ABSTRACT
Nuclear lamins are the main components of the nuclear lamina at the nuclear periphery, providing mechanical support to the nucleus. However, recent findings suggest that lamins also reside in the nuclear interior, as a distinct and dynamic pool with critical roles in transcriptional regulation. In our work we found a functional and evolutionary conserved crosstalk between Lamin A/C and the Polycomb group (PcG) of proteins, this being required for the maintenance of the PcG repressive functions. Indeed, Lamin A/C knock-down causes PcG foci dispersion and defects in PcG-mediated higher order structures, thereby leading to impaired PcG mediated transcriptional repression. By using ad-hoc algorithms for image analysis and PLA approaches we hereby show that PcG proteins are preferentially located in the nuclear interior where they interact with nucleoplasmic Lamin A/C. Taken together, our findings suggest that nuclear components, such as Lamin A/C, functionally interact with epigenetic factors to ensure the correct transcriptional program maintenance.
Introduction
The nuclear lamina, the inner part of the nuclear envelope (NE), is made up of a complex meshwork of proteins, collectively called lamins, which provide the mechanical support to the nucleus and the entire cell.Citation1 Lamins are classified as type V filament proteins and in vertebrates are divided into A and B types, based on sequence homologies. In particular, Lamin A and C, the major isoforms of A-type lamins, are encoded by the LMNA gene as alternative splice variants, while LMNB1 and LMNB2 genes encode the 2 major B-type lamins: Lamin B1 and Lamin B2, respectively.Citation1
Intriguingly, in the last few years, in addition to their well-established role in structural stability, lamins have been found to be involved in several cellular processes from cell differentiation to spatial organization of chromatin and gene expression.Citation2 Seminal studies have described lamin proteins as being tightly associated with heterochromatic and transcriptionally repressed large genomic regions, called Lamina-associated domains (LADs). These domains show variable lengths from 0.1 to 10 Mb and generally create an environment that maintains genes repressedCitation3,4 and marked by H3K9me2, H3K9me3 or H3K27me3.Citation5-8 Although A- and B-type lamins share common protein structural features, recent studies have revealed that they differ in several aspects from gene expression to nuclear distribution and function. In mammals the expression of A-and B-type lamins is developmentally regulated, resulting in cell type-specific arrangement of lamins. In particular, while at least one B-type lamin is expressed in every cell, A-type lamins are principally expressed after birth in differentiated cells.Citation9,10 Interestingly, recent findings have indicated that lamin A:B ratio, more than their individual concentration, plays a pivotal role in cell differentiation.Citation11 One hypothesis suggests that mechanical signals from the extracellular environment can be physically transmitted by the cytoskeleton to the nucleus by changing the lamin A:B ratioCitation12 and, in turn, cell-specific gene expression.Citation1 This hypothesis is supported by the observation that A and B-type lamins also differ in their nuclear distribution, where they probably bind and regulate distinct genomic regions. In fact, although both A and B-type lamins are predominantly present at the inner nuclear membrane, Lamin A also exists in lower concentration as a detergent-soluble pool within the nucleoplasm.Citation13,14 Nucleoplasmic lamins have been also identified within whole tissues,Citation15 excluding that nuclear interior localization of Lamin A was an artifact of in vitro cell cultures. Despite such evidence, in recent years, the nucleoplasmic localization of A-type lamins has been much debated, with some scientists proposing a nucleoplasmic pool of Lamin A composed of intermediates of Lamin A complexes after disassembly of nuclear lamin in mitosis.Citation16 Live imaging studies in cells at different cell cycle stages have solved this controversy by showing a stable, nucleoplasmic