras E, Gariboldi MB. Targeting Mitochondrial ROS production to reverse the epithelial-mesenchymal transition in breast cancer cells. Curr Issues Mol Biol. 2022;44(11):5277–5293. doi:10.3390/cimb44110359.
- Xu X, Liu Y, Luan J, Liu R, Wang Y, Liu Y, Xu A, Zhou B, Han F, Shang W. et al. Effect of downregulated citrate synthase on oxidative phosphorylation signaling pathway in HEI-OC1 cells. Proteome Sci. 2022;20(1):14. doi:10.1186/s12953-022-00196-0.
- Chang J, Jung HJ, Jeong SH, Kim HK, Han J, Kwon HJ. A mutation in the mitochondrial protein UQCRB promotes angiogenesis through the generation of mitochondrial reactive oxygen species. Biochem Biophys Res Commun. 2014;455(3–4):290–297. doi:10.1016/j.bbrc.2014.11.005.
- Yu Z, Zhang Y, Liu N, Yuan J, Lin L, Zhuge Q, Xiao J, Wang X. Roles of neuroglobin binding to mitochondrial complex III subunit cytochrome c1 in oxygen-glucose deprivation-induced neurotoxicity in primary neurons. Mol Neurobiol. 2016;53(5):3249–3257. doi:10.1007/s12035-015-9273-4.
- Zheng J, Xu M, Walker V, Yuan J, Korologou-Linden R, Robinson J, Huang P, Burgess S, Au Yeung SL, Luo S. et al. Evaluating the efficacy and mechanism of metformin targets on reducing Alzheimer’s disease risk in the general population: a Mendelian randomisation study. Diabetologia. 2022;65(10):1664–1675. doi:10.1007/s00125-022-05743-0.
- Chen R, Yi Y, Xiao W, Zhong B, Shu Y, Zhang L, Zeng Y. Label-free liquid chromatography–mass spectrometry proteomic analysis of urinary identification in diabetic vascular dementia in a Han Chinese population. Front Aging Neurosci. 2021;13:619945. doi:10.3389/fnagi.2021.619945.
- Yu H, Wang D, Zou L, Zhang Z, Xu H, Zhu F, Ren X, Xu B, Yuan J, Liu J. et al. Proteomic alterations of brain subcellular organelles caused by low-dose copper exposure: implication for Alzheimer’s disease. Arch Toxicol. 2018;92(4):1363–1382. doi:10.1007/s00204-018-2163-6.
- Qi YJ, Lu YR, Shi LG, Demmers JAA, Bezstarosti K, Rijkers E, Balesar R, Swaab D, Bao A-M. Distinct proteomic profiles in prefrontal subareas of elderly major depressive disorder and bipolar disorder patients. Transl Psychiatry. 2022;12(1):275. doi:10.1038/s41398-022-02040-7.
- Henningsen K, Palmfeldt J, Christiansen S, Baiges I, Bak S, Jensen ON, Gregersen N, Wiborg O. Candidate hippocampal biomarkers of susceptibility and resilience to stress in a rat model of depression. Molecular & Cellular Proteomics: MCP. 2012;11(7):M111.016428. doi:10.1074/mcp.M111.016428.
- Wei K, Wei Y, Wang Y, Wei X. Amelioration effects and regulatory mechanisms of different tea active ingredients on DSS-Induced colitis. J Agric Food Chem. 2023;71(44):16604–16617. doi:10.1021/acs.jafc.3c04524.
- Lei W, Cheng Y, Gao J, Liu X, Shao L, Kong Q, Zheng N, Ling Z, Hu W. Akkermansia muciniphila in neuropsychiatric disorders: friend or foe? Front Cell Infect Microbiol. 2023;13:1224155. doi:10.3389/fcimb.2023.1224155.
- Huang F, Marungruang N, Martinsson I, Camprubí Ferrer L, Nguyen TD, Gondo TF, Karlsson EN, Deierborg T, Öste R, Heyman-Lindén L. et al. A mixture of Nordic berries improves cognitive function, metabolic function and alters the gut microbiota in C57Bl/6J male mice. Front Nutr. 2023;10:1257472. doi:10.3389/fnut.2023.1257472.
- Xu R, Zhang Y, Chen S, Zeng Y, Fu X, Chen T, Luo S, Zhang X. The role of the probiotic akkermansia muciniphila in brain functions: insights underpinning therapeutic potential. Crit Rev Microbiol. 2023;49(2):151–176. doi:10.1080/1040841X.2022.2044286.
- Higarza SG, Arboleya S, Arias JL, Gueimonde M, Arias N. Akkermansia muciniphila and environmental enrichment reverse cognitive impairment associated with high-fat high-cholesterol consumption in rats. Gut Microbes. 2021;13(1):1–20. doi:10.1080/19490976.2021.1880240.
