 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Pre-mRNA splicing is essential for the regulation of gene expression in eukaryotes and is fundamental in development and cancer, and involves the selection of a consensus sequence that defines the 5′ splice site (5′SS). Human introns harbor multiple sequences that conform to the 5′SS consensus, which are not used under normal growth conditions. Under heat shock conditions, splicing at such intronic latent 5′SSs occurred in thousands of human transcripts, resulting in pre-maturely terminated aberrant proteins. Here we performed a survey of the C. elegans genome, showing that worm's introns contain latent 5′SSs, whose use for splicing would have resulted in pre-maturely terminated mRNAs. Splicing at these latent 5′SSs could not be detected under normal growth conditions, while heat shock activated latent splicing in a number of tested C. elegans transcripts. Two scenarios could account for the lack of latent splicing under normal growth conditions (i) Splicing at latent 5′SSs do occur, but the nonsense mRNAs thus formed are rapidly and efficiently degraded (e.g. by NMD); and (ii) Splicing events at intronic latent 5′SSs are suppressed. Here we support the second scenario, because, nematode smg mutants that are devoid of NMD-essential factors, did not show latent splicing under normal growth conditions. Hence, these experiments together with our previous experiments in mammalian cells, indicate the existence of a nuclear quality control mechanism, termed Suppression Of Splicing (SOS), which discriminates between latent and authentic 5′SSs in an open reading frame dependent manner, and allows splicing only at the latter. Our results show that SOS is an evolutionary conserved mechanism, probably shared by most eukaryotes.
Abbreviations
| 5′ | = | SS |
| 5′ | = | splice site |
| NMD | = | nonsense mediated decay |
| PTC | = | premature termination codon |
| SOS | = | suppression of splicing |
Introduction
The accuracy of pre-mRNA splicing is attributed to a number of trans-acting factors as well as to cis-acting sequence elements. The latter include 5′ and 3′ splice sites (SSs), a branch point, a polypyrimidine tract and exonic and intronic enhancers and silencers.Citation1-3 Most pre-mRNAs undergo alternative splicing, in which different combinations of exons encoded in a single gene are spliced to produce different mRNAs encoding functionally diverse proteins.Citation4-7 Thus, alternative splicing is a major contributor to the versatility of the proteome. Both alternative and constitutive splicing operate through the combinatorial interplay of positive and negative regulatory signals present in the pre-mRNA (e.g., splicing enhancers and silencers) that are recognized by trans-acting factors, the most studied of which are members of the hnRNP and SR protein families.Citation8-10
A key step in both constitutive and alternative pre-mRNA splicing involves the recognition and selection of a consensus sequence AG/GTRAGT that defines the 5′SS (in mammals, where R denotes purine and “/” denotes the splice junction).Citation1 Based on alignment of EST data bases, splicing involving alternative 5′SSs account to only ∼8% of the alternative splicing events that are conserved between the human and the mouse genomes.Citation11 However, this fraction is much smaller than that expected from sequence alone – as reflected in a survey of a database of human genes, demonstrating that the abundance in bona fide introns of 5′SS consensus sequences that are not involved in splicing (hence termed latent 5′SSs), exceeds the number of authentic 5′SSs by a factor of 6–9.Citation12,13 The survey also revealed that more then 95% of these intronic latent 5′SSs are preceded by at least one stop codon in the reading frame of the upstream exonic sequence. Because premature termination codons (PTC)-containing transcripts are considered toxic to cells, as their translation to truncated proteins may cause loss of function or exert dominant negative effects, the above bioinformatics findings invoked a nuclear quality control mechanism, termed stop codon-mediated suppression of splicing (SOS). This mechanism was proposed to suppress the use of latent 5′SSs, that otherwise would generate transcripts with PTCs, thereby preventing their potential deleterious effects. Support for a nuclear recognition of a PTC harboring pre-mRNA and suppression of splicing from such transcripts was observed in a number of studies,Citation14-16 including a more recent study that showed nuclear retention of unspliced PTC harboring transcripts at transcription sites.Citation17
The possibility that the apparent lack of latent mRNAs (namely, mRNAs resulting from splicing employing intronic latent 5′SSs having an upstream in-frame stop codon), could have been attributed to the degradation of such mRNAs by the nonsense mediated mRNA decay (NMD) Citation18-22 was also considered and experimentally tested. It was thus shown that SOS was not affected (i.e. latent splicing was not elicited in the tested gene transcripts) upon abrogation or bypassing the NMD pathway by a number of ways: Inhibition of translationCitation23-25 and, in particular, inhibition of the pioneer round of translation [which had been shown to be essential for NMDCitation26] by expressing a dominant negative mutant of the translation initiation factor eIF2-α, did not elicit latent splicing.Citation25 Furthermore, RNAi of the NMD genes hUpf1 and hUpf2, or the expression of 3 mutants of hUpf1 that abrogated NMD,Citation27 did not elicit latent splicing.Citation24 Because mammalian NMD and all its known branches appear to converge at a common requirement for Upf1,Citation28-30 the results cited above show that the apparent suppression of latent splicing could not be attributed to NMD. We also ruled out degradation of latent mRNA by a yet unknown RNA degradation mechanism by showing that constructs in which we forced formation of latent mRNA, through its expression from a plasmid harboring the already spliced variant, express latent mRNA at levels only slightly lower than the level of authentic mRNA expressed from a plasmid harboring the already spliced DNA at the authentic 5′SS.Citation31
Latent splicing can be activated by removal of in frame stop codons between the latent and the upstream authentic 5′SSCitation23,24 and by mutating the initiation codon sequence.Citation25,31 In accordance with these findings, SOS requires the initiator-tRNA,Citation31 which acts in a manner that is independent of its role in protein translation, as an SOS trans-factor that marks the AUG to establish a reading frame. Another way for activating latent splicing is by heat shock, as was shown for a number of minigenes.Citation13,32 Recently, using a splicing-sensitive microarray, we showed that heat-induced activation of latent splicing is widespread across the human genome, thus highlighting the possibility that latent splicing may underlie certain diseases.Citation13 Furthermore, analyses of data from the GEO revealed significant activation of latent splicing, which occurred in cancers such as breast cancer and gliomas. Such changes were found in thousands of gene transcripts representing a wide variety of functional groups, among them are genes involved in cell proliferation and differentiation. Because most latent splice sites are preceded by at least one in frame stop codon, their use would result in pre-maturely terminated aberrant mRNAs and proteins, which could cause deleterious effects on normal cell functions.Citation13
Here we searched for latent 5′SSs in C.elegans, in which the splice signals are highly conserved and are similar to those in humans. Also, similar spliceosomal components were found in the worm, including proteins homologous to human alternative splicing factors.Citation33,34 It should be noted that the stress response system is also conserved in C.elegans.Citation35,36 First, we show in this study, by a bioinformatics survey, that latent 5′SSs are found in nematodes. Second, we show that under heat treatment splicing from latent sites is activated in a number of candidate genes, selected from the bioinformatics survey. We further show that suppression of latent splicing from the tested candidate genes is not specific to a developmental stage and occurs throughout the different stages of development, while after heat shock, latent splicing is activated in all developmental stages. Finally, we show that the lack of latent splicing under normal growth conditions cannot be attributed to NMD, because a number of nematode mutant smg strains, lacking essential NMD factors, did not express the latent splicing isoform under normal growth conditions. These findings further confirm that SOS is independent of NMD. Furthermore, they show that SOS is functioning also in nematodes and is thus conserved between species, indicating the importance and generality of this mechanism.