GFP-Lamin A in interphase cells.Citation17 More recently, several efforts have been made to understand the molecular role of nucleoplasmic Lamin A. Experimental limitations such as the low abundance and the difficulty in separating nucleoplasmic from peripheral Lamin A have made these studies highly complex. However, researchers have managed to avoid these obstacles by focusing their studies on the nuclear interior components interacting with nucleoplasmic Lamin A. One example is the Lamina-associated polypeptide 2alpha (LAP2 α), the only isoform of LAP2 protein lacking the transmembrane domain (reviewed inCitation18). LAP2 α directly interacts with Lamin and is a key factor in regulating the nucleo-plasmic localization of A-type lamins.Citation19 In fact, knock out mice lacking LAP2 α show an absence of nucleoplasmic A-type lamins. Re-expression into LAP2 α-deficient cells of full length LAP2 α but not of a lamin binding-defective LAP2 α mutant rescues the nucleoplasmic Lamin A/C.Citation20 Functionally, lack of LAP2 α and the subsequent delocalization of Lamin A leads to hyperproliferation of progenitor tissue cells, ultimately leading to cardiac and muscle differentiation defects.Citation20-23 This suggests that nucleoplasmic Lamin/Lap2alpha complexes play a key role in tissue homeostasis maintenance, regulating the balance between proliferation and differentiation.Citation24 Recent genome wide studies have further supported the nucleoplasmic Lamin A's functional role, revealing that Lamin A/C enriched DNA sequences are not always confined to the nuclear periphery and suggesting that Lamin A dependent transcriptional regulation can also take place in the nuclear interior.Citation5,7,25 Furthermore, microscopy and combined chip-DamID experiments have revealed that the same Lamin Associated Domain (LAD) exhibits distinct epigenetic signatures when located in the nuclear interior or in the periphery,Citation5 suggesting that nucleoplasmic or peripheral Lamin A can regulate the same LAD and that the engagement of different epigenetic factors is dependent on the nuclear localization. Although the scientific community is making great efforts to elucidate the role of Lamin A/C in the chromatin organization, it has still not explored how A-type lamins interact with the network of epigenetic players involved in the regulation of chromatin conformation. Our research provided new insights into Lamin studies, describing for the first time a functional interaction between Lamin A and key epigenetic regulators governing essential cellular processes: the Polycomb group (PcG) of proteins.
The Polycomb group of proteins (PcG)
PcG proteins are transcriptional repressors that play central roles in cell differentiation, development and cell-identity maintenance.Citation26 They are present predominantly in the nucleus as multimeric protein complexes named Polycomb repressive complexes (PRCs), able to post-translationally modify histones and silence target genes. In mammals, the best-characterized complexes are PRC1 and PRC2, that can act synergistically or independently of each other.Citation26 The PRC1 complex contains a mix of proteins from distinct families including Chromobox homolog (Cbx), Polyhomeotic (PH), Ring1, PcG RING fingers (PcGFs), YY1-associated factor 2 (YAF2) and RYBP;Citation27 PRC2 is composed of Enhancer of Zeste1/2 (Ezh1/2), retinoblastoma-associated protein 46 and 48 (RbAp46/48), Embryonic Ectoderm Development (EED) and Suppressor of Zeste 12 (SUZ12).Citation28 PRC1 is responsible for lysine 119 mono-ubiquitination of histone H2A (H2A119Ub1), catalyzed by the subunits Ring1a and b; while the histone methyl-transferase Ezh2 di- and tri- methylates the Lys 27 of histone H3 (H3K27me2/3). Despite the traditional hierarchical model of PRC recruitment on chromatin depicting PRC2 dependent deposition of H3K27me3 as the epigenetic mark recognized and bound by the PRC1 complex, emerging findings have shown that PRC recruitment relies on several alternative locus-specific mechanisms involving other subunits and noncoding RNA.Citation26
