Abstract
Alternative pre-mRNA splicing in higher eukaryotes enhances transcriptome complexity and proteome diversity. Its regulation is mediated by a complex RNA-protein network that is essential for the maintenance of cellular and tissue homeostasis. Disruptions to this regulatory network underlie a host of human diseases and contribute to cancer development and progression. The splicing kinases are an important family of pre-mRNA splicing regulators, , which includes the CDC-like kinases (CLKs), the SRSF protein kinases (SRPKs) and pre-mRNA splicing 4 kinase (PRP4K/PRPF4B). These splicing kinases regulate pre-mRNA splicing via phosphorylation of spliceosomal components and serine-arginine (SR) proteins, affecting both their nuclear localization within nuclear speckle domains as well as their nucleo-cytoplasmic shuttling. Here we summarize the emerging evidence that splicing kinases are dysregulated in cancer and play important roles in both tumorigenesis as well as therapeutic response to radiation and chemotherapy.
Abbreviations
| SR | = | serine arginine |
| SRPK | = | serine arginine protein kinase |
| CLK | = | CDC-like kinase |
| PRP4K | = | pre-mRNA processing factor 4 kinase |
| snRNP | = | small nuclear ribonucleic protein. |
Introduction
Increased complexity of alternative splicing during evolution and the expansion of the SR-protein kinases
When comparing single-cell eukaryotes like the yeast S. cerevisae (∼6,000 genes),Citation1 to metazoans of increasing complexity, there appears to be a general relationship between increased complexity of the organism and the number of genes. After the human genome was sequenced in 2001, it was found that our genome contains approximately 23,000 genes, a much lower number than expected.Citation2 The human genome is larger than the genome of the fly D. melanogaster (∼14,000 genes) and comparable to the genome of the worm C. elegans (∼20,000 genes).Citation3,4 At the same time, it was discovered that genes containing introns encode many possible transcripts, which arise by alternative mRNA splicing and allow organisms with a similar number of genes to have more complex and diverse proteomes as a result of mRNA splicing. The potential of alternative mRNA splicing to increase protein diversity is most clearly illustrated by the extreme example of the fly axonal guidance gene Down syndrome cell adhesion molecule 1 (Dscam1), which is predicted to produce up to ∼38,000 possible alternative transcripts.Citation5
Pre-mRNA splicing allows increased protein diversity and cellular complexity between species and also provides the plasticity for one cell to alter its protein complement dynamically in response to cellular stress or developmental cues. As one would expect, the mechanisms of pre-mRNA splicing are tightly regulated to maintain cellular and tissue homeostasis, and errors in splicing underlie a host of genetic diseases and can contribute to cancer development and progression. In fact, it is estimated that 22% of disease causing mutations affect splicingCitation6 (for review of splicing defects resulting in disease, see:Citation7-9). Although there are many dozens of splicing factors, many of which are serine arginine (SR)-rich, ostensibly their functions in splicing are regulated by several serine/threonine kinases. These kinases share a general preference for phosphorylating SR-rich proteins and collectively are referred to as SR protein specific kinases, or simply splicing kinases. Therefore, it is perhaps not surprising to note that during evolution there appears to be a concomitant increase in the diversity and number of isoforms of these kinases. This occurs in lock-step with increasing gene complexity in terms of alternative splicing between single-cell eukaryotes like S. cerevisiae, which encode very few intron-containing genes and a single bona fide SR-protein kinase Sky1, to complex metazoans like humans, whose genome encodes many intron containing genes and multiple paralogs of at least 3 classes of splicing kinases (). In humans, the 3 classes of splicing kinases include the serine-arginine protein kinases (SRPK1/2/3), the CDC-like kinases (CLK1/2/3/4), and the pre-mRNA processing factor 4 kinase (PRP4K). Each class of splicing kinase has a distinct cellular localization, which may be based in part on their different roles in splicing regulation ().
Figure 1. Splicing kinase diversity across species. A table of the known and putative splicing kinases in baker's yeast (S. cerevisiae (Sc)), fission yeast (S. pombe (Sb), the worm C. elegans (Ce), the fly D. melanogaster (Dm) and humans (Hs), is shown on the left. On the right, a phylogenetic tree showing the evolutionary relationships between the various splicing kinase families and their homologs in yeast, worms, flies and in humans. The phylogenetic tree was created based on amino acid composition of the splicing homologs using the web resource: phylogeny.limrr.fr.
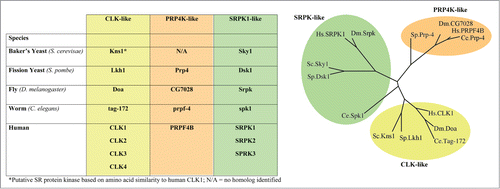
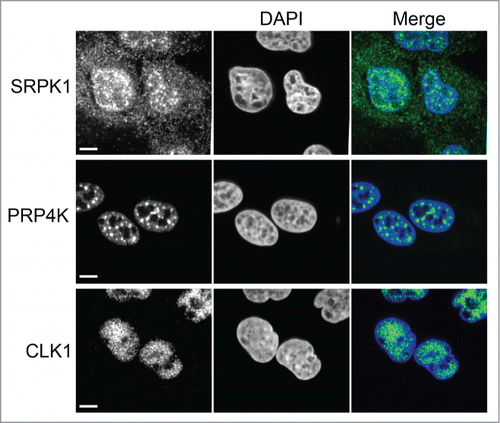
Figure 2. Splicing kinase cellular localization. Human osteosarcoma U2OS cells were analyzed by immunofluorescence confocal microscopy using an anti-SRPK1, anti-PRP4K or anti-CLK antibody (green). Nuclei were stained with DAPI (blue). Scale bar = 5 microns.
One of the first splicing kinases to be described in the literature is the SRSF protein kinase 1 (SRPK1), which was identified by Gui et al. in 1994 when the authors purified and cloned a cell cycle regulated kinase which was responsible for redistribution of SR proteins from a nuclear speckle localization in interphase cells, to a more ubiquitous nucleoplasm localization in mitotic cells.Citation10,11 SRPK2 and SRPK3 were later identified based on sequence homology with SRPK1.Citation12,13 SRPK2, much like SRPK1, was shown to regulate splicing through SR protein phosphorylationCitation12 while SRPK3 was identified for its role in normal muscle growth and homeostasis.Citation13
CDC-like kinase 1 (CLK1) was identified as a splicing kinase in 1996 when a yeast 2 hybrid screen using Clk/sty (Clk1) kinase as bait identified 5 SR proteins as binding partners.Citation14 The authors went on to show that one of the interacting SR proteins, ASF/SF2 (SRSF1), was phosphorylated within its RS domain by Clk/sty, and that overexpression of Clk/sty, much like SRPK1, caused a redistribution of SR proteins from nuclear speckles, to a ubiquitous nucleoplasm localization.Citation14
Pre-mRNA processing factor 4 kinase (PRP4K)(also known as PRPF4B), a lesser-known splicing kinase, was first linked to splicing in 1991 when a temperature sensitive library of Schizosaccharomyces pombe mutants were screened for splicing defects.Citation15 At the restrictive temperature, yeast carrying a temperature sensitive mutation in prp4 accumulated un-spliced pre-mRNA. Subsequent characterization of the prp4 gene revealed that the splicing factor encoded by the gene contained the characteristic sequence that defines a serine/threonine protein kinase, making prp4 the first kinase shown to play a role in splicing.Citation16 The mammalian homolog of prp4 (PRP4K) has been shown to interact with pre-mRNA splicing factors PRP6 and Suppressor-of-White-Apricot (SFSWAP/SRSF8) and copurify with the U5 snRNP.Citation17 Furthermore, PRP4K has been shown to be a key regulator of U4/U6-U5 tri-snRNP assembly through the phosphorylation of PRP6 and PRP31.Citation18
In humans, pre-mRNA splicing and the expression of the splicing kinases are perturbed in cancer. In this review, we will discuss the conserved roles of these kinases in pre-mRNA splicing and their emerging roles in tumorigenesis and treatment response.