Results
Alternative latent 5′SSs are abundant within introns of C. elegans– Bioinformatics survey
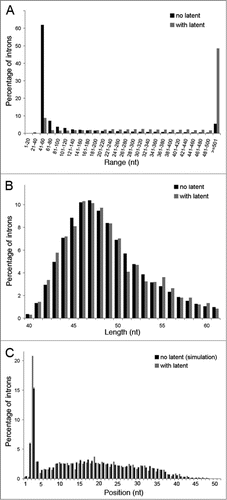
To search for latent 5′SSs in C. elegans introns, we first conducted a genome wide computational analysis. The introns of C. elegans are generally much shorter than their human counterparts, as 50% of them are between 40 and 61 nucleotides in length. Therefore, the probability of finding a latent 5′SS within an intron is much lower in C. elegans than in humans; in addition, they may not exist at all due to elimination by negative selection. To search for latent 5′SSs in C. elegans, genomic sequences and gene structure data were downloaded from the NCBI website (updated on 6/10/2006, based on Wormbase version WS160) followed by filtering out known alternatively spliced genes (see Methods for details). The resulting database contained 78,938 introns located within 16,412 genes. We then assigned a log-likelihood ratio score for each 8 consecutive bases within the intronic sequences. This score reflects the likelihood of a site to be a 5′SS relative to that of being a random intronic sequence. A threshold of 4 (namely, ∼55-fold more likely to be a 5′SS than a random sequence) was chosen, such that 73.66% of the authentic 5′SSs in the same database exceeded this threshold. About one quarter of the introns (18,607) had at least one latent 5′SS with a score that exceeded this threshold (). In total, the 18,607 introns contained 42,283 latent 5′SSs, amounting to an average of 2.27 latent sites per intron.
Table 1. Occurrences of latent 5′ splice sites (5′SSs) in C. elegans introns
Our previous analysis of the human genome showed that at least one in frame stop codon resided upstream of 98% of the latent 5′SSs identified in the database.Citation12,13,37 Similarly here, at least one in frame stop codon was found in 90% of the sequences between the latent 5′SSs identified in the C. elegans non-UTR introns and the respective upstream authentic 5′SSs (). The significance of this finding is that splicing at latent 5′SSs (latent splicing) would introduce stop codons into the reading frame of most of the resulting mRNAs, indicating that SOS might be operational in C. elegans.
In order to assess the significance of the association between latent 5′SSs and the presence of upstream in frame stop codons, we used the group of introns without latent sites as a background model. Finding a latent site and then searching for an upstream in frame stop codon was simulated on introns without latent sites and simulation results were compared to our real data (Figure S1). In order to avoid bias in our simulation procedure, only a sub-group of introns, with lengths ranging between 40 and 61 bases, was used, overcoming the difference in length distribution between introns with and without latent 5′SSs (). This sub-group of introns kept the same length distribution and latent 5′SS (real or simulated) position distribution between our real and simulated data (, respectively). This way we avoided the bias of finding an upstream in frame stop codon in favor of our real data simply because of its longer length. The results for this sub-group of introns are given in , where 41.2% of the most upstream latent 5′SSs found in each intron introduce an upstream in frame stop codon upon activation. However, in the background group of introns without latent 5′SSs, only 36.6% of the simulated latent 5′SSs had an upstream in frame stop codon, a significantly lower percentage than for the real data, P < 0.0001 (fisher exact test).
Table 2. Enrichment of stop codons upstream to latent 5′ splice sites (5′SSs)
Figure 1. Simulation of search for latent 5′SSs in introns lacking latent sites. C. elegans introns of the data base were divided into introns lacking latent 5′SSs and those with at least one latent 5′SS. (A) Length distribution of introns lacking latent 5′SSs (black) and introns having latent sites (gray). The length distribution is presented in groups of 20 nt. The last right column depicts introns longer than 500 nt. Introns having latent sites clearly tend to be much longer than the ones lacking such sites. (B) Length distribution of introns lacking latent 5′SSs (black), and having such sites (gray), for introns of length 40–61 nt is depicted. (C) Distribution of the location of the latent 5′SSs (measured from the authentic 5′SS, in nt) in introns of 40–61 nt length (gray), compared with the locations selected by the simulation for introns lacking latent 5′SSs (black). For the simulation, the average percentage, of 10,000 repeats, as well as the standard deviation, are presented. It is clear that choosing the group of introns of length 40–61 nt overcomes the length bias that exists in the general introns population, and that the simulation mimics well the real situation.
We conclude that latent 5′SSs do exist in the nematode C. elegans and that their suppression is probably achieved by a similar mechanism to the mammalian nonsense-mediated suppression of splicing.
Heat shock elicits latent splicing in C. elegans endogenous genes
The bioinformatics survey for latent 5′SSs in C. elegans revealed that, similar to the human genome, potential latent sites are abundant in the worm's genome, and that in frame stop codons occur upstream of over 90% of them. Yet, splicing at these alternative 5′SSs, in normally growing worms, has not been reported. These findings have indicated that SOS is also functioning in the nematode. To test this prediction we asked whether heat treatment, which was previously shown to elicit latent splicing in mammalian cells (presumably by abrogating SOS),Citation13,32 would affect C. elegans in the same way. To this end we selected a number of candidate genes, whose latent 5′SSs differ from each other with respect to their score, distance from the upstream authentic 5′SS, and intron length (). In the experiment described in , C. elegans worms were grown at various temperatures (23–34°C), and total RNA was extracted and analyzed by semi quantitative RT-PCR. The primers used were a sense intronic primer and an antisense primer from the downstream exon, which amplify the pre-mRNA and the latent RNA of the respective RNA transcripts, as indicated in . Actin mRNA was used as a loading control in these experiments.