- Yan J, Sheng L, Li H. Akkermansia muciniphila: is it the holy grail for ameliorating metabolic diseases? Gut Microbes. 2021;13(1). doi:10.1080/19490976.2021.1984104.
- Ullah R, Ali G, Baseer A, Irum Khan S, Akram M, Khan S, Ahmad N, Farooq U, Kanwal Nawaz N, Shaheen S. et al. Tannic acid inhibits lipopolysaccharide-induced cognitive impairment in adult mice by targeting multiple pathological features. Int Immunopharmacol. 2022;110:108970. doi:10.1016/j.intimp.2022.108970.
- Lee Y, Ju X, Cui J, Zhang T, Hong B, Kim YH, Ko Y, Park J, Choi CH, Heo JY. et al. Mitochondrial dysfunction precedes hippocampal IL-1β transcription and cognitive impairments after low-dose lipopolysaccharide injection in aged mice. Heliyon. 2024;10(7):e28974. doi:10.1016/j.heliyon.2024.e28974.
- Liang Y, Kang X, Zhang H, Xu H, Wu X. Knockdown and inhibition of hippocampal GPR17 attenuates lipopolysaccharide-induced cognitive impairment in mice. J Neuroinflammation. 2023;20(1):271. doi:10.1186/s12974-023-02958-9.
- Wu Y, Yuan Q, Ma Y, Zhou X, Wang G, Wang S, Li S, Shi J, Wang D. Dietary intervention with the gut microbial metabolite urolithin a attenuates lipopolysaccharide-induced neuroinflamm and cognitive deficits via the Sirt1/acetyl-NF-κB signaling pathway. Mol Nutr Food Res. 2023;67(13):e2200401. doi:10.1002/mnfr.202200401.
- Zeini S, Davoodian N, Kazemi H, Shareghi Brojeni M, Ghani E, Arab Firouzjaei M, Atashabparvar A. Resveratrol prevents cognitive impairment and hippocampal inflammatory response induced by lipopolysaccharide in a mouse model of chronic neuroinflammation. Physiol Behav. 2024;278:114508. doi:10.1016/j.physbeh.2024.114508.
- Öz M, Erdal H. A TNF-α inhibitor abolishes sepsis-induced cognitive impairment in mice by modulating acetylcholine and nitric oxide homeostasis, BDNF release, and neuroinflammation. Behav Brain Res. 2024;466:114995. doi:10.1016/j.bbr.2024.114995.
- Muhammad T, Ikram M, Ullah R, Rehman SU, Kim MO. Hesperetin, a citrus flavonoid, attenuates LPS-Induced neuroinflammation, apoptosis and memory impairments by modulating TLR4/NF-κB signaling. Nutrients. 2019;11(3):11. doi:10.3390/nu11030648.
- Qiu F, Liu Y, Liu Y, Zhao Z, Zhou L, Chen P, Du Y, Wang Y, Sun H, Zeng C. et al. CD137L inhibition ameliorates hippocampal neuroinflammation and behavioral deficits in a mouse model of sepsis-associated encephalopathy. Neuromolecular Med. 2023;25(4):616–631. doi:10.1007/s12017-023-08764-z.
- Jiang C, Caskurlu A, Ganesh T, Dingledine R. Inhibition of the prostaglandin EP2 receptor prevents long-term cognitive impairment in a model of systemic inflammation. Brain Behav Immun Health. 2020;8:100132. doi:10.1016/j.bbih.2020.100132.
- Liu L, Kelly MG, Yang XR, Fernandez TG, Wierzbicki EL, Skrobach A, Doré S. Nrf2 deficiency exacerbates cognitive impairment and reactive microgliosis in a lipopolysaccharide-induced neuroinflammatory mouse model. Cell Mol Neurobiol. 2020;40(7):1185–1197. doi:10.1007/s10571-020-00807-4.
- Huang WY, Liu KH, Lin S, Chen TY, Tseng CY, Chen HY, Wu HM, Hsu KS. NADPH oxidase 2 as a potential therapeutic target for protection against cognitive deficits following systemic inflammation in mice. Brain Behav Immun. 2020;84:242–252. doi:10.1016/j.bbi.2019.12.006.
- Connell E, Le Gall G, Pontifex MG, Sami S, Cryan JF, Clarke G, Müller M, Vauzour D. Microbial-derived metabolites as a risk factor of age-related cognitive decline and dementia. Mol Neurodegener. 2022;17(1):43. doi:10.1186/s13024-022-00548-6.