Splicing kinases in spliceosomal assembly and mRNA splicing
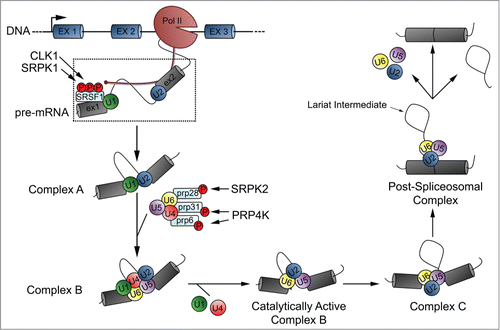
Transcribed pre-mRNA must be spliced to remove introns prior to nuclear export and translation. This process is carried out by the spliceosome; a large macromolecular machine composed of 5 small nuclear ribonucleic proteins (snRNPs) and numerous protein cofactors.Citation19 Spliceosome assembly is a complex, multistep process as illustrated in . The first step involves recognition of the 5′ and 3′ splice sites located on adjacent exons by U1 and U2 snRNP respectively; a process which is mediated by the C-terminal domain of polymerase II. Binding of the U1 snRNP to the 5′ splice site is mediated by SRSF1, but only when the RS-domain of SRSF1 is hyper-phosphorylated by CLK1 and SRPK1.Citation20,21 Once U1 and U2 snRNP have bound their target splice site, they interact with each other to form the pre-spliceosome (complex A). The next step in assembly involves the binding of pre-assembled U4/U6-U5 tri-snRNP to the pre-spliceosome to form complex B; a reaction catalyzed by pre-mRNA processing factors PRP28, PRP6 and PRP31, among others. PRP28 association with the tri-snRNP is dependent on its phosphorylation by SRPK2Citation22 while PRP6 and PRP31 association is dependent on their phosphorylation by PRP4K.Citation17,18 Loss of these phosphorylation events have been shown to inhibit association of the pre-mRNA processing factor with the tri-snRNP and, ultimately, tri-snRNP association with complex A.18 Complex B next undergoes a series of rearrangements resulting in the release of U1 and U4 snRNPs, creating a catalytically active complex B. Once catalytically active, the complex carries out the first of 2 splicing reactions, to form complex C containing free exon from the 5′ splice site, and the intron-exon lariat intermediate from the 3′ splice site. Complex C, after a series of rearrangements, carries out the second splicing reaction resulting in a post-spliceosomal complex containing the 2 spliced exons and the lariat intron. Finally, the remaining U2, U5 and U6 snRNPs are released from the transcript to be re-used in additional rounds of splicing.
Figure 3. The role of splicing kinases in spliceosome assembly and pre-mRNA splicing. Splicing kinases are involved in 2 distinct steps in spliceosome assembly. First, CLK1 and SRPK1 are required to hyper-phosphorylate SRSF1. This is essential for SRSF1 binding to the 5′ splice site which, in turn, recruits the U1 snRNP. Second, SRPK2 and PRP4K are required to phosphorylate PRP28 and PRP31/PRP6 respectively. These phosphorylation events are required to mediate tri-snRNP association with complex A.
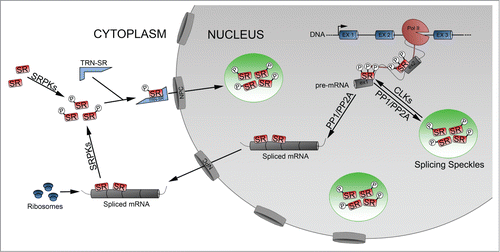
Splicing kinases in SR protein shuttling
SR proteins are a family of RNA binding proteins containing a characteristic arginine/serine rich domain (RS domain).Citation23,24 The first SR proteins (SRSF1 and SRSF2) were identified as essential regulators of constitutive splicing for their ability to promote U1 and U2 snRNP binding to the 5′ and 3′ splice site, respectively.Citation25-29 In addition to their role in constitutive splicing, SR-proteins play an essential role in the regulation of alternative splicing by promoting splice site selection through the binding of cis-acting splicing regulatory elements (SREs). SREs are classified as exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs) or intronic splicing silencers (ISSs). These designations depend on where in the pre-mRNA the SR protein is binding (exon or intron), and what the net effect of binding is on splicing (enhance or inhibit) (reviewed in Citation30-32). Adding even further to the complexity of splicing regulation, binding of SR proteins to SREs can be antagonized by the heterogeneous nuclear ribonucleoparticles (hnRNP) A/B family of proteins.Citation33 As a result, disruptions in the molar ratio of SR protein to hnRNP antagonist in the nucleus can have profound effects on splicing.Citation34-36
In addition to regulating U1 snRNP binding to the 5′ splice site and tri-snRNP assembly, splicing kinases are able to regulate splicing through the phosphorylation dependent shuttling of SR-proteins (). Unphosphorylated SR proteins in the cytoplasm are phosphorylated by SRPK1 and targeted for nuclear import via the SR protein import receptor, transportin-SR (TRN-SR; also known as transportin-SR2 (TRN-SR2) and transportin 3 (TNPO3)).Citation37,38 Once in the nucleus phosphorylated SR proteins become enriched in interchromatin granules called splicing speckle domains. To be recruited from speckles to nascent pre-mRNA, where they act to regulate splicing, SR proteins require additional phosphorylation eventsCitation39 which are mediated by the splicing kinase CLK1.Citation40 Studies suggest that the extent to which SR proteins are phosphorylated can impact their cellular function. For example, hypo-phosphorylation of the RS domain of SRSF1 promotes interaction with its own RNA recognition domain (RRM), while hyper-phosphorylation releases the RS domain and promotes RRM binding with the U1snRNP, promoting spliceosomal assembly.Citation21 Therefore, step-wise phosphorylation mediated by SRPK1 and CLK1 is essential in the regulation of splicing. Once splicing is complete hyper-phosphorylated SR proteins bound to mRNA are dephosphorylated by nuclear phosphatases (protein phosphatase 1 (PP1) and 2A (PP2A)) and either recycled to the cytoplasm as a chaperone for mRNA exportCitation41,42 where they also play a role in regulating translation of specific transcriptsCitation43 or re-phosphorylated and returned to speckles to await the next round of splicing. Thus, the splicing kinases connect the cell biology of splicing speckles domains to the biochemistry of pre-mRNA splicing by providing a mechanism for the shuttling of splicing factors to and from these domains to sites of splicing through reversible phosphorylation events.
Regulation of SR Protein Phosphorylation
SR-protein phosphorylation by the splicing kinases plays an essential role in splice site selection, subcellular localization, mRNA transport and translation, as described above. Regulation of these phosphorylation events is mediated in part by the distinct cellular localization and substrate specificity of the splicing kinases. For example, the RS domain of SR-protein SRSF1 contains an N-terminal stretch of Arg-Ser repeats (termed “RS1”) and a C-terminal stretch of Ser-Pro repeats (termed “RS2”). Cytoplasmic SRPK1 phosphorylates RS1 at multiple sites using a directional and processive mechanism. An acidic docking grove distal to the active site of SRPK1 binds RS1 leading to the “priming” phosphorylation of a single site in RS2. This site then binds a basic site within SRPK1 which serves to advance the RS repeat sequence through the docking grove and toward the kinase domain resulting in sequential phosphorylation events.Citation44 The phosphorylation of SRSF1 enhances the interaction with TRN-SR allowing it to transport SRSF1 into the nucleus where it assembles into nuclear speckles.Citation40,44 Nuclear CLK1 then phosphorylates RS2 causing SRSF1 dispersion from speckles and changes in alternative splicing.Citation14,45 CLK1 substrate specificity has been shown to be mediated, at least in part, by nature and extent of CLK1 autophosphorylation. It has been shown that CLK1 specificity for SRSF1 phosphorylation is sensitive to Tyr, but not Ser/Thr autophospharylation whereas its specificity for SC35 displays the opposite pattern.Citation46 In this example, sequential phosphorylation of SRSF1 by SRPK1 and CLK1 is regulated both by the cellular localization of the splicing kinases and by the unique specificity of SRPK1 for structural features within SRSF1 and of CLK1 for Ser-Pro dipeptide phosphorylation.Citation47 Interestingly, PRP4K has also been shown to phosphorylate SRSF1 in vitro,Citation48 is nuclear and resides predominately in splicing speckle domains (), suggesting PRP4K may too be involved in the sequential phosphorylation of SRSF1.