Table 3. Details of validated latent 5′ splice sites (5′SSs)
Figure 2. Splicing at latent 5′SSs is observed in C. elegans after heat shock. Semiquantitative RT-PCR analyses of RNA expressed from C. elegans grown under normal growth conditions and under different stress conditions. (A) Effect of temperature and stress duration (lanes 2–7) on latent splicing expressed from jph-1, compared to normal growth conditions (23°C, lane 1). Symbols on the left represent pre-mRNA and latent RNA. Open boxes, exons; lines, introns; narrow boxes, latent exons. The PCR analyses were performed using primer pairs from the latent exon (sense) and the downstream exon (antisense), as indicated by arrows. RT-PCR analysis of actin was used as a loading control. (B, C) Analogous RT-PCR analyses of latent splicing expressed from unc-47 (B), and goa-1 (C), after heat shock (34°C, 3 h), compared to normal growth conditions (23°C). D. RT-PCR analysis of 3-fold serial dilutions of RNA extracted from N2 worms grown under normal growth conditions, using RNA levels ranging from x9 lower to x3 larger than those used in A-C. The PCR analyses were performed using the primer pairs from the specified latent exon (sense) and the downstream exon (antisense) of jph-1, unc-47 and goa-1, as indicated in A-C, respectively, and represent the respective levels of unspliced transcripts. The results represent at least 3 independent experiments.
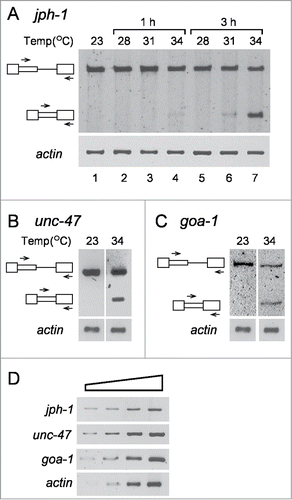
We first tested the effect of heat shock on the expression of the junctophilin -1 (jph-1) gene, a member of the junctophilin gene family. The protein encoded by this gene is a component of junctional complexes between the plasma membrane and the endoplasmic/sarcoplasmic reticulum, and according to our bioinformatics survey jph-1 has a latent 5′SS in intron 5 ().
RT-PCR analysis of total RNA extracted from worms grown at the normal temperature (23°C) showed a PCR product arising from the jph-1 pre-mRNA, but no PCR product arising from latent mRNA was observed (, lane 1). Similar results were observed when worms grown at 23°C were shifted to temperatures up to 31°C for 1 h (, lanes 2 and 3) or 28°C for 3 h (, lane 5). However, small amounts of a PCR band, representing splicing at the latent 5′SS of intron 5, were observed in worms treated at 34°C for 1 h or 31°C for 3 h (, lanes 4 and 6, respectively), and after shifting the temperature to 34°C for 3 h the level of latent splicing was comparable to that of the pre-mRNA (, lane 7). The identity of these bands as representing splicing from the latent site was confirmed by sequencing. From now on, unless stated otherwise, heat treatment will be analyzed at 34°C for 3 h. These studies show that the splicing pattern of jph-1 transcript is affected by heat shock, resulting in alternative splicing from the latent site.
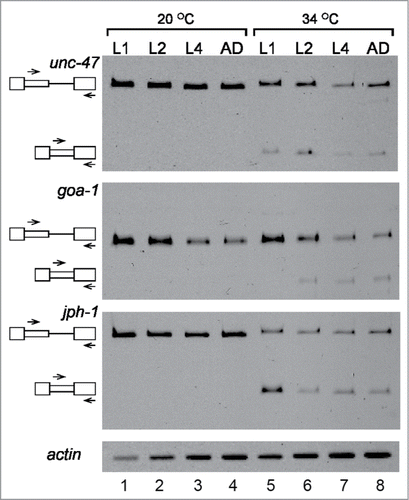
Figure 3. Latent splicing is suppressed during the developmental stages of C. elegans. RT-PCR analyses of latent splicing expressed from the endogenous genes: goa-1, jph-1 and unc-47, during the different stages of worm development: larval stages L1, L2, L4, and adult (AD). Worms were analyzed under normal temperature (20°C, lanes 1–4), or after heat shock (34°C, 3 h, lanes 5–8). RT-PCR analysis of actin was used as a loading control. Symbols on the left represent pre-mRNA and latent RNA. Open boxes, exons; lines, introns; narrow boxes, latent exons. The primer pairs used are indicated by arrows. The results represent at least 2 independent experiments.
The activation of latent splicing of jph-1 transcript after heat shock, prompted us to test for the effect of heat shock on latent splicing of additional nematode genes. The next candidate was the uncoordinated - 47 (unc-47) gene, which had a latent 5′SS in intron 4. This gene encodes a transmembrane vesicular GABA transporter. UNC-47 is required in GABAergic neurons for all GABA neurotransmission, specifically for loading of GABA into synaptic vesicles, where UNC-47 is localized. When splicing of unc-47 pre-mRNA was analyzed by RT-PCR, no latent splicing was observed under normal growth conditions, while latent splicing was observed after heat shock (, 23°C and 34°C, respectively).
Another gene, which showed a potential latent 5′SS (in intron 1), according to the bioinformatics search, was the goa-1 gene. The goa-1 gene - Guanine nucleotide binding protein (G-protein), α subunit - encodes an ortholog of the heterotrimeric G protein α subunit Go (Go/Gi class) that affects normal locomotion, egg-laying, and male mating. As can be seen in , in worms grown at 23°C no splicing from the latent site of goa-1 was observed, while after heat treatment at 34°C, latent splicing was detected. represents 3-fold serial dilutions of RNA extracted from untreated worms to demonstrate that the RT-PCR was semi quantitative. These three genes (jph-1, unc-47 and goa-1), which showed activation of splicing from the latent sites after heat treatments, were chosen for further experimental analyses.