In the nucleus Polycomb proteins form microscopically visible foci called Polycomb bodies, localized close to the pericentric heterochromatin.Citation29 PcG bodies are formed in the early embryos and progressively increase in size and number during embryogenesis, mirroring the repressive function of PcG proteins.Citation30 This suggests that the formation of PcG bodies is necessary for a correct maintenance of PcG repressive programs. Consistently, it has been shown that the coordinated action of PcG proteins is evolutionarily required to form specific, cell cycle regulated, multi-looped DNA structures where all the PcG targets are clustered.Citation31-36 The formation of PcG dependent higher order chromatin structures and their influence on transcriptional repression were recently confirmed by super-resolution microscopy.Citation37
We found a previously unanticipated functional and evolutionary conserved interplay between intra-nuclear Lamin A/C and PcG proteins, this being required for a correct PcG-mediated transcriptional repression and PcG nuclear compartmentalization.Citation38
Nucleoplasmic Lamin A/C interacts with PcG proteins
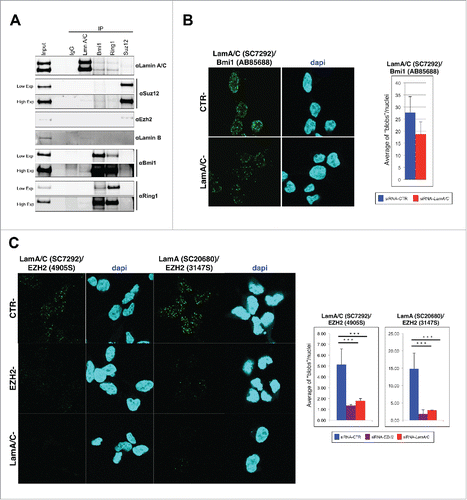
By co-immunoprecipitation (co-IP) and proximity ligation assay (PLA) experiments we previously showed that Lamin A/C interacts with Polycomb Repressive Complexes (PRC) in murine muscular cells.Citation38 Here we show that these interactions are evolutionary conserved. By performing co-IP in human 293 cells, we found that the endogenous Lamin A/C co-precipitated members of both Polycomb complexes PRC1 and PRC2 (). Similarly, by immunoprecipitating components of both PRC1 and PRC2 complexes, we were able to detect Lamin A/C (). Of note, the stoichiometry of the identified interactions indicated that only a small portion of the PcG complexes is associated with Lamin A/C, suggesting that nucleoplasmic Lamin A/C interacts with Polycomb complexes. Proximity ligation assay (PLA) analysis in 293 cells corroborated these findings, showing several spots of protein-protein interactions in the nuclear interior obtained by using different combinations of antibodies against Lamin A/C and PRC components (). Importantly, knock-down of PcG subunits or Lamin A/C strongly reduces the detected associations, proving the reliability of the assay.
Figure 1. Polycomb proteins and Lamin A/C interact endogenously. (A) Western blot analysis of co-IP performed in 293. Nuclear extracts immunoprecipitated with Suz12, Bmi1, Ring1b or Lamin A/C antibodies together with inputs were immunoblotted and hybridized with indicated antibodies. An unrelated antibody (murine IgG) was used as negative control. Two panels with low and high exposures of Suz12, Ring1b and Bmi1 were shown. (B and C) Representative fields of confocal microscopy images of PLA experiments performed on 293 cells transfected with indicated siRNAs. Each fluorescent dot, ‘blob’, represents the co-localization of Lamin A/C and Bmi1 (B) or Ezh2 (C). Data are reported for 3 different combinations of antibodies. Quantification of the blobs is represented in the graphs beside. The average of blobs/nuclei in the diagram corresponds to the quantification of 2 independent experiments, n > 244, 91 and 267, respectively (from left to right). Two-tailed t-test was applied for statistical analysis. Standard error of the mean is indicated. Asterisks indicate statistically relevant differences: (α = 0.05). * p < 0.05, *** p < 0.001.
PcG proteins are preferentially localized in the nuclear interior
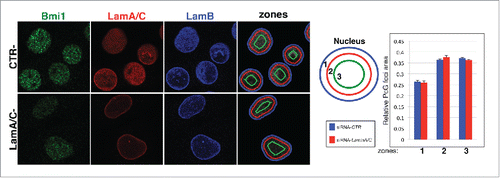
To further investigate the putative preferential positioning of PcG proteins in the nuclear interior, we evaluated the sub-nuclear localization of PcG aggregates relative to the nuclear periphery on single plane images of C2C12 nuclei, by measuring the area occupied by PcG foci in 3 concentric nuclear zones of equal surface area (). We used our MCV algorithm to select nuclei and to automatically segment PcG foci on a large number of images.Citation38 We found that PcG foci are mostly localized in the interior part of the nucleus, being excluded from nuclear periphery (); this confirms that PcG-Lamin A/C interplay takes place in the nucleoplasm.