Alternative splicing in tumorigenesis
Alternative splicing occurs in an estimated 95% of human gene transcripts enhancing transcriptome complexity and proteome diversity in higher eukaryotes.Citation49 The most frequent alternative splicing event is the choice to include or skip an exon, termed a cassette exon. Other events involve the inclusion of one of 2 mutually exclusive exons, the use of alternative 3′ and 5′ splice sites, intron retention and the use of alternative promoters or poly(A) sites.Citation50 Alternative splicing also occurs in the 3′ and 5′ untranslated regions (UTR) of mRNA which can alter mRNA stability and/or translation efficiency.Citation51 Dysregulation of alternative splicing can render an mRNA transcript inactive by introducing a stop codon, alter the protein-coding function or even result in the transcript encoding a protein of opposing function. Given the striking ability through which alternative splicing can alter the proteome, it is perhaps not surprising that this system is frequently manipulated throughout the process of tumorigenesis.Citation52-54 In fact, changes in alternative splicing have been found to affect nearly every aspect of tumor biology including metabolism, apoptosis, cell cycle control, invasion, metastasis and angiogenesis (reviewed in Citation55-58).
While the molecular mechanism of alternative splicing has been shown to play a prominent role in tumorigenesis, there is limited knowledge of the regulation of these alternative splicing events observed in human cancers. One emerging mechanism of regulation is through altered expression of the splicing kinases, stemming from the observation that several of these kinases are overexpressed in various human cancers (SRPK1: breast,Citation59 colon,Citation59 pancreatic,Citation59 lung,Citation60 melanoma,Citation61 prostate.Citation62 SRPK2: acute myelogenous leukemia,Citation63 lung.Citation60 CLK1-4: erythroleukemia.Citation64 CLK2: breastCitation65). The remainder of this review will focus on the 3 families of splicing kinases and the specific roles they have been identified to play in tumorigenesis and the response to chemotherapy.
Splicing kinases and their role in tumorigenesis and therapeutic response to chemotherapy SRPK Family
SRPK1, perhaps the most widely studied splicing kinase, has been shown to directly regulate pathways essential to the development, growth and dissemination of cancer. A number of proteins have been shown to be directly phosphorylated by SRPK1 including SC35,Citation11 SRp20,Citation11 SRp55,Citation11 SRSF1,Citation11 Tra2β1Citation66 and RBM4.Citation67 Of these, the most well characterized with respect to tumorigenesis are SRSF1 and RBM4. SRSF1 is a prototypical member of the SR protein family which, in addition to its role in splicing regulation, has also been shown to regulate nuclear export,Citation42 mRNA stability,Citation68 miRNA processing,Citation69 translation,Citation43 and nonsense-mediated mRNA decay.Citation70 While the oncogenic potential of SRSF1 is likely due to a combination of the above mentioned functions, it is the splicing function which has been most extensively studied. Several SRPK1-SRSF1 mediated alternative splice events have been linked to tumorigenesis (reviewed in Citation71), but for the purpose of this review, we will focus on 3 splice events each effecting different aspects of tumorigenesis.
Rac1, a member of the Rho GTPase family, is involved in the regulation of cytoskeletal reorganization and cell motility.Citation72 An alternative splice variant of Rac1 (Rac1b) was identified in colorectal tumors that contain an alternative exon (3b) and is thought to maintain the GTPase in an active GTP-bound state.Citation73 Expression of Rac1b has been shown to induce epithelial-to-mesenchimal transition in cultured cells,Citation74 induce cell cycle progression and promote survival under conditions of serum-starvation.Citation75 Importantly, SRSF1 has been shown to promote inclusion of exon 3b in RAC1 pre-mRNACitation76; an event which requires SRPK1 mediated phosphorylation to shuttle SRSF1 to the nucleus. Consequently, knockdown of SRPK1 has been shown to decrease SRSF1 shuttling to the nucleus and reduce expression of the oncogenic Rac1b splice variant.Citation77
SRPK1-SRSF1 mediated alternative splicing has also been shown to regulate angiogenesis, the formation of new blood vessels, via vascular endothelial growth factor (VEGF) splicing control. VEGF alternative splicing leads to anti-angiogenic and pro-angiogenic mRNA isoforms which differ in the 3′ splice site of exon 8. Splicing at the proximal 3′ splice site results in pro-angiogenic splice isoforms (including VEGF165)Citation78 while splicing at the distal 3′ splice site gives rise to a family of isoforms with anti-angiogenic properties (including VEGF165b).Citation79 SRSF1 binds VEGF pre-mRNA in a region near the proximal 3′ splice site of exon 8, promoting use of this splice site, resulting in increased production of the pro-angiogenic VEGF splice isoforms.Citation80,81 Much like in Rac1 alternative splicing, SRSF1 binding to VEGF pre-mRNA is dependent on its nuclear localization, mediated by SRPK1 phosphorylation. Interestingly, recent studies have taken this model one step further to show that in Denys Drash Syndrome podocytes, mutations in the WT1 tumors suppressor gene prevents WT1-mediated transcriptional repression of SRPK1. Up-regulation of SRPK1 led to increased hyper-phosphorylation, and nuclear translocation, of SRSF1 which pushed the VEGF165/VEGF165b ratio in favor of the pro-angiogenic VEGF165, resulting in the formation of highly vascularized tumors using a colorectal tumor xenograft model.Citation82 Importantly, inhibition of SRPK1 using the small molecule inhibitors SRPIN340 or SPHINX increased expression of the anti-angiogenic VEGF165b isoform in colorectalCitation82 and prostateCitation62 cancer cell lines in vitro, while SPHINX treatment in a mouse model of prostate cancer led to smaller tumors with decreased microvessel density.Citation62 These studies indicate that small molecule inhibition of SRPK1 may prove to be an effective anti-angiogenic therapy for some cancers.
The Myeloid Cell Leukemia 1 (MCL1) gene is a member of the BCL2 family of apoptosis regulating genes and was first identified in differentiating myeloid leukemia cells.Citation83 MCL1 has 2 alternatively spliced variants, the full-length MCL1L which is anti-apoptotic, and an exon 2 skipped variant, MCL1S, which can form a dimer with, and antagonize, the anti-apoptotic effects of MCL1L.Citation84 In breast cancer, SRSF1 expression has been shown to increase stability and translational efficiency of the anti-apoptotic MCL1L isoform.Citation85 While it is likely that SRPK1 is mediating the nuclear localization of SRSF1and thus promoting splicing of the anti-apoptotic MCL1L in a manner similar to that seen during RAC1 and VEGF splicing regulation, another mechanism of SRPK1 regulation has also been proposed. A recent study has shown that phosphorylation of RBM4 by SRPK1 targets RNA binding protein RBM4 to the cytoplasm, inhibiting its binding to MCL1 and preventing an exon 2 skipping event.Citation86 Therefore, in the case of MCL1, SRPK1 may promote splicing of the anti-apoptotic isoform though at least 2 distinct mechanisms.