Suppression of splicing from latent 5′SSs occurs in all stages of worm development
The C. elegans undergoes through 4 larval developmental stages, L1-L4, before turning into an adult worm, a process accompanied by changes in gene expression and in response to stress.Citation38 To examine if suppression of latent splicing is dependent on the developmental stage of C. elegans, we analyzed the expression of latent splicing in transcripts of the jph-1, goa-1 and unc-47 genes during the different stages of development. For this aim worms were synchronized, and RNA was extracted from worms at stages L1, L2, L4 and adults (AD) grown at 20°C, and after heat treatment at 34°C for 3 h. RT-PCR analyses of the splicing pattern of unc-47, goa-1 and jph-1 (), revealed that under normal growth conditions, no latent splicing was observed in any of these transcripts at any of the developmental stages (lanes 1–4). However, after heat shock, latent splicing was activated in all these 3 gene transcripts (lanes 5–8). These experiments show that the lack of expression of latent splicing from the tested candidate genes is not specific to a developmental stage and occurs throughout the different stages of development, starting at L1 stage and continues through L2, L4 and adult worms.
Latent splicing is observed when a PTC upstream to a latent site is removed
It was hypothesized that splicing from a latent 5′SS is suppressed to avoid the introduction of a PTC into the resultant mRNA. Indeed, latent splicing was elicited in all mutants, in which all in frame stop codons residing upstream of the latent 5′SSs had been eliminated either by point mutations or by frame shifting.Citation23-25 Here we used the same approach, by generating transgenic strains using 2 minigene constructs of the unc-47 gene, which has a latent 5′SS with 4 upstream in frame stop codons in intron 4 (). The wild-type minigene (unc-47.wt) was constructed by joining the 5′ untranslated region (5′UTR) of unc-47 and the 5′ 90 nt of exon 1 (Ex 1) to exon 4 (Ex 4) intron 4 (Int 4) and exon 5 (Ex 5), and this sequence was inserted upstream of the GFP reporter in the pAD009 vector. In the mutant minigene (unc-47.mut), the 4 in frame stop codons residing between the authentic and latent 5′SS of intron 4 (marked by a stop sign in ) were eliminated by frame shifting. The unc-47 minigene plasmids were each introduced by bombardmentCitation39 into worm strain DP38, defective in lower body movement [unc119(-)]. Selection of strains harboring the unc-47 minigene constructs was based on gain of lower body movement due to unc-119 presence in the original plasmid. To confirm integration of the minigenes into the worm genome, worms were grown for several generations and analyzed by PCR on genomic DNA for the presence of the inserted unc-47 minigene. As shown in , 4 transgenic strains were isolated and analyzed: (i) Two strains each harboring the unc-47.wt minigene – unc-47 (wt1); unc-47 (wt2); and (ii) 2 strains each harboring the unc-47.mut minigene that lacks the PTCs – unc-47 (mut1); unc-47 (mut2); DP38 worms that failed to harbor the unc-47 minigene sequences were used as control.
Table 4. Splicing of mutant strains expressing unc-47 minigenes
Figure 4. Analysis of latent splicing in transgenic strains. (A) Schemes of 2 minigene constructs prepared from the unc-47 gene: unc-47.wt, which harbors the unc-47 intron 4 latent site, and has 4 PTCs between the authentic and latent site; unc-47.mut, lacks the PTCs. Both minigenes were cloned in pAD009 vector and transgenic strains were prepared and selected. (B) Upper panel: RT-PCR analysis of unc-47 splice variants expressed from the unc-47 minigene in 2 wild type transgenic strains (unc-47 (wt1), unc-47 (wt2), lanes 2, 3, respectively), 2 mutant strains in which the PTCs were removed (unc-47 (mut1), unc-47 (mut2), lanes 4, 5, respectively) and a strain lacking the unc-47 minigene (lane 1). All worms were grown at 23°C. Lower panels: RT-PCR analysis of transcripts expressed from endogenous goa-1 and jph-1 in the 4 transgenic strains, grown at 23°C, revealed that suppression of latent splicing in the endogenous goa-1 and jph-1 transcripts was not affected by the genetic manipulation. RT-PCR analysis of actin was used as a loading control. Symbols on the left represent pre-mRNA and latent RNA. Open boxes, exons; lines, introns; narrow boxes, latent exons. The results represent at least 3 independent experiments.
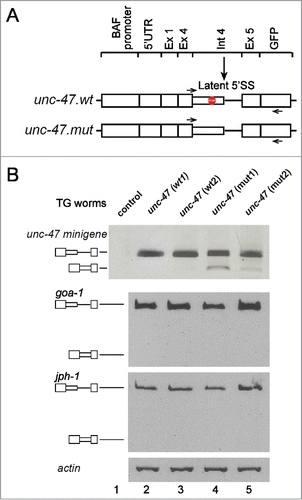
To analyze expression from the integrated unc-47 minigene in the new transgenic strains, a primer pair specific for the minigene (intron 4 sense primer and a GFP antisense primer) () was used in RT-PCR analysis. The analysis revealed (, upper panel) that under normal growth conditions latent splicing was not observed in the unc-47 (wt1) and unc-47 (wt2) strains (lanes 2 and 3), while a weak signal of latent splicing was observed in the unc-47 (mut1) and unc-47 (mut2) strains, in which the in-frame stop codons have been removed (lanes 4, 5). The DP38 (control) strain, that lacks the unc-47 minigene sequence, showed no PCR products (lane 1), confirming that the RT-PCR analysis specifically analyzed the transcripts derived from the unc-47 minigene constructs.
It should be noted that in these transgenic strains, the splicing pattern of the endogenous goa-1 and jph-1 transcripts was not affected by the transformation (, lower panels). Under normal growth conditions no latent splicing was observed in the goa-1 and jph-1 transcripts in any of the transgenic strains (lanes 2–5).