Figure 2. Intra-nuclear localization of PcG proteins depend on Lamin A/C. (A) Representative confocal microscopy images of C2C12 myoblasts transfected with indicated siRNA. PcG foci distribution is evaluated in a 3-zone assay using the focal plane in which the PcG foci have the highest intensity. Each cross-section was divided into 3 concentric zones of equal surface. A random distribution gives 33% per zone. (B) Quantification of PcG distribution described in.Citation27 PcG foci area in the indicated zone relative to total area of PcG foci measured in the whole nucleus. The average in the graph corresponds to the quantification of 4 independent experiments. n > 352.
Lamin A/C-Polycomb axis in muscle differentiation
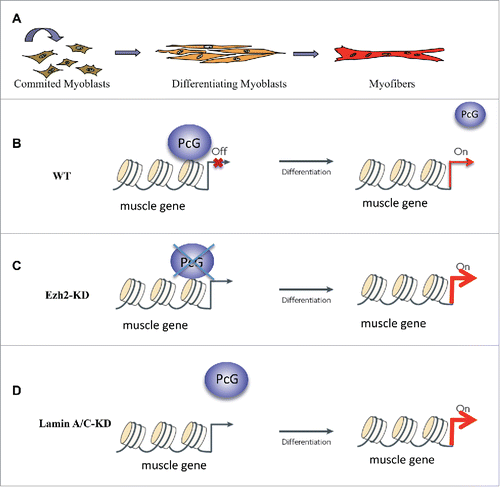
We previously showed that reduction of Lamin A/C determines an “erosion” of PcG bodies due to dispersion of PcG proteins in the nucleus; this, in turn, is accompanied by an aberrant transcriptional reactivation of some PcG targets, ultimately leading to an anticipation of the myogenic program.Citation38 Interestingly, after knock down of Lamin A/C, although we were able to observe a higher reduction of nucleoplasmic Lamin A/C in respect to peripheral Lamin A/C (), no differences in the nuclear localization of the remaining PcG foci were found, further suggesting that peripheral Lamin A/C does not interact with PcG proteins (). Our work reveals the importance of intra-nuclear PcG architecture for PcG function, underlying that nucleoplasmic LaminA/C acts as a scaffold for PcG foci stability. It remains to be elucidated what is the determinant of the PcG foci localization in the nucleoplasm. One possible candidate is the Lap2 α protein, whose expression is physiologically downregulated during myoblast differentiation. This event determines a reduction in nucleoplasmic Lamin A/C,Citation23 in line with the concomitant PcG target activation.Citation38 This hypothesis was recently supported by an interesting work showing that nucleoplasmic Lamin A/C and Lap2 α co-occupy genomic regions enriched with the PcG-dependent histone mark H3K27me3.Citation39 Importantly, in Lap2alpha KO, Lamin A/C is redistributed in the nucleus and the H3K27me3 marks increased in regions which gained Lamin A/C binding,Citation39 suggesting a coordinated Lamin A/C -PcG proteins crosstalk guided, in wt, by Lap2 α. In line with this hypothesis, we think that the interplay between Lamin A/C-PcG-Lap2 α could dynamically regulate physiological skeletal muscle differentiation. We found that the reduction of endogenous Lamin A/C levels accelerates the myogenic program in an evolutionary conserved manner, phenocopying Ezh2 depletioCitation38,40 (). Hence, the Lamin dependent premature muscle differentiation is due to a detachment of Ezh2 from chromatin and the concomitant transcription of PcG regulated muscle genes (). Interestingly, it has been reported that the processing rate and localization of Lamin A/C are regulated during myoblast differentiation, dropping when myoblasts achieve the confluence, at the onset of muscle differentiation.Citation41 On the other hand, the downregulation of Lap2 α during myogenesis is accompanied by a reduction of the nucleoplasmic pool of A-type lamins.Citation21 These dynamics suggest a physiological role for the reduction of Lamin A/C and Lap2alpha in the induction of the differentiation process. In line with this evidence, the enforced reduction of Lamin A/C before the onset of myogenesis, which mimics physiological conditions, is probably responsible for the delocalization of PcG proteins from muscle specific genes and consequent transcriptional activation of the myogenic program.