In addition to playing several roles in tumorigenesis, SRPK1 is also implicated in the therapeutic response to cisplatin. Cisplatin is a platinum-based chemotherapy drug, among the most commonly used to target human cancers. Intrinsic or acquired cellular resistance to cisplatin is common, limiting the therapeutic efficacy and requiring increasing doses of drug to treat recurring cancers. Cisplatin resistance is correlated with down-regulated SRPK1 expression in testicular germ cell tumors and ovarian cancers.Citation87,88 Furthermore, silencing of SRPK1 induces cisplatin sensitivity in multiple epithelial cell types including colon, breast, pancreatic and ovarian cancers and is accompanied by increased apoptosis, reduced cell proliferation, slower cell cycle progression and decreased anchorage-dependent growth in vitro.Citation59,89
While SRPK1 has received the most attention for its role in tumorigenesis, SRPK3 has recently been described to promote tumorigenicity in rhabdomyosarcoma (RMS) as a regulator of MEF2C alternative splicing.Citation90 MEF2C, a member of the myocyte enhancer factor 2 (MEF2) family of proteins, plays a key role in synaptic formation and muscle differentiation.Citation91 MEF2C has 3 alternative splice variants which appear to perform distinct functions in myogenesis and neurogenesis.Citation92-94 In particular, MEF2Cα2, the isoform containing the alternative α2 exon, has been shown to be required for differentiation of skeletal muscle cells and is frequently downregulated in RMS cells.Citation95 It has recently been demonstrated that SRPK3, which has been shown to be upregulated during myogenesis,Citation13 is required for the isoform switch between MEF2Cα1 and MEF2Cα2. In RMS, SRPK3 is down-regulated preventing the isoform switch and failure of myogenic precursors to differentiate into normal muscle.Citation90
The body of evidence surrounding the SRPK family of splicing kinases in tumorigenesis has made it clear that alterations in SR protein phosphorylation can have a significant impact on cancer development. As a result, recent studies have begun to focus on other splicing kinases to determine their possible roles in tumourigenesis and/or therapeutic response.
CLK Family
The splicing factor 45 (SPF45), first identified as a member of the spliceosome complex,Citation96 is known to promote exon 6 skipping in Fas pre-mRNA.Citation97 This exon encodes the transmembrane domain of the Fas death receptor, and its deletion results in the formation of a soluble Fas protein molecule.Citation98 Interestingly, expression of the soluble Fas molecule has been shown to prevent Fas mediated cell death, presumably by binding to Fas ligand (FasL), preventing FasL from binding to membrane-bound Fas and activating the apoptotic pathway. Given that evasion of apoptosis is a hallmark of cancer, it is not surprising that elevated levels of soluble Fas have been found in a variety of cancers.Citation99,100 A recent study has shown that CLK1 directly phosphorylates SPF45 on 8 serine residues, and that this phosphorylation led to the stabilization of SPF45 protein levels, and regulated exon 6 skipping in Fas pre-mRNA.Citation101 Furthermore, SPF45 overexpression induced cell migration and invasion in ovarian cancer cells,Citation101 suggesting CLK1 mediated stabilization of SPF45 could impact multiple aspects of tumor progression.
CLK2, a member of the CLK family of splicing kinases, has recently been shown to function as an oncogene in breast cancer.Citation65 A lentiviral shRNA cell viability screen was carried out that targeted 26 genes which the authors found to be commonly overexpressed or amplified in breast tumors or breast cancer cell lines based on SNP array analysis.Citation65 Of the 26 targets, knock-down of CLK2 was identified for its ability to inhibit breast cancer cell growth in vitro, and later confirmed to inhibit tumorigenesis in a mouse xenograft model. However, the knock-down of CLK2 was also shown to promote metastasis and invasion of breast cancer cells in vivo by inducing alternative splice patterns characteristic of the epithelial-to-mesenchymal transition (EMT). Specifically, knockdown of CLK2 was shown to regulate the alternative splicing of ENAH, an actin cytoskeletal protein, which contains a small coding exon (11a) which is included in the mRNA of epithelial cells, but excluded in mesenchymal cells.Citation65 Loss of CLK2 promoted the exclusion of exon 11a. Thus, CLK2 expression levels could represent an important marker for EMT during breast cancer progression.
PRP4K
PRP4K, the least studied of the 3 splicing kinases, has recently begun to emerge as an important regulator of therapeutic response. In addition to its role in splicing, PRP4K has been implicated in the regulation of mitosis as expression of a dominant truncated form of PRP4K in S. pombe was shown to induce mitotic aberrations.Citation102 Consequently, mammalian PRP4K was shown to be a regulator of the mitotic spindle assembly checkpoint (SAC) through its ability to recruit checkpoint proteins MPS1, MAD1 and MAD2 to the kinetochore.Citation103 This finding has important implications for therapeutic response to taxanes, a family of anti-cancer agents that depend on SAC activity for cell killing.Citation104 The taxanes (docetaxel, paclitaxel and cabazitaxel) function by binding to and stabilizing mictrotubules, resulting in a disruption of microtubule dynamics. As a cell progresses through mitosis, taxanes inhibit the ability of sister chromatids to properly segregate which triggers activation of the SAC and arrests the cell in prometaphase.Citation105 If the checkpoint cannot be satisfied, its prolonged activation will result in mitotic cell death.Citation106 Not surprisingly, disruption of SAC function has been has been shown to increase cellular resistance to taxanes in a number of different cancer models.Citation107-112 Consistent with these models, we have shown that PRP4K functions downstream of the receptor tyrosine kinase HER2 to regulate paclitaxel response in breast and ovarian cancer, presumably by altering SAC activity, and that its expression is decreased in ovarian cancer patients that have relapsed from taxane treatment.Citation113 Importantly, we also demonstrated that, among ovarian cancer patients with low HER2 expressing tumors, PRP4K expression can be used as a predictive marker to identify patients likely to benefit from taxane therapy.Citation113
In addition to promoting resistance to microtubule targeting chemotherapeutics, disruptions in the SAC has been suggested to facilitate tumorigenesis by inducing chromosomal instability (CIN), a hallmark of human neoplasia.Citation114,115 Under normal conditions, the SAC becomes activated in response to one or more unattached chromosomes, blocking progression to anaphase. When checkpoint components are mutated, or expression levels decreased, un-attached chromosomes are unable to activate the SAC and become mis-segregated, leading to increased aneuploidy.Citation116-118 While it is still unclear as to whether or not a weakened checkpoint is sufficient to drive tumorigenesis, there is significant evidence suggesting it can facilitate tumorigenesis, especially in collaboration with a weakened tumor suppressor such as a BRCA2 deficiency.Citation119,120 Therefore, it is tempting to speculate that loss of PRP4K expression would not only increase cellular resistance to taxanes, but could also facilitate tumorigenesis by inducing CIN.
PRP4K, in addition to its role in regulating taxane response, has also been shown to regulate the cellular response to ionizing radiation (IR) and the phytochemical curcumin.Citation121,122 In the human colorectal carcinoma cell line HCT-15, treatment with curcumin or IR led to a decrease in PRP4K expression and an increase in reactive oxygen species (ROS) production; a well-known mediator of apoptosis (reviewed in Citation123). Importantly, overexpression of PRP4K was shown to prevent ROS production and provide cellular protection from apoptosis in response to both treatments, possibly through the activation of an anti-oxidant enzyme system.Citation121,122 This data suggests that tumors with high PRP4K expression may show increased resistance to radiation therapy due to increased ROS scavenging. With both increased and decreased PRP4K expression shown to be associated with resistance to different anti-cancer therapies, knowing PRP4K expression levels at the time of diagnosis may prove to be useful in choosing an appropriate treatment modality.
Regulation of tumorigenic alternative splice events by other kinases
While this review focuses on the classic SR-protein kinases, it is important to note that they are not the only kinases involved in the regulation of alternative splicing with implications in tumorigenesis. For example, the RNA binding protein SAM68 (Src-associated in mitosis, with a molecular weight of 68 kD) has been shown to regulate a number of alternative splice events with implications in tumor progression and metastasis. This includes the splicing of cyclin D1 (CCND1), a proto-oncogene frequently deregulated in human cancers.Citation124,125 Alternative splicing of the CCND1 gene produces a variant transcript (CyclinD1b) which retains intron 4.Citation126 CyclinD1b displays increased oncogenic potentialCitation126,127 and its upregulation correlates with poor prognosis in multiple types of tumors.Citation128 Retention of intron 4 has been shown to be mediated by SAM68 which binds the proximal region of intron 4, inhibiting U1snRNP recruitment.Citation129 Importantly, the phosphorylation of SAM68 by Erk1/2 increases its binding affinity to the CCND1 transcript enhancing intron 4 retention.Citation129 SAM68 has also been implicated in the regulation of EMT through its ability to repress alternative-splicing activated nonsense-mediated mRNA decay of SRSF1.Citation130 SAM68 promotes the retention of intron 4 in SRSF1 pre-mRNA which inhibits its degradation by nonsense-mediated decay and increases SRSF1 protein levels. Increased SRSF1 impacts a number of tumorigenic alternative splice events, including stimulating the skipping of exon 11 in the proto-onocgene RON, producing a constitutively active isoform that promotes an invasive cellular phenotype.Citation131 While SAM68 phosphorylation by Erk1/2 provides an example of tumorigenic alternative splicing regulated by non-classical SR-protein specific kinases, numerous other kinases have been shown to phosphorylate splice factors with implications in cancer (reviewed in Citation132).