SOS is distinct from NMD
It could have been argued that splicing from the latent sites did occur, yet the PTC containing mRNAs were completely degraded by the NMD pathway. We have previously shown that upon downregulation of hUpf1 and hUpf2, genes essential for NMD, latent splicing was not observed in transcripts regulated by SOS in mammalian cells.Citation24 We also showed that inhibition of translation and especially inhibition of the pioneer round of translation, required for NMD, did not abrogate SOS.Citation24,25 Furthermore, we ruled out degradation of latent mRNA by a yet unknown RNA degradation mechanism by showing that constructs in which we forced formation of latent mRNA through its expression from a plasmid harboring the already spliced DNA, express latent mRNA at levels only slightly lower than that of authentic mRNA expressed from a plasmid harboring the already spliced DNA at the authentic site.Citation31 Here we have taken advantage of the availability of C. elegans mutant strains in which NMD genes have been functionally deleted, in order to test the effect on SOS. Nematodes have 7 NMD genes encoding for the proteins – SMG-1 to SMG -7: SMG-1 (phosphatidylinositol 3-kinase-related kinase) which phosphorylates SMG-2; SMG-2 is the ortholog of human hUpf1 and is conserved in human, yeast and worm; SMG-3 is the ortholog of hUpf2 and SMG-4 is the ortholog of hUpf3; SMG-5, SMG-6 and SMG-7 are required for efficient SMG-2 dephosphorylation.Citation40-45 Upf1, Upf2 and Upf3 are the core components of the NMD pathway, while Upf1 is the key player in NMD and is required for all known pathways of NMD.Citation18,20,21 Here, we have analyzed 3 mutant strains in which NMD is abrogated: (i) strain TR1331, smg-1(r861) I;Citation43,46 (ii) strain TR1421, smg-2(r908) unc-54(r293) I – in which SMG-2 is deleted;Citation40 (iii) strain TR1696, smg-3(r930) IV unc-54 (r293) I – in which SMG-3 is abrogated.Citation44,47 The smg mutant strains were obtained from the Caenorhabditis Genetics Center (CGC) at the University of Minnesota. All smg (-) strains used here were shown to express none of the encoded proteins.Citation40,43,44 We also tested the expression of a known NMD target, rpl-12 (PTC) splice variant,Citation48 in these mutant strains (). Primers from flanking exons were used to amplify both an open reading frame preserving variant (rpl-12), and the NMD target, PTC-containing, alternative variant (rpl-12 (PTC)). Under different normal growth temperatures (20˚C and 23˚C), the expression of rpl-12 (PTC) was elevated in the mutant strains relative to that in the N2 wild type strain, showing that NMD was indeed abrogated.
Figure 5. Latent 5′SSs in C. elegans are silenced under normal growth conditions even in mutant strains lacking NMD essential genes. Worms from the wild type strain (N2) and 3 smg(-) strains (TR1331, SMG-1-null; TR1421, SMG-2-null; TR1696, SMG-3-null), in which NMD essential genes were abrogated, were grown under normal growth conditions (23°C), or were heat-shocked (34°C, 3h). (A) RT-PCR analysis of splicing expressed from the rpl-12 pre-mRNA, a known substrate of NMD, in N2 wild type strain and the 3 smg(-) mutant strains, showing that NMD is indeed abrogated in the 3 smg(-) mutant strains. rpl-12, an open reading frame preserving variant; rpl-12 (PTC), a variant containing a PTC, the NMD substrate. (B) RT-PCR analysis of splicing of the goa-1, jph-1 and unc-47 pre-mRNAs expressed from the WT (N2) strain, and the 3 smg(-) strains. At 23°C (lanes 1–4), only bands representing pre-mRNA and no bands representing latent RNA are observed. After heat treatment (34°C, lanes 5–8) latent splicing is activated. Actin PCR was used as a loading control. (C) RT-PCR analysis of 3-fold serial dilutions of RNA extracted from N2 worms grown under normal growth conditions, using RNA levels ranging from x9 lower to x3 larger than those used in A-B. The PCR analyses were performed using the primer pairs from the specified latent exon (sense) and the downstream exon (antisense) of jph-1, unc-47 and goa-1, as indicated in B, respectively, and represent the respective levels of unspliced transcripts. The results represent at least 3 independent experiments.
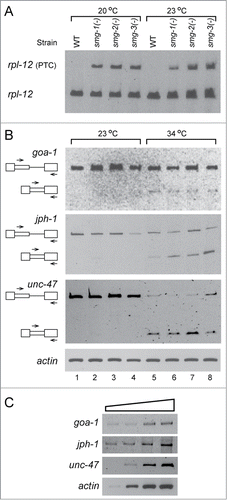
We tested the effect of abrogation of NMD on the expression of latent splicing. As shown in , under normal growth conditions (23˚C), splicing from the latent site of the endogenous goa-1 gene transcript is not observed in all the 3 NMD mutant strains [smg-1(-), smg-2 (-) and smg-3 (-)] (lanes 2–4, respectively). Similar results, showing no latent splicing, was observed for each of the smg(-) mutants grown at either 16˚C, or 20˚C (data not shown). No latent splicing was observed also when we analyzed the expression of the jph-1 and unc-47 genes in the 3 smg(-) mutant strains (, lanes 2–4), namely, no latent splicing was observed under normal growth conditions. represents 3-fold serial dilutions of RNA extracted from untreated worms to demonstrate that the RT-PCR is semiquantitative.
After heat treatment at 34˚C for 3 hours, latent splicing from the jph-1, goa-1 and unc-47 gene transcripts appeared in all 3 smg (-) mutant strains (, lanes 6–8). Results in the smg (-) mutant strains were comparable to those in the N2 wild type strain (, lanes 1 and 5). Because under normal growth conditions latent splicing is not observed in the worm mutant strains, in which NMD was abrogated, we can conclude that the lack of latent splicing in the N2 wild type strain, grown under normal growth conditions, is not due to degradation by the NMD pathway, but rather to suppression of splicing from the latent 5′SSs. Furthermore, these results support our earlier conclusionsCitation24,25 that SOS is distinct from NMD.
Discussion
In our bioinformatics search, we found that ∼24% of the worm's introns contained latent 5′SSs and ∼90% of those had an in frame stop codon upstream to the latent site. For the statistical analysis we focused on introns ranging in size between 40–61 nt, and found that 1668 introns contained latent sites, of which 728 had at least one upstream in frame stop codon. We further showed a significant correlation between latent sites and an appearance of the upstream in frame stop codon, with an expected p-value P < 0.0001. These findings show that the abundance of latent 5′SSs is conserved between species, indicating the importance and generality of this phenomenon.
To experimentally test the occurrence of SOS in nematodes, we analyzed 3 gene transcripts (goa-1, jph-1, and unc-47), that according to the bioinformatics survey have potential latent 5′SSs, each having a better score than its upstream authentic 5′SS. No latent splicing was observed under normal growth conditions in these transcripts. However, similar to abrogation of SOS by heat shock in mammalian cells,Citation13,32 activation of splicing at the latent site was observed upon heat treatment in the 3 worm gene transcripts analyzed. The lack of latent splicing under normal growth conditions was not restricted to a specific developmental stage (). Furthermore, upon heat treatment latent splicing was observed in all analyzed stages of development.