Figure 3. Nucleoplasmic LaminA/(C) affects muscle differentiation through PcG functions. (A) Schematic representation of muscle differentiation in vitro. (B) In physiological conditions PcG proteins total levels are reduced at the onset of myogenesis. This determines a displacement from muscle promoters and a consequent transcriptional activation of target genes. (C) Depletion of PcG proteins induces de-repression of muscle genes and premature muscle differentiation. (D) Depletion of Lamin A/C, although does not change overall PcG proteins levels, leads to an acceleration of myogenesis due to a premature displacement of PcG proteins from muscle-specific genes promoters.
Future directions
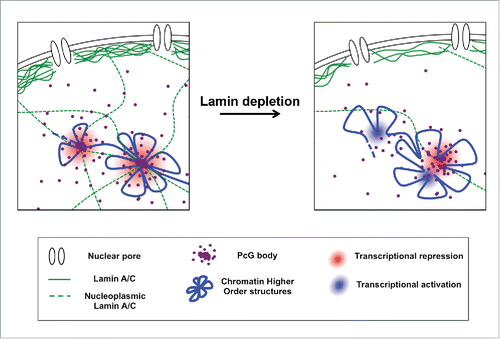
Recently, super-resolution imaging studies have shown that the chromatin is organized in different 3D conformations, depending on its epigenetic state.Citation42 Intriguingly, PcG repressed domains show a special chromatin folding, with a high degree of chromatin intermixing and exclusion of neighboring active chromatin, suggesting that PcG proteins play an active role in the packaging of repressed chromatin. These observations were confirmed and corroborated by other studies showing that PcG protein aggregation facilitates long-range interactions.Citation35,37 We provided new insights into PcG biology, describing for the first time a functional interaction between Lamin A and PcG proteins and demonstrating that an intact intra-nuclear Lamin A/C is necessary to provide a framework for the stability of PcG proteins aggregations and to ensure the maintenance of PcG-mediated higher order nuclear structures and epigenetic transcriptional programsCitation38 (). In the near future, studies aimed at understanding the role of lamin proteins in regulating chromatin higher order structures and epigenetic functions could open up alternative approaches in the cure of lamin-associated diseases, where Lamin A shows a nuclear mislocalization leading to aberrant cellular processes.Citation43-45
Figure 4. Molecular mechanism underlying PcG/Lamin A interplay. An intact intra-nuclear Laminv A/C is required for the stability of PcG foci aggregation. Reduction of Lamin A/C levels determines an erosion of PcG foci caused by PcG protein dispersion. This is accompanied by a relaxation of PcG-mediated higher-order chromatin structure that acquires a conformation more prone to transcriptional reactivation.
Experimental procedures
Co-immunoprecipitation (co-IP)
coIP on nuclear extracts was performed using standard procedures. In brief, nuclear extracts were prepared after cytosolic extraction with cytosolic lysis buffer (10 mM Hepes, pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 10% glycerol, 0.2% NP-40, 1 mM DTT, and protease inhibitors) for 10 min on ice and a 3/8-in syringe with a 25G needle. Nuclei pellets were then lysed in nuclei lysis buffer (10 mM Hepes, pH 7.5, 1.5 mM MgCl2, 300 mM KCl, 10% glycerol, 0.2% NP-40, 1 mM DTT, and protease inhibitors) for 30 min at 4°C. 0.5 to 1 mg of nuclear extract was immunoprecipitated overnight at 4°C with 5 μg corresponding antibodies at a final concentration of 150 mM KCl in nuclei lysis buffer. Immuno-precipitates were incubated with magnetic protein G beads (10004D; Dynabeads; Invitrogen) for 2 h at 4°C. Beads were extensively washed with wash buffer (10 mM Hepes, pH 7.5, 1.5 mM MgCl2, 150 mM KCl, 0.2% NP-40, and 10% glycerol) and eluted in Tris-EDTA. Finally, beads were re-suspended in Laemmli buffer and boiled at 95°C for 5 min, and the supernatant was analyzed by Western blot.