Concluding Remarks
In light of recent evidence implicating the splicing kinases as major regulators of tumorigenic alternative splice events, drugs which modulate splicing kinase activity are actively being studied as potential anti-cancer agents. As an example, SRPK1 small molecule inhibitors have already been shown to promote the splicing of the anti-angiogenic VEGF isoform in vitro and in vivo in prostate cancer cells which have elevated SRPK1 and SRSF1 expression.Citation62 It has yet to be determined if SRPK1 inhibition alters other tumorigenic splice events like Rac1 and MCL−1, but the potential for a single drug to target multiple aspects of tumorigenesis holds promise. Of course, SRPK1-mediated regulation is not limited to pro-tumorigenic pre-mRNA splicing events, raising concern over potential “off-target” effects of SRPK1 inhibition. High throughput next generation sequencing will allow for a more comprehensive understanding of changes to the transcriptome in response to SRPK1 inhibition, and provide insight into these potential off-target effects. In fact, an understanding of transcriptome changes in response to alterations in all splicing kinases would help identify which kinase, or combination of kinases, represents the best therapeutic target, which kinases to avoid due to potential off target effects, and the degree of redundancy between kinases which could be a potential mechanism of resistance for splicing kinase inhibition.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was funded by a Canadian Breast Cancer Foundation (CBCF)-Atlantic operating grant awarded to GD. GD is a Senior Scientist of the Beatrice Hunter Cancer Research Institute (BHCRI), ACH was supported by a postdoctoral fellowship from CBCF-Atlantic, and DPC was supported by a CIBC Graduate Scholarship in Medical Research trainee award from the BHCRI with funds provided by CBCF – Atlantic, and The Canadian Cancer Society, Nova Scotia Division as part of The Terry Fox Foundation Strategic Health Research Training (STIHR) Program in Cancer Research at the Canadian Institutes of Health Research (CIHR).
References
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science 1996; 274:546, 63-7; http://dx.doi.org/10.1126/science.274.5287.546
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature 2001; 409:860-921; PMID:11237011; http://dx.doi.org/10.1038/35057062
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science 2000; 287:2185-95; PMID:10731132; http://dx.doi.org/10.1126/science.287.5461.2185
- Consortium CeS. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 1998; 282:2012-8; PMID:9851916; http://dx.doi.org/10.1126/science.282.5396.2012
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 2000; 101:671-84; PMID:10892653; http://dx.doi.org/10.1016/S0092-8674(00)80878-8
- Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci U S A 2011; 108:11093-8; PMID:21685335; http://dx.doi.org/10.1073/pnas.1101135108
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell 2009; 136:777-93; PMID:19239895; http://dx.doi.org/10.1016/j.cell.2009.02.011
- Havens MA, Duelli DM, Hastings ML. Targeting RNA splicing for disease therapy. Wiley interdisciplinary Rev RNA 2013; 4:247-66; PMID:23512601; http://dx.doi.org/10.1002/wrna.1158
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Gen Dev 2003; 17:419-37; PMID:12600935; http://dx.doi.org/10.1101/gad.1048803
- Gui JF, Tronchere H, Chandler SD, Fu XD. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc Natl Acad Sci U S A 1994; 91:10824-8; PMID:7526381; http://dx.doi.org/10.1073/pnas.91.23.10824
- Gui JF, Lane WS, Fu XD. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 1994; 369:678-82; PMID:8208298; http://dx.doi.org/10.1038/369678a0
- Wang HY, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu XD. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol 1998; 140:737-50; PMID:9472028; http://dx.doi.org/10.1083/jcb.140.4.737
- Nakagawa O, Arnold M, Nakagawa M, Hamada H, Shelton JM, Kusano H, Harris TM, Childs G, Campbell KP, Richardson JA, et al. Centronuclear myopathy in mice lacking a novel muscle-specific protein kinase transcriptionally regulated by MEF2. GenDev 2005; 19:2066-77; PMID:16140986; http://dx.doi.org/10.1101/gad.1338705
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J 1996; 15:265-75; PMID:8617202
- Rosenberg GH, Alahari SK, Kaufer NF. prp4 from Schizosaccharomyces pombe, a mutant deficient in pre-mRNA splicing isolated using genes containing artificial introns. Mol Gen Genet 1991; 226:305-9; http://dx.doi.org/10.1007/BF00273617
- Alahari SK, Schmidt H, Kaufer NF. The fission yeast prp4+ gene involved in pre-mRNA splicing codes for a predicted serine/threonine kinase and is essential for growth. Nucleic Acids Res 1993; 21:4079-83; PMID:8371982; http://dx.doi.org/10.1093/nar/21.17.4079
- Dellaire G, Makarov EM, Cowger JM, Longman D, Sutherland HGE, Luhrmann R, Torchia J, Bickmore WA. Mammalian PRP4 Kinase Copurifies and Interacts with Components of Both the U5 snRNP and the N-CoR Deacetylase Complexes. Mol Cell Biol 2002; 22:5141-56; PMID:12077342; http://dx.doi.org/10.1128/MCB.22.14.5141-5156.2002
- Schneider M, Hsiao HH, Will CL, Giet R, Urlaub H, Luhrmann R. Human PRP4 kinase is required for stable tri-snRNP association during spliceosomal B complex formation. Nat structural Mol Biol 2010; 17:216-21; PMID:20118938; http://dx.doi.org/10.1038/nsmb.1718
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009; 136:701-18; PMID:19239890; http://dx.doi.org/10.1016/j.cell.2009.02.009
- Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci U S A 1997; 94:1148-53; PMID:9037021; http://dx.doi.org/10.1073/pnas.94.4.1148
- Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci U S A 2011; 108:8233-8; PMID:21536904; http://dx.doi.org/10.1073/pnas.1017700108
- Mathew R, Hartmuth K, Mohlmann S, Urlaub H, Ficner R, Luhrmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat StructMol Biol 2008; 15:435-43; PMID:18425142; http://dx.doi.org/10.1038/nsmb.1415
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. GenDev 1992; 6:837-47; PMID:1577277; http://dx.doi.org/10.1101/gad.6.5.837
- Fu XD. The superfamily of arginine/serine-rich splicing factors. Rna 1995; 1:663-80; PMID:7585252
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 1990; 343:437-41; PMID:2137203; http://dx.doi.org/10.1038/343437a0
- Ge H, Zuo P, Manley JL. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell 1991; 66:373-82; PMID:1855257; http://dx.doi.org/10.1016/0092-8674(91)90626-A
- Fu XD, Maniatis T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc Natl Acad Sci U S A 1992; 89:1725-9; PMID:1531875; http://dx.doi.org/10.1073/pnas.89.5.1725
- Krainer AR, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 1991; 66:383-94; PMID:1830244; http://dx.doi.org/10.1016/0092-8674(91)90627-B
- Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 1994; 368:119-24; PMID:8139654; http://dx.doi.org/10.1038/368119a0