Our previous studies of SOS in mammalian cells have shown that a PTC between an authentic 5′SS and a downstream latent 5′SS is sufficient to suppress splicing from the latent 5′SS, because elimination of the PTC by either point or frame-shift mutation activated latent splicing.Citation23,24 We have analyzed the effect of removing the PTCs on latent splicing in worms, by generating mutant strains stably transfected with either WT or PTC lacking mutant minigenes of unc-47. RT-PCR analysis revealed that no latent splicing was observed in the WT strains. In the mutant strains, in which the latent 5′SS was converted to an alternative 5′SS by removal of the PTCs, only low levels of latent splicing were observed. It should be pointed out that several parameters affect the level of splicing from that “newly generated” alternative 5′SS. Among them are the resemblance of the 5′SS sequence to that of the consensus; the presence of exonic splicing enhancers (ESEs) and silencers (ESS), as well as intronic splicing enhancers (ISEs) and silencers (ISSs) within the pre-mRNA; the relative location of the enhancers and silencers within the transcript; and the structure of the pre-mRNA. All these factors together, in concert with the splicing factors that recognize them, play a role in the extent of alternative splicing from that site. In any event, our experiments show that under normal growth conditions, latent splicing was not observed from the 3 endogenous candidate genes analyzed here (goa-1, jph-1, and unc-47) and also from the strains harboring the WT minigne of unc-47. When the minigene construct was mutated to remove the stop codons a low level of splicing from the downstream 5′SS was obtained.
As discussed in the Introduction, the possibility of attributing the lack of latent splicing, in mammalian cells grown under normal conditions, to NMD was extensively tested and consequently rejected.Citation23-25,31,49,50 In this study we took advantage of the availability of C. elegans mutant strains that are devoid of NMD essential factors and lack a NMD mechanism. The rationale for this undertaking was that if lack of latent splicing were attributed to NMD, it could have been expected that latent splicing would be detected in these mutant strains, even when grown at normal temperature. As shown here (), under normal growth conditions (23˚C), latent splicing is not observed from the endogenous jph-1, goa-1 and unc-47 gene transcripts in each of the 3 NMD mutant strains [smg-1(-), smg-2 (-) and smg-3 (-)], and similar results were obtained also at 16˚C and 20˚C. This study adds a further proof to our earlier findings that SOS is distinct from NMD.Citation23-25,31,49,50 Furthermore, our recent experimentsCitation31 strongly suggest that the possibility of attributing the apparent lack of latent splicing to any kind of hUpf1-independent RNA surveillance mechanism is highly unlikely.
We can therefore conclude, that like in mammals, SOS is also functioning in worms, and the lack of splicing from the latent 5′SSs in both mammalian cells and worms is due to regulation by the SOS nuclear quality control mechanism. The finding that SOS is conserved between species indicates the importance and generality of this mechanism. This study further shows that SOS functions in an organism. Because of the complexity of eukaryotic post-transcriptional RNA processing events, these processes are prone to errors that may be harmful or even fatal. Therefore, quality control or proof-reading mechanisms such as the SOS mechanism are required to ensure that fully processed mRNAs function properly. Further studies are required to elucidate the trans acting-factors of this mechanism.
Materials and Methods
C. elegans strains and stress conditions
All strains were grown using standard protocols.Citation51 The wild type strain N2 and the NMD-deficient strains TR1331 smg-1(r861) I, TR1421 smg-2(r908) unc-54(r293) I and TR1696 unc-54(r293) I; smg-3(r930) IV were obtained from the Caenorhabditis Genetics Center (CGC) at the University of Minnesota. Strains generated in this work were unc-47 (wt1) and unc-47 (wt2), by bombarding the construct unc-47.wt into unc-119(ed3) worms, and unc-47 (mut1) and unc-47 (mut2), by bombarding the construct unc-47.mut into unc-119(ed3) worms.Citation39
Heat shock was done by growing the worms under normal conditions (16°C to 23°C) until feeding bacteria were nearly gone (for mixed-staged worms) or until the desired developmental stage, then placing the NGM plates in an incubator at the respective temperature (ranging from 28°C to 38°C) for either 1 h or 3 h. The standard procedure for activation of latent splicing was determined as 3 h at 34°C.
Cloning of bombardment constructs
The constructs unc-47.wt and unc-47.mut were based on the endogenous unc-47 gene, by PCR on genomic DNA of the wild type N2 strain. Both constructs were generated by fusing the 5′UTR and beginning of exon 1 to the end of exon 4, together with intron 4 and the beginning of exon 5. Primers for generating the 5′ fragment were:
5′ ATGTGTCGACACACACACGCTATTTGAAG 3′ and 5′ CTGAATTCGCTCCTGATTCCAGTAGTCC 3′, for both constructs. Primers for the 3′ fragment were 5′ TTGAATTCTCAATCTGATCATGGTCCTT 3′ for the unc-47.wt construct and 5′ ATGAATTCGTCAATCTGCATCATGGTCCT 3′ for the unc-47.mut construct, together with the same reverse primer 5′ ATTGGTACCAATTGAGCAGGATTTTTCAT 3′ for both constructs. PCR products were cleaved with the appropriate restriction enzymes (SalI and EcoRI for the 5′ fragment, and EcoRI and Acc65I for the 3′ fragment) and ligated into the pAD009 vector (a kind gift from Prof. Yosef Gruenbaum) after cleavage with SalI and Acc65I. The difference between the forward primers for the creation of the 3′ fragment of the 2 constructs creates a frameshift in the unc-47.mut construct that clears all in-frame stop codons within the latent exon. Cloning was confirmed by DNA sequencing. Approximately 4 μg of each plasmid were used for bombardment as described.Citation39
Preparation of total RNA, RT-PCR and single-worm PCR analyses
Genomic DNA from single worms of strains unc-47 (wt1), unc-47 (wt2), unc-47 (mut1) and unc-47 (mut2) was obtained essentially as describedCitation52 and used for PCR with primers 5′ GTGTTTCAGAAACCGAGC 3′ and 5′ CTGAACTTGT GGCCGTTT 3′ in order to verify the integration of the bombardment constructs into the worm's genome. For RT-PCR analyses worms were washed off the NGM plates with DEPC-treated water, centrifuged and washed again with DEPC-treated water in order to remove residual bacteria. After centrifugation worms were mixed with 1 ml of extraction mix and total RNA was extracted as described.Citation53 Reverse transcription was done with 1 μg to 3 μg of DNase-treated total RNA as previously described.Citation53 Semi-quantitative RT-PCR was done by 3-fold serial dilutions of the DNase-treated total RNA and taking equal volumes from the different dilutions to the RT reaction. Amounts of RNA were equivalent to 3:1, 1:1, 1:3 and 1:9 relative to the amount used for the indicative RT-PCR. Primer sequences and conditions for PCRs against act-1, goa-1, jph-1, rpl-12, endogenous unc-47 and unc-47 from the bombardment constructs are given in Tables S1 and S2, respectively. The identity of all PCR products was confirmed by sequencing.