PLA experiments
Coverslips were fixed with 4% paraformaldehyde in PBS for 10 min. Cells were permeabilized with 0.5% Triton X-100 in PBS and blocked with 1% BSA in PBS for 1 h at RT. Detection of protein interactions was performed using the Duolink system (Sigma-Aldrich) according to the manufacturer's instructions. PLA blob quantification was performed using Cell Profiler 2.0. Nuclei were detected using the Otsu method with a global 2-class threshold strategy, and foci were detected using the Otsu method with a per-object 3-class threshold strategy.
PcG foci quantification
PcG foci quantification was performed with an automated pipeline described in.Citation38 For the analysis of PcG localization respect to the nuclear periphery, each nucleus area was divided in 3 concentric regions of the same size using matlab (bwmorph shrink operation). The amount of polycomb proteins in each region was measured by counting the pixels contained.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Prof. Martin Bennett for “proofreading work” of the manuscript. We thank Chiara Mozzetta, the Italian network of Laminopathies and members of the laboratory for stimulating discussions and constructive criticisms.
Funding
CL and GO are supported by grants from the Italian Ministry of Research and University (Futuro in Ricerca RBFR106S1Z_001) and the flagship CNR project, (Epigen).
References
- Gruenbaum Y, Foisner R. Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem 2015; 84:131-64; PMID:25747401; http://dx.doi.org/10.1146/annurev-biochem-060614-034115
- Bianchi A, Lanzuolo C. Into the chromatin world: Role of nuclear architecture in epigenome regulation. AIMS Biophysics 2015; 2:585-612; http://dx.doi.org/10.3934/biophy.2015.4.585
- Collas P, Lund EG, Oldenburg AR. Closing the (nuclear) envelope on the genome: How nuclear lamins interact with promoters and modulate gene expression. Bioessays 2014; 36:75-83; PMID:24272858; http://dx.doi.org/10.1002/bies.201300138
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res 2013; 23:270-80; PMID:23124521; http://dx.doi.org/10.1101/gr.141028.112
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell 2013; 153:178-92; PMID:23523135; http://dx.doi.org/10.1016/j.cell.2013.02.028
- Kind J, van Steensel B. Genome-nuclear lamina interactions and gene regulation. Curr Opin Cell Biol 2010; 22:320-5; PMID:20444586; http://dx.doi.org/10.1016/j.ceb.2010.04.002
- Lund E, Oldenburg A, Delbarre E, Freberg C, Duband-Goulet I, Eskeland R, Buendia B, Collas P. Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes. Genome Res 2013; 23(10):1580-9; PMID:23861385; http://dx.doi.org/10.1101/gr.159400.113
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 2006; 38:1005-14; PMID:16878134; http://dx.doi.org/10.1038/ng1852
- Kubben N, Voncken JW, Konings G, van Weeghel M, van den Hoogenhof MM, Gijbels M, van Erk A, Schoonderwoerd K, van den Bosch B, Dahlmans V, et al. Post-natal myogenic and adipogenic developmental: defects and metabolic impairment upon loss of A-type lamins. Nucleus 2011; 2:195-207; PMID:21818413; http://dx.doi.org/10.4161/nucl.2.3.15731
- Stewart C, Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell 1987; 51:383-92; PMID:3311384; http://dx.doi.org/10.1016/0092-8674(87)90634-9
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 2013; 341:1240104; PMID:23990565; http://dx.doi.org/10.1126/science.1240104
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol 2006; 26:3738-51; PMID:16648470; http://dx.doi.org/10.1128/MCB.26.10.3738-3751.2006