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 2005; 6:386-98; PMID:15956978; http://dx.doi.org/10.1038/nrm1645
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. Rna 2008; 14:802-13; PMID:18369186; http://dx.doi.org/10.1261/rna.876308
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Ann Rev Biochem 2003; 72:291-336; PMID:12626338; http://dx.doi.org/10.1146/annurev.biochem.72.121801.161720
- Caceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 1994; 265:1706-9; PMID:8085156; http://dx.doi.org/10.1126/science.8085156
- Hanamura A, Caceres JF, Mayeda A, Franza BR, Jr., Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. Rna 1998; 4:430-44; PMID:9630249
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 1992; 68:365-75; PMID:1531115; http://dx.doi.org/10.1016/0092-8674(92)90477-T
- Yang X, Bani MR, Lu SJ, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci U S A 1994; 91:6924-8; PMID:8041722; http://dx.doi.org/10.1073/pnas.91.15.6924
- Kataoka N, Bachorik JL, Dreyfuss G. Transportin-SR, a nuclear import receptor for SR proteins. J Cell Biol 1999; 145:1145-52; PMID:10366588; http://dx.doi.org/10.1083/jcb.145.6.1145
- Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc Natl Acad Sci U S A 2001; 98:10154-9; PMID:11517331; http://dx.doi.org/10.1073/pnas.181354098
- Misteli T, Caceres JF, Clement JQ, Krainer AR, Wilkinson MF, Spector DL. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol 1998; 143:297-307; PMID:9786943; http://dx.doi.org/10.1083/jcb.143.2.297
- Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol Cell 2005; 20:77-89; PMID:16209947; http://dx.doi.org/10.1016/j.molcel.2005.08.025
- Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell 2001; 7:899-905; PMID:11336712; http://dx.doi.org/10.1016/S1097-2765(01)00233-7
- Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell 2003; 11:837-43; PMID:12667464; http://dx.doi.org/10.1016/S1097-2765(03)00089-3
- Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. GenDev 2004; 18:755-68; PMID:15082528; http://dx.doi.org/10.1101/gad.286404
- Ngo JC, Giang K, Chakrabarti S, Ma CT, Huynh N, Hagopian JC, Dorrestein PC, Fu XD, Adams JA, Ghosh G. A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol Cell 2008; 29:563-76; PMID:18342604; http://dx.doi.org/10.1016/j.molcel.2007.12.017
- Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, Sumi K, Yomoda J, Murray MV, Kimura H, et al. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Bio Chem 2004; 279:24246-54; PMID:15010457; http://dx.doi.org/10.1074/jbc.M314298200
- Prasad J, Manley JL. Regulation and substrate specificity of the SR protein kinase Clk/Sty. Mol Cell Biol 2003; 23:4139-49; PMID:12773558; http://dx.doi.org/10.1128/MCB.23.12.4139-4149.2003
- Aubol BE, Plocinik RM, Hagopian JC, Ma CT, McGlone ML, Bandyopadhyay R, Fu XD, Adams JA. Partitioning RS domain phosphorylation in an SR protein through the CLK and SRPK protein kinases. J Mol Biol 2013; 425:2894-909; PMID:23707382; http://dx.doi.org/10.1016/j.jmb.2013.05.013
- Kojima T, Zama T, Wada K, Onogi H, Hagiwara M. Cloning of human PRP4 reveals interaction with Clk1. J Biol Chem 2001; 276:32247-56; PMID:11418604; http://dx.doi.org/10.1074/jbc.M103790200
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40:1413-5; PMID:18978789; http://dx.doi.org/10.1038/ng.259
- Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 2010; 11:345-55; PMID:20376054; http://dx.doi.org/10.1038/nrg2776
- Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends in Genet : TIG 2006; 22:119-22; PMID:16430990; http://dx.doi.org/10.1016/j.tig.2006.01.001
- Sette C. Alternative splicing programs in prostate cancer. Int J cell Biol 2013; 2013:458727; PMID:23983695
- Lapuk A, Marr H, Jakkula L, Pedro H, Bhattacharya S, Purdom E, Hu Z, Simpson K, Pachter L, Durinck S, et al. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res : MCR 2010; 8:961-74; PMID:20605923; http://dx.doi.org/10.1158/1541-7786.MCR-09-0528
- Brosseau JP, Lucier JF, Nwilati H, Thibault P, Garneau D, Gendron D, Durand M, Couture S, Lapointe E, Prinos P, et al. Tumor microenvironment-associated modifications of alternative splicing. Rna 2014; 20:189-201; PMID:24335142; http://dx.doi.org/10.1261/rna.042168.113
- Venables JP. Aberrant and alternative splicing in cancer. Cancer Res 2004; 64:7647-54; PMID:15520162; http://dx.doi.org/10.1158/0008-5472.CAN-04-1910
- Ghigna C, Valacca C, Biamonti G. Alternative splicing and tumor progression. Curr Gen 2008; 9:556-70; PMID:19516963; http://dx.doi.org/10.2174/138920208786847971
- Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene 2014; 33:5311-8; PMID:24336324; http://dx.doi.org/10.1038/onc.2013.533
- Ladomery M. Aberrant alternative splicing is another hallmark of cancer. Int J cell Biol 2013; 2013:463786; PMID:24101931; http://dx.doi.org/10.1155/2013/463786
- Hayes GM, Carrigan PE, Miller LJ. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res 2007; 67:2072-80; PMID:17332336; http://dx.doi.org/10.1158/0008-5472.CAN-06-2969
- Gout S, Brambilla E, Boudria A, Drissi R, Lantuejoul S, Gazzeri S, Eymin B. Abnormal expression of the pre-mRNA splicing regulators SRSF1, SRSF2, SRPK1 and SRPK2 in non small cell lung carcinoma. PloS one 2012; 7:e46539; PMID:23071587; http://dx.doi.org/10.1371/journal.pone.0046539
- Gammons MV, Lucas R, Dean R, Coupland SE, Oltean S, Bates DO. Targeting SRPK1 to control VEGF-mediated tumour angiogenesis in metastatic melanoma. British J Cancer 2014; 111:477-85; PMID:25010863; http://dx.doi.org/10.1038/bjc.2014.342
- Mavrou A, Brakspear K, Hamdollah-Zadeh M, Damodaran G, Babaei-Jadidi R, Oxley J, Gillatt DA, Ladomery MR, Harper SJ, Bates DO, et al. Serine-arginine protein kinase 1 (SRPK1) inhibition as a potential novel targeted therapeutic strategy in prostate cancer. Oncogene 2014; PMID:25381816
- Jang SW, Yang SJ, Ehlen A, Dong S, Khoury H, Chen J, Persson JL, Ye K. Serine/arginine protein-specific kinase 2 promotes leukemia cell proliferation by phosphorylating acinus and regulating cyclin A1. Cancer Res 2014; 68:4559-70; PMID:18559500; http://dx.doi.org/10.1158/0008-5472.CAN-08-0021
- Garcia-Sacristan A, Fernandez-Nestosa MJ, Hernandez P, Schvartzman JB, Krimer DB. Protein kinase clk/STY is differentially regulated during erythroleukemia cell differentiation: a bias toward the skipped splice variant characterizes postcommitment stages. Cell Res 2005; 15:495-503; PMID:16045812; http://dx.doi.org/10.1038/sj.cr.7290319
- Yoshida T, Kim JH, Carver K, Su Y, Weremowicz S, Mulvey L, Yamamoto S, Brennan C, Mei S, Long H, et al. CLK2 Is an Oncogenic Kinase and Splicing Regulator in Breast Cancer. Cancer Res 2015; 75:1516-26; PMID:25670169; http://dx.doi.org/10.1158/0008-5472.CAN-14-2443
- Jamros MA, Aubol BE, Keshwani MM, Zhang Z, Stamm S, Adams JA. Intra-domain cross talk regulates serine-arginine protein kinase 1 dependent phosphorylation and splicing function of transformer 2 beta 1. J Biol Chem 2015; PMID:26013829; http://dx.doi.org/10.1074/jbc.M115.656579