- Davis C, Bryan J, Hodgson J, Murphy K. Definition of the Mediterranean diet; a literature review. Nutrients. 2015;7(11):9139–9153. doi:10.3390/nu7115459.
- Boutron I, Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Browne WJ, Clark A, Cuthill IC, Dirnagl U. et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLOS Biol. 2020;18(7):18. doi:10.1371/journal.pbio.3000410.
- Laukens D, Brinkman BM, Raes J, De Vos M, Vandenabeele P, Normark BH. Heterogeneity of the gut microbiome in mice: guidelines for optimizing experimental design. FEMS Microbiol Rev. 2016;40(1):117–132. doi:10.1093/femsre/fuv036.
- Hoyles L, Pontifex MG, Rodriguez-Ramiro I, Anis-Alavi MA, Jelane KS, Snelling T, Solito E, Fonseca S, Carvalho AL, Carding SR. et al. Regulation of blood–brain barrier integrity by microbiome-associated methylamines and cognition by trimethylamine N-oxide. Microbiome. 2021;9(1). doi:10.1186/s40168-021-01181-z.
- Denninger JK, Smith BM, Kirby ED. Novel object recognition and object location behavioral testing in mice on a budget. J Vis Exp. 2018;(141). doi:10.3791/58593-v.
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi:10.1128/AEM.00062-07.
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41(D1):D590–D6. doi:10.1093/nar/gks1219.
- Tran TTT, Corsini S, Kellingray L, Hegarty C, Le Gall G, Narbad A, Müller M, Tejera N, O’Toole PW, Minihane A-M. et al. APOE genotype influences the gut microbiome structure and function in humans and mice: relevance for Alzheimer’s disease pathophysiology. FASEB J. 2019;33(7):8221–8231. doi:10.1096/fj.201900071R.
- Pirone A, Ciregia F, Lazzarini G, Miragliotta V, Ronci M, Zuccarini M, Zallocco L, Beghelli D, Mazzoni MR, Lucacchini A. et al. Proteomic profiling reveals specific molecular hallmarks of the pig claustrum. Mol Neurobiol. 2023;60(8):4336–4358. doi:10.1007/s12035-023-03347-2.
- Barbalace MC, Zallocco L, Beghelli D, Ronci M, Scortichini S, Digiacomo M, Macchia M, Mazzoni MR, Fiorini D, Lucacchini A. et al. Antioxidant and neuroprotective activity of extra virgin olive oil extracts obtained from quercetano cultivar trees grown in different areas of the Tuscany region (Italy). Antioxidants. 2021;10(3):10. doi:10.3390/antiox10030421.
- Giusti L, Angeloni C, Barbalace M, Lacerenza S, Ciregia F, Ronci M, Urbani A, Manera C, Digiacomo M, Macchia M. et al. A proteomic approach to uncover neuroprotective mechanisms of oleocanthal against oxidative stress. Int J Mol Sci. 2018;19(8):2329. doi:10.3390/ijms19082329.
- Lanas A, Tacconelli S, Contursi A, Piazuelo E, Bruno A, Ronci M, Marcone S, Dovizio M, Sopeña F, Falcone L. et al. Biomarkers of response to low-dose aspirin in familial adenomatous polyposis patients. Cancers. 2023;15(9):15. doi:10.3390/cancers15092457.
- Xin L, Qiao R, Chen X, Tran H, Pan S, Rabinoviz S, Bian H, He X, Morse B, Shan B. et al. A streamlined platform for analyzing tera-scale DDA and DIA mass spectrometry data enables highly sensitive immunopeptidomics. Nat Commun. 2022;13(1). doi:10.1038/s41467-022-30867-7.
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D13. doi:10.1093/nar/gky1131.
- Xie F, Wang J, Zhang B. RefFinder: a web-based tool for comprehensively analyzing and identifying reference genes. Funct Integr Genomics. 2023;23(2):23. doi:10.1007/s10142-023-01055-7.
- Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques P-É, Li S, Xia J. et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49(W1):W388–W96. doi:10.1093/nar/gkab382.
- Ni Y, Yu G, Chen H, Deng Y, Wells PM, Steves CJ, Ju F, Fu J. M2IA: a web server for microbiome and metabolome integrative analysis. Bioinformatics. 2020;36(11):3493–3498. doi:10.1093/bioinformatics/btaa188.
- Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45(W1):W180–w8. doi:10.1093/nar/gkx295.
- You Y, Liang D, Wei R, Li M, Li Y, Wang J, Wang X, Zheng X, Jia W, Chen T. et al. Evaluation of metabolite-microbe correlation detection methods. Anal Biochem. 2019;567:106–111. doi:10.1016/j.ab.2018.12.008.