- Hozak P, Sasseville AM, Raymond Y, Cook PR. Lamin proteins form an internal nucleoskeleton as well as a peripheral lamina in human cells. J Cell Sci 1995; 108(Pt 2):635-44; PMID:7769007
- Kolb T, Maass K, Hergt M, Aebi U, Herrmann H. Lamin A and lamin C form homodimers and coexist in higher complex forms both in the nucleoplasmic fraction and in the lamina of cultured human cells. Nucleus 2011; 2:425-33; PMID:22033280; http://dx.doi.org/10.4161/nucl.2.5.17765
- Naetar N, Foisner R. Lamin complexes in the nuclear interior control progenitor cell proliferation and tissue homeostasis. Cell Cycle 2009; 8:1488-93; PMID:19377295; http://dx.doi.org/10.4161/cc.8.10.8499
- Lehner CF, Furstenberger G, Eppenberger HM, Nigg EA. Biogenesis of the nuclear lamina: in vivo synthesis and processing of nuclear protein precursors. Proc Natl Acad Sci U S A 1986; 83:2096-9; PMID:3515346; http://dx.doi.org/10.1073/pnas.83.7.2096
- Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol 2000; 151:1155-68; PMID:11121432; http://dx.doi.org/10.1083/jcb.151.6.1155
- Dechat T, Vlcek S, Foisner R. Review: lamina-associated polypeptide 2 isoforms and related proteins in cell cycle-dependent nuclear structure dynamics. J Struct Biol 2000; 129:335-45; PMID:10806084; http://dx.doi.org/10.1006/jsbi.2000.4212
- Dechat T, Korbei B, Vaughan OA, Vlcek S, Hutchison CJ, Foisner R. Lamina-associated polypeptide 2alpha binds intranuclear A-type lamins. J Cell Sci 2000; 113(Pt 19):3473-84; PMID:10984438
- Naetar N, Korbei B, Kozlov S, Kerenyi MA, Dorner D, Kral R, Gotic I, Fuchs P, Cohen TV, Bittner R, et al. Loss of nucleoplasmic LAP2alpha-lamin A complexes causes erythroid and epidermal progenitor hyperproliferation. Nat Cell Biol 2008; 10:1341-8; PMID:18849980; http://dx.doi.org/10.1038/ncb1793
- Gotic I, Leschnik M, Kolm U, Markovic M, Haubner BJ, Biadasiewicz K, Metzler B, Stewart CL, Foisner R. Lamina-associated polypeptide 2alpha loss impairs heart function and stress response in mice. Circ Res 2010; 106:346-53; PMID:19926876; http://dx.doi.org/10.1161/CIRCRESAHA.109.205724
- Gotic I, Schmidt WM, Biadasiewicz K, Leschnik M, Spilka R, Braun J, Stewart CL, Foisner R. Loss of LAP2 alpha delays satellite cell differentiation and affects postnatal fiber-type determination. Stem Cells 2010; 28:480-8; PMID:20039368; http://dx.doi.org/10.1002/stem.292
- Markiewicz E, Ledran M, Hutchison CJ. Remodelling of the nuclear lamina and nucleoskeleton is required for skeletal muscle differentiation in vitro. J Cell Sci 2005; 118:409-20; PMID:15654018; http://dx.doi.org/10.1242/jcs.01630
- Gesson K, Vidak S, Foisner R. Lamina-associated polypeptide (LAP)2alpha and nucleoplasmic lamins in adult stem cell regulation and disease. Semin Cell Dev Biol 2014; 29:116-24; PMID:24374133; http://dx.doi.org/10.1016/j.semcdb.2013.12.009
- Kind J, van Steensel B. Stochastic genome-nuclear lamina interactions: Modulating roles of Lamin A and BAF. Nucleus 2014; 5:124-30; PMID:24717229; http://dx.doi.org/10.4161/nucl.28825
- Lanzuolo C, Orlando V. Memories from the Polycomb Group Proteins. Annu Rev Genet 2012; 46:561-89; PMID:22994356; http://dx.doi.org/10.1146/annurev-genet-110711-155603
- Luis NM, Morey L, Di Croce L, Benitah SA. Polycomb in stem cells: PRC1 branches out. Cell Stem Cell 2012; 11:16-21; PMID:22770239; http://dx.doi.org/10.1016/j.stem.2012.06.005
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011; 469:343-9; PMID:21248841; http://dx.doi.org/10.1038/nature09784