Bioinformatics analyses
The October 2006 GenBank version of the worm′s genome (based on WS160 version of WormBase) was used to obtain genes' sequence and structure. Genes with more than one CDS and genes with non-gt/ag introns were filtered out in order to create a simpler database. The authentic 5′SSs in the remaining database were used to calculate a position specific score matrix (PSSM) that defined the probabilities of seeing each nucleotide in each position of a sequence that is a 5′SS (Table S3). Intronic 8-mer sequences in this database were scanned using a sliding window (1-nucleotide step) and predicted as latent 5′SSs when they conformed to the 5′SS consensus sequence, namely were ∼55 fold more likely to arise from the 5′SSs PSSM than from a random intronic sequence.Citation54 The log-likelihood ratio score was calculated as follows: where ni is the nucleotide seen at the i-th position, P(ni|SS) is the frequency of that nucleotide at the i-th position of authentic 5′SSs, as taken from the PSSM, and P(ni|BG) is the frequency of that nucleotide in C. elegans intronic sequences in general (given as Table S4). For each predicted latent site, the impact of its activation on the open reading frame, namely introducing a PTC or not, was calculated (see ) and numerous characteristics were recorded (such as the location along the gene, the distance from the authentic 5′SS, etc., see ).
Assessing the statistical significance of the association between the presence of a latent 5′SS and the presence of an upstream in frame stop codon was done by simulation, focusing on introns of length between 40 nt to 61 nt. Mimicking the search for latent 5′SSs and the presence of an upstream in frame stop codon, the simulation process randomly chose a similarly sized group of introns without any latent site and searched for an in frame stop codon from their authentic 5′SS to a point randomly chosen according to the latent sites position distribution in our database. An illustration of the simulation process is given in Figure S1. The average percentage of introns with an in frame stop codon found in 10,000 repeats of the simulation process was then compared to the percentage of latent exons that harbor an in frame stop codon by the Fisher exact test.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary File.pdf
Download PDF (135.1 KB)Acknowledgments
We thank Prof. Yosef Gruenbaum and members of his laboratory for advise, support and share of know how in worms handling and in the generation of mutant strains.
Funding
This work was supported in part by grants from the Israel Cancer Research Fund (J.S and R.S.), and the Helen and Milton Kimmelman Center for Biomolecular Structure and Assembly at the Weizmann Institute of Science (J.S.).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- Burge CB, Tuschl TH, Sharp PA. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland RF, Cech TR, Atkins JF, eds. The RNA World, second edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press, 1999:525-60.
- Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet 2002; 36:333-60; PMID:12429696
- Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol 2011; 3:a003707; PMID:21441581
- Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol 2004; 22:535-46; PMID:15122293; http://dx.doi.org/10.1038/nbt964
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene 2005; 344:1-20; PMID:15656968; http://dx.doi.org/10.1016/j.gene.2004.10.022
- Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci 2006; 119:2635-41; PMID:16787944; http://dx.doi.org/10.1242/jcs.03053
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 2007; 8:749-61; PMID:17726481; http://dx.doi.org/10.1038/nrg2164
- Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci 2000; 25:106-10; PMID:10694877; http://dx.doi.org/10.1016/S0968-0004(00)01549-8
- Bourgeois CF, Lejeune F, Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol 2004; 78:37-88; PMID:15210328; http://dx.doi.org/10.1016/S0079-6603(04)78002-2
- Sanford JR, Ellis J, Caceres JF. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem Soc Trans 2005; 33:443-6; PMID:15916537; http://dx.doi.org/10.1042/BST0330443]
- Sugnet CW, Kent WJ, Ares M, Jr., Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac Symp Biocomput 2004:66-77; PMID:14992493
- Miriami E, Motro U, Sperling J, Sperling R. Conservation of an open-reading frame as an element affecting 5' splice site selection. J Struct Biol 2002; 140:116-22; PMID:12490159; http://dx.doi.org/10.1016/S1047-8477(02)00539-7
- Nevo Y, Kamhi E, Jacob-Hirsch J, Amariglio N, Rechavi G, Sperling J, Sperling R. Genome-wide activation of latent donor splice sites in stress and disease. Nucleic Acids Res 2012; 40:10980-94; PMID:23002147; http://dx.doi.org/10.1093/nar/gks834
- Mühlemann O, Mock-Casagrande CS, Wang J, Li S, Custodio N, Carmo-Fonseca M, Wilkinson MF, Moore MJ. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol Cell 2001; 8:33-44; PMID:11511358; http://dx.doi.org/10.1016/S1097-2765(01)00288-X
- Aoufouchi S, Yelamos J, Milstein C. Nonsense mutations inhibit RNA splicing in a cell-free system: recognition of mutant codon is independent of protein synthesis. Cell 1996; 85:415-22; PMID:8616896; http://dx.doi.org/10.1016/S0092-8674(00)81119-8
- Gersappe A, Burger L, Pintel DJ. A premature termination codon in either exon of minute virus of mice P4 promoter-generated pre-mRNA can inhibit nuclear splicing of the intervening intron in an open reading frame-dependent manner. J Biol Chem 1999; 274:22452-8; PMID:10428819; http://dx.doi.org/10.1074/jbc.274.32.22452
- de Turris V, Nicholson P, Orozco RZ, Singer RH, Mühlemann O. Cotranscriptional effect of a premature termination codon revealed by live-cell imaging. RNA New York, NY 2011; 17:2094-107; PMID:22028363; http://dx.doi.org/10.1261/rna.02918111
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett 2007; 581:2845-53; PMID:17531985; http://dx.doi.org/10.1016/j.febslet.2007.05.027
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 2007; 76:51-74; PMID:17352659; http://dx.doi.org/10.1146/annurev.biochem.76.050106.093909
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev 2007; 21:1833-56; PMID:17671086; http://dx.doi.org/10.1101/gad.1566807
- Stalder L, Muhlemann O. The meaning of nonsense. Trends in cell biology 2008; 18:315-21; PMID:18524595; http://dx.doi.org/10.1016/j.tcb.2008.04.005
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci 2010; 67:677-700; PMID:19859661; http://dx.doi.org/10.1007/s00018-009-0177-1
- Li B, Wachtel C, Miriami E, Yahalom G, Friedlander G, Sharon G, Sperling R, Sperling J. Stop codons affect 5' splice site selection by surveillance of splicing. Proc Natl Acad Sci USA 2002; 99:5277-82.