- Lin JC, Hsu M, Tarn WY. Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proc Natl Acad Sci U S A 2007; 104:2235-40; PMID:17284590; http://dx.doi.org/10.1073/pnas.0611015104
- Lemaire R, Prasad J, Kashima T, Gustafson J, Manley JL, Lafyatis R. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. GenDev 2002; 16:594-607; PMID:11877379; http://dx.doi.org/10.1101/gad.939502
- Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell 2010; 38:67-77; PMID:20385090; http://dx.doi.org/10.1016/j.molcel.2010.02.021
- Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol Cell 2004; 16:597-607; PMID:15546619; http://dx.doi.org/10.1016/j.molcel.2004.10.031
- Das S, Krainer AR. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res 2014; 12:1195-204; PMID:24807918; http://dx.doi.org/10.1158/1541-7786.MCR-14-0131
- Jiang P, Enomoto A, Takahashi M. Cell biology of the movement of breast cancer cells: intracellular signalling and the actin cytoskeleton. Cancer Lett 2009; 284:122-30; PMID:19303207; http://dx.doi.org/10.1016/j.canlet.2009.02.034
- Jordan P, Brazao R, Boavida MG, Gespach C, Chastre E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene 1999; 18:6835-9; PMID:10597294; http://dx.doi.org/10.1038/sj.onc.1203233
- Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 2005; 436:123-7; PMID:16001073; http://dx.doi.org/10.1038/nature03688
- Matos P, Jordan P. Expression of Rac1b stimulates NF-kappaB-mediated cell survival and G1/S progression. Exp Cell Res 2005; 305:292-9; PMID:15817154; http://dx.doi.org/10.1016/j.yexcr.2004.12.029
- Goncalves V, Matos P, Jordan P. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum Mol Genet 2009; 18:3696-707; PMID:19602482; http://dx.doi.org/10.1093/hmg/ddp317
- Goncalves V, Henriques A, Pereira J, Neves Costa A, Moyer MP, Moita LF, Gama-Carvalho M, Matos P, Jordan P. Phosphorylation of SRSF1 by SRPK1 regulates alternative splicing of tumor-related Rac1b in colorectal cells. Rna 2014; 20:474-82; PMID:24550521; http://dx.doi.org/10.1261/rna.041376.113
- Houck KA, Ferrara N, Winer J, Cachianes G, Li B, Leung DW. The vascular endothelial growth factor family: identification of a fourth molecular species and characterization of alternative splicing of RNA. Mol Endocrinol 1991; 5:1806-14; PMID:1791831; http://dx.doi.org/10.1210/mend-5-12-1806
- Bates DO, Cui TG, Doughty JM, Winkler M, Sugiono M, Shields JD, Peat D, Gillatt D, Harper SJ. VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, is down-regulated in renal cell carcinoma. Cancer Res 2002; 62:4123-31; PMID:12124351
- Nowak DG, Woolard J, Amin EM, Konopatskaya O, Saleem MA, Churchill AJ, Ladomery MR, Harper SJ, Bates DO. Expression of pro- and anti-angiogenic isoforms of VEGF is differentially regulated by splicing and growth factors. J Cell Sci 2008; 121:3487-95; PMID:18843117; http://dx.doi.org/10.1242/jcs.016410
- Rennel ES, Varey AH, Churchill AJ, Wheatley ER, Stewart L, Mather S, Bates DO, Harper SJ. VEGF(121)b, a new member of the VEGF(xxx)b family of VEGF-A splice isoforms, inhibits neovascularisation and tumour growth in vivo. Br J cancer 2009; 101:1183-93; PMID:19707198; http://dx.doi.org/10.1038/sj.bjc.6605249
- Amin EM, Oltean S, Hua J, Gammons MV, Hamdollah-Zadeh M, Welsh GI, Cheung MK, Ni L, Kase S, Rennel ES, Symonds KE, et al. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell 2011; 20:768-80; PMID:22172722; http://dx.doi.org/10.1016/j.ccr.2011.10.016
- Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci U S A 1993; 90:3516-20; PMID:7682708; http://dx.doi.org/10.1073/pnas.90.8.3516
- Bae J, Leo CP, Hsu SY, Hsueh AJ. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J Biol Chem 2000; 275:25255-61; PMID:10837489; http://dx.doi.org/10.1074/jbc.M909826199
- Gautrey HL, Tyson-Capper AJ. Regulation of Mcl-1 by SRSF1 and SRSF5 in cancer cells. PloS one 2012; 7:e51497; PMID:23284704; http://dx.doi.org/10.1371/journal.pone.0051497
- Lin JC, Lin CY, Tarn WY, Li FY. Elevated SRPK1 lessens apoptosis in breast cancer cells through RBM4-regulated splicing events. Rna 2014; 20:1621-31; PMID:25140042; http://dx.doi.org/10.1261/rna.045583.114
- Schenk PW, Boersma AW, Brandsma JA, den Dulk H, Burger H, Stoter G, Brouwer J, Nooter K. SKY1 is involved in cisplatin-induced cell kill in Saccharomyces cerevisiae, and inactivation of its human homologue, SRPK1, induces cisplatin resistance in a human ovarian carcinoma cell line. Cancer Res 2001; 61:6982-6; PMID:11585720
- Schenk PW, Stoop H, Bokemeyer C, Mayer F, Stoter G, Oosterhuis JW, Wiemer E, Looijenga LH, Nooter K. Resistance to platinum-containing chemotherapy in testicular germ cell tumors is associated with downregulation of the protein kinase SRPK1. Neoplasia 2004; 6:297-301; PMID:15256051; http://dx.doi.org/10.1593/neo.03406
- Odunsi K, Mhawech-Fauceglia P, Andrews C, Beck A, Amuwo O, Lele S, Black JD, Huang RY. Elevated expression of the serine-arginine protein kinase 1 gene in ovarian cancer and its role in Cisplatin cytotoxicity in vitro. PloS one 2012; 7:e51030
- Zhang M, Zhu B, Davie J. Alternative splicing of MEF2C pre-mRNA controls its activity in normal myogenesis and promotes tumorigenicity in rhabdomyosarcoma cells. J Biol Chem 2015; 290:310-24; PMID:25404735; http://dx.doi.org/10.1074/jbc.M114.606277
- Potthoff MJ, Arnold MA, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol Cell Biol 2007; 27:8143-51; PMID:17875930; http://dx.doi.org/10.1128/MCB.01187-07
- Hakim NH, Kounishi T, Alam AH, Tsukahara T, Suzuki H. Alternative splicing of Mef2c promoted by Fox-1 during neural differentiation in P19 cells. Genes cells 2010; 15:255-67; http://dx.doi.org/10.1111/j.1365-2443.2009.01378.x
- Infantino V, Convertini P, Menga A, Iacobazzi V. MEF2C exon alpha: role in gene activation and differentiation. Gene 2013; 531:355-62; PMID:24008018; http://dx.doi.org/10.1016/j.gene.2013.08.044
- McDermott JC, Cardoso MC, Yu YT, Andres V, Leifer D, Krainc D, Lipton SA, Nadal-Ginard B. hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Mol Cell Biol 1993; 13:2564-77; PMID:8455629
- MacQuarrie KL, Yao Z, Fong AP, Diede SJ, Rudzinski ER, Hawkins DS, Tapscott SJ. Comparison of genome-wide binding of MyoD in normal human myogenic cells and rhabdomyosarcomas identifies regional and local suppression of promyogenic transcription factors. Mol Cell Biol 2013; 33:773-84; PMID:23230269; http://dx.doi.org/10.1128/MCB.00916-12
- Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet 1998; 20:46-50; PMID:9731529; http://dx.doi.org/10.1038/1700
- Corsini L, Bonnal S, Basquin J, Hothorn M, Scheffzek K, Valcarcel J, Sattler M. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat StructMol Biol 2007; 14:620-9; PMID:17589525; http://dx.doi.org/10.1038/nsmb1260
- Cheng J, Zhou T, Liu C, Shapiro JP, Brauer MJ, Kiefer MC, Barr PJ, Mountz JD. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science 1994; 263:1759-62; PMID:7510905; http://dx.doi.org/10.1126/science.7510905