- Cheutin T, Cavalli G. Polycomb silencing: from linear chromatin domains to 3D chromosome folding. Curr Opin Genet Dev 2014; 25:30-7; PMID:24434548; http://dx.doi.org/10.1016/j.gde.2013.11.016
- Cheutin T, Cavalli G. Progressive Polycomb Assembly on H3K27me3 Compartments Generates Polycomb Bodies with Developmentally Regulated Motion. PLoS Genet 2012; 8:e1002465; PMID:22275876; http://dx.doi.org/10.1371/journal.pgen.1002465
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. Polycomb-Dependent Regulatory Contacts between Distant Hox Loci in Drosophila. Cell 2011; 144:214-26; PMID:21241892; http://dx.doi.org/10.1016/j.cell.2010.12.026
- Lanzuolo C, Lo Sardo F, Orlando V. Concerted epigenetic signatures inheritance at PcG targets through replication. Cell Cycle 2012; 11:1296-300; PMID:22421150; http://dx.doi.org/10.4161/cc.19710
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol 2007; 9:1167-74; PMID:17828248; http://dx.doi.org/10.1038/ncb1637
- Lo Sardo F, Lanzuolo C, Comoglio F, De Bardi M, Paro R, Orlando V. PcG-mediated higher-order chromatin structures modulate replication programs at the drosophila BX-C. PLoS Genet 2013; 9:e1003283; PMID:23437006; http://dx.doi.org/10.1371/journal.pgen.1003283
- Schoenfelder S, Sugar R, Dimond A, Javierre BM, Armstrong H, Mifsud B, Dimitrova E, Matheson L, Tavares-Cadete F, Furlan-Magaril M, et al. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat Genet 2015; 47:1179-86; PMID:26323060; http://dx.doi.org/10.1038/ng.3393
- Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, Nieuwland M, Simonis M, de Laat W, van Lohuizen M, van Steensel B. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet 2011; 7:e1001343; PMID:21455484; http://dx.doi.org/10.1371/journal.pgen.1001343
- Wani AH, Boettiger AN, Schorderet P, Ergun A, Munger C, Sadreyev RI, Zhuang X, Kingston RE, Francis NJ. Chromatin topology is coupled to Polycomb group protein subnuclear organization. Nat Commun 2016; 7:10291; PMID:26759081; http://dx.doi.org/10.1038/ncomms10291
- Cesarini E, Mozzetta C, Marullo F, Gregoretti F, Gargiulo A, Columbaro M, Cortesi A, Antonelli L, Di Pelino S, Squarzoni S, et al. Lamin A/C sustains PcG protein architecture, maintaining transcriptional repression at target genes. J Cell Biol 2015; 211:533-51; PMID:26553927; http://dx.doi.org/10.1083/jcb.201504035
- Gesson K, Rescheneder P, Skoruppa MP, von Haeseler A, Dechat T, Foisner R. A-type lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2alpha. Genome Res 2016; e-pub ahead of print. PMID:26798136
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 2004; 18:2627-38; PMID:15520282; http://dx.doi.org/10.1101/gad.1241904
- Capanni C, Del Coco R, Squarzoni S, Columbaro M, Mattioli E, Camozzi D, Rocchi A, Scotlandi K, Maraldi N, Foisner R, et al. Prelamin A is involved in early steps of muscle differentiation. Exp Cell Res 2008; 314:3628-37; PMID:18951892; http://dx.doi.org/10.1016/j.yexcr.2008.09.026
- Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, Fudenberg G, Imakaev M, Mirny LA, Wu CT, Zhuang X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 2016; 529(7586):418-22; PMID:26760202; http://dx.doi.org/10.1038/nature16496
- Lanzuolo C. Epigenetic alterations in muscular disorders. Comp Funct Genomics 2012; 2012:256892; PMID:22761545; http://dx.doi.org/10.1155/2012/256892
- McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res 2013; 23:260-9; PMID:23152449; http://dx.doi.org/10.1101/gr.138032.112
- Ostlund C, Bonne G, Schwartz K, Worman HJ. Properties of lamin A mutants found in Emery-Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J Cell Sci 2001; 114:4435-45; PMID:11792809