- Wachtel C, Li B, Sperling J, Sperling R. Stop codon-mediated suppression of splicing is a novel nuclear scanning mechanism not affected by elements of protein synthesis and NMD. RNA 2004; 10:1740-50; PMID:15388876; http://dx.doi.org/10.1261/rna.7480804
- Kamhi E, Yahalom G, Kass G, Hacham Y, Sperling R, Sperling J. AUG sequences are required to sustain nonsense-codon-mediated suppression of splicing. Nucleic Acids Res 2006; 34:3421-33; PMID:16855285; http://dx.doi.org/10.1093/nar/gkl390
- Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev 2004; 18:745-54; PMID:15059963; http://dx.doi.org/10.1101/gad.1170204
- Mendell JT, ap Rhys CMJ, Dietz HC. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 2002; 298:419-22; PMID:12228722; http://dx.doi.org/10.1126/science.1074428
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell 2005; 20:65-75; PMID:16209946; http://dx.doi.org/10.1016/j.molcel.2005.08.012
- Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, 2nd, Wilkinson MF. An alternative branch of the nonsense-mediated decay pathway. EMBO J 2007; 26:1820-30; PMID:17363904; http://dx.doi.org/10.1038/sj.emboj.7601628
- Matsuda D, Hosoda N, Kim YK, Maquat LE. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat Struct Mol Biol 2007; 14:974-9; PMID:17873884; http://dx.doi.org/10.1038/nsmb1297
- Kamhi E, Raitskin O, Sperling R, Sperling J. A potential role for initiator-tRNA in pre-mRNA splicing regulation. Proc Natl Acad Sci USA 2010; 107:11319-24; PMID:20534564; http://dx.doi.org/10.1073/pnas.0911561107
- Miriami E, Sperling J, Sperling R. Heat shock affects 5′ splice site selection, cleavage and ligation of CAD pre-mRNA in hamster cells, but not its packaging in lnRNP particles. Nucleic Acids Res 1994; 22:3084-91; PMID:7915031; http://dx.doi.org/10.1093/nar/22.15.3084
- Zahler AM. Alternative splicing in C. elegans. WormBook 2005:1-13; PMID:18050427
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J 2009; 417:15-27; PMID:19061484; http://dx.doi.org/10.1042/BJ20081501
- Kultz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol 2005; 67:225-57; PMID:15709958
- Prahlad V, Morimoto RI. Integrating the stress response: lessons for neurodegenerative diseases from C. elegans. Trend Cell Biol 2009; 19:52-61; PMID:19112021; http://dx.doi.org/10.1016/j.tcb.2008.11.002
- Miriami E, Sperling R, Sperling J, Motro U. Regulation of splicing: The importance of being translatable. RNA 2004; 10:1-4; PMID:14681577; http://dx.doi.org/10.1261/rna.5112704
- Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 2010; 11:545-55; PMID:20628411; http://dx.doi.org/10.1038/nrm2938
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 2001; 157:1217-26; PMID:11238406
- Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Molecular and Cellular Biology 1999; 19:5943-51; PMID:10454541
- Cali BM, Kuchma SL, Latham J, Anderson P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics 1999; 151:605-16; PMID:9927455
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 2003; 12:1187-200; PMID:14636577; http://dx.doi.org/10.1016/S1097-2765(03)00443-X
- Grimson A, O'Connor S, Newman CL, Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans. Mol Cell Biol 2004; 24:7483-90; PMID:15314158; http://dx.doi.org/10.1128/MCB.24.17.7483-7490.2004
- Johns L, Grimson A, Kuchma SL, Newman CL, Anderson P. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol Cell Biol 2007; 27:5630-8; PMID:17562857; http://dx.doi.org/10.1128/MCB.00410-07
- Mango SE. Stop making nonSense: the C. elegans smg genes. Trends Genet 2001; 17:646-53; PMID:11672865; http://dx.doi.org/10.1016/S0168-9525(01)02479-9
- Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 1989; 123:301-13; PMID:2583479
- Pulak R, Anderson P. Messenger-RNA Surveillance by the Caenorhabditis-Elegans Smg Genes. Genes Dev 1993; 7:1885-97; PMID:8104846; http://dx.doi.org/10.1101/gad.7.10.1885
- Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev 2000; 14:2173-84; PMID:10970881; http://dx.doi.org/10.1101/gad.819900
- Sperling J, Sperling R. Nuclear surveillance of RNA polymerase II transcripts. RNA Biol 2008; 5:220-4; PMID:18971637; http://dx.doi.org/10.4161/rna.7162
- Shefer K, Sperling J, Sperling R. The supraspliceosome-a multi-task-machine for regulated pre-mRNA processing in the cell nucleus. Comput Struct Biotechnol J 2014; 11:113-22; PMID:25408845; http://dx.doi.org/10.1016/j.csbj.2014.09.008
- Lewis JA, Fleming JT. Basic culture methods. Methods Cell Biol 1995; 48:3-29; PMID:8531730; http://dx.doi.org/10.1016/S0091-679X(08)61381-3
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics 1992; 131:609-24; PMID:1321065
- Margalit A, Brachner A, Gotzmann J, Foisner R, Gruenbaum Y. Barrier-to-autointegration factor–a BAFfling little protein. Trends Cell Biol 2007; 17:202-8; PMID:17320395; http://dx.doi.org/10.1016/j.tcb.2007.02.004
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res 1984; 12:505-19; PMID:6364039