- Kondera-Anasz Z, Mielczarek-Palacz A, Sikora J. Soluble Fas receptor and soluble Fas ligand in the serum of women with uterine tumors. Apoptosis 2005; 10:1143-9; PMID:16151647; http://dx.doi.org/10.1007/s10495-005-1018-9
- Sheen-Chen SM, Chen HS, Eng HL, Chen WJ. Circulating soluble Fas in patients with breast cancer. World J surgery 2003; 27:10-3; PMID:12557031; http://dx.doi.org/10.1007/s00268-002-6378-5
- Liu Y, Conaway L, Rutherford Bethard J, Al-Ayoubi AM, Thompson Bradley A, Zheng H, Weed SA, Eblen ST. Phosphorylation of the alternative mRNA splicing factor 45 (SPF45) by Clk1 regulates its splice site utilization, cell migration and invasion. Nucleic acids Res 2013; 41:4949-62; PMID:23519612; http://dx.doi.org/10.1093/nar/gkt170
- Gross T, Lutzelberger M, Weigmann H, Klingenhoff A, Shenoy S, Kaufer NF. Functional analysis of the fission yeast Prp4 protein kinase involved in pre-mRNA splicing and isolation of a putative mammalian homologue. Nucleic acids Res 1997; 25:1028-35; PMID:9102632; http://dx.doi.org/10.1093/nar/25.5.1028
- Montembault E, Dutertre S, Prigent C, Giet R. PRP4 is a spindle assembly checkpoint protein required for MPS1, MAD1, and MAD2 localization to the kinetochores. J Cell Biol 2007; 179:601-9; PMID:17998396; http://dx.doi.org/10.1083/jcb.200703133
- Wang X, Cheung HW, Chun AC, Jin DY, Wong YC. Mitotic checkpoint defects in human cancers and their implications to chemotherapy. Frontiers in bioscience : a journal and virtual library 2008; 13:2103-14; PMID:17981695; http://dx.doi.org/10.2741/2827
- Yu H. Regulation of APC-Cdc20 by the spindle checkpoint. Curr Opin Cell Biol 2002; 14:706-14; PMID:12473343; http://dx.doi.org/10.1016/S0955-0674(02)00382-4
- Vogel C, Hager C, Bastians H. Mechanisms of mitotic cell death induced by chemotherapy-mediated G2 checkpoint abrogation. Cancer Res 2007; 67:339-45; PMID:17210716; http://dx.doi.org/10.1158/0008-5472.CAN-06-2548
- Anand S, Penrhyn-Lowe S, Venkitaraman AR. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell 2003; 3:51-62; PMID:12559175; http://dx.doi.org/10.1016/S1535-6108(02)00235-0
- Noguchi S. Predictive factors for response to docetaxel in human breast cancers. Cancer Sci 2006; 97:813-20; PMID:16805818; http://dx.doi.org/10.1111/j.1349-7006.2006.00265.x
- Lens SM, Wolthuis RM, Klompmaker R, Kauw J, Agami R, Brummelkamp T, Kops G, Medema RH. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J 2003; 22:2934-47; PMID:12805209; http://dx.doi.org/10.1093/emboj/cdg307
- Chen JG, Yang CP, Cammer M, Horwitz SB. Gene expression and mitotic exit induced by microtubule-stabilizing drugs. Cancer Res 2003; 63:7891-9; PMID:14633718
- Sudo T, Nitta M, Saya H, Ueno NT. Dependence of paclitaxel sensitivity on a functional spindle assembly checkpoint. Cancer Res 2004; 64:2502-8; PMID:15059905; http://dx.doi.org/10.1158/0008-5472.CAN-03-2013
- Tian XP, Qian D, He LR, Huang H, Mai SJ, Li CP, Huang XX, Cai MY, Liao YJ, Kung HF, et al. The telomere/telomerase binding factor PinX1 regulates paclitaxel sensitivity depending on spindle assembly checkpoint in human cervical squamous cell carcinomas. Cancer Lett 2014; 353:104-14; PMID:25045845; http://dx.doi.org/10.1016/j.canlet.2014.07.012
- Corkery DP, Le Page C, Meunier L, Provencher D, Mes-Masson AM, Dellaire G. PRP4K is a HER2-regulated modifier of taxane sensitivity. Cell Cycle 2015; 14:1059-69; PMID:25602630; http://dx.doi.org/10.1080/15384101.2015.1007775
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature 1997; 386:623-7; PMID:9121588; http://dx.doi.org/10.1038/386623a0
- Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer 2005; 5:773-85; PMID:16195750; http://dx.doi.org/10.1038/nrc1714
- Weaver BA, Bonday ZQ, Putkey FR, Kops GJ, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol 2003; 162:551-63; PMID:12925705; http://dx.doi.org/10.1083/jcb.200303167
- Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol 2003; 160:341-53; PMID:12551952; http://dx.doi.org/10.1083/jcb.200211048
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 2001; 409:355-9; PMID:11201745; http://dx.doi.org/10.1038/35053094
- Lee H, Trainer AH, Friedman LS, Thistlethwaite FC, Evans MJ, Ponder BA, Venkitaraman AR. Mitotic checkpoint inactivation fosters transformation in cells lacking the breast cancer susceptibility gene, Brca2. Mol Cell 1999; 4:1-10; PMID:10445022; http://dx.doi.org/10.1016/S1097-2765(00)80182-3
- Rao CV, Yang YM, Swamy MV, Liu T, Fang Y, Mahmood R, Jhanwar-Uniyal M, Dai W. Colonic tumorigenesis in BubR1+/-ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci U S A 2005; 102:4365-70; PMID:15767571; http://dx.doi.org/10.1073/pnas.0407822102
- Shehzad A, Park JW, Lee J, Lee YS. Curcumin induces radiosensitivity of in vitro and in vivo cancer models by modulating pre-mRNA processing factor 4 (Prp4). Chemico-Biol Interact 2013; 206:394-402; PMID:24144778; http://dx.doi.org/10.1016/j.cbi.2013.10.007
- Shehzad A, Lee J, Huh TL, Lee YS. Curcumin induces apoptosis in human colorectal carcinoma (HCT-15) cells by regulating expression of Prp4 and p53. Mol Cell 2013; 35:526-32; PMID:23686430; http://dx.doi.org/10.1007/s10059-013-0038-5
- Buttke TM, Sandstrom PA. Oxidative stress as a mediator of apoptosis. Immunol Today 1994; 15:7-10; PMID:8136014; http://dx.doi.org/10.1016/0167-5699(94)90018-3
- Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene 2006; 25:1620-8; PMID:16550162; http://dx.doi.org/10.1038/sj.onc.1209371
- Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer 2011; 11:558-72; PMID:21734724; http://dx.doi.org/10.1038/nrc3090
- Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene 1995; 11:1005-11; PMID:7675441
- Li Z, Jiao X, Wang C, Shirley LA, Elsaleh H, Dahl O, Wang M, Soutoglou E, Knudsen ES, Pestell RG. Alternative cyclin D1 splice forms differentially regulate the DNA damage response. Cancer Res 2010; 70:8802-11; PMID:20940395; http://dx.doi.org/10.1158/0008-5472.CAN-10-0312
- Wang Y, Dean JL, Millar EK, Tran TH, McNeil CM, Burd CJ, Henshall SM, Utama FE, Witkiewicz A, Rui H, et al. Cyclin D1b is aberrantly regulated in response to therapeutic challenge and promotes resistance to estrogen antagonists. Cancer Res 2008; 68:5628-38; PMID:18632615; http://dx.doi.org/10.1158/0008-5472.CAN-07-3170
- Paronetto MP, Cappellari M, Busa R, Pedrotti S, Vitali R, Comstock C, Hyslop T, Knudsen KE, Sette C. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res 2010; 70:229-39; PMID:20028857; http://dx.doi.org/10.1158/0008-5472.CAN-09-2788
- Valacca C, Bonomi S, Buratti E, Pedrotti S, Baralle FE, Sette C, Ghigna C, Biamonti G. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J Cell Biol 2010; 191:87-99; PMID:20876280; http://dx.doi.org/10.1083/jcb.201001073
- Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell 2005; 20:881-90; PMID:16364913; http://dx.doi.org/10.1016/j.molcel.2005.10.026
- Naro C, Sette C. Phosphorylation-mediated regulation of alternative splicing in cancer. Int J Cell Biol 2013; 2013:151839; PMID:24069033; http://dx.doi.org/10.1155/2013/151839