SUMMARY
Polycomb group (PcG) repress, whereas Trithorax group (TrxG) activate transcription for tissue development and cellular proliferation, and misregulation of these factors is often associated with cancer. ENL (MLLT1) and AF9 (MLLT3) are fusion partners of Mixed Lineage Leukemia (MLL), TrxG proteins, and are factors in Super Elongation Complex (SEC). SEC controls transcriptional elongation to release RNA polymerase II, paused around transcription start site. In MLL rearranged leukemia, several components of SEC have been found as MLL-fusion partners and the control of transcriptional elongation is misregulated leading to tumorigenesis in MLL-SEC fused Leukemia. It has been suggested that unexpected collaboration of ENL/AF9-MLL and PcG are involved in tumorigenesis in leukemia. Recently, we found that the collaboration of ENL/AF9 and PcG led to a novel mechanism of transcriptional switch from elongation to repression under ATM-signaling for genome integrity. Activated ATM phosphorylates ENL/AF9 in SEC, and the phosphorylated ENL/AF9 binds BMI1 and RING1B, a heterodimeric E3-ubiquitin-ligase complex in Polycomb Repressive complex 1 (PRC1), and recruits PRC1 at transcriptional elongation sites to rapidly repress transcription. The ENL/AF9 in SEC- and PcG-mediated transcriptional repression promotes DSB repair near transcription sites. The implication of this is that the collaboration of ENL/AF9 in SEC and PcG ensures a rapid response of transcriptional switching from elongation to repression to neighboring genotoxic stresses for DSB repair. Therefore, these results suggested that the collaboration of ENL/AF9 and PcG in transcriptional control is required to maintain genome integrity and may be link to the MLL-ENL/AF9 leukemia.
ENL/AF9 in elongation and PcG in repression
ENL (MLLT1) protein and its paralog AF9 (MLLT3) are fusion partners of the mixed-lineage leukemia (MLL) protein,Citation21,28,34,51 and are components of the transcriptional Super Elongation Complex (SEC).Citation14,21-23,44,55,58 In SEC, ENL or AF9 (ENL/AF9) have been reported to connect SEC with other transcriptional factors and to regulate transcriptional elongation.Citation21,23,58 ENL/AF9 has a YEATS-domain in the N-terminus and an ANC1 homology domain (AHD) in the C-terminus. The YEATS domain binds directly with histone and PAF1, a component of the PAF complex, suggesting that ENL/AF9 delivers SEC to Pol II on a chromatin template through the PAF complex, and that SEC and PAF complexes cooperatively promote transcriptional elongation.Citation14,21,23,57,58 The AHD of ENL/AF9 directly binds with factors in transcriptional activation, DOT1L (a histone methyl transferase that promotes transcription) and AF4 (another component of SEC), and CBX8 (a factor in PcG).Citation11,14,15,20,23,24,28,42,44,48,55,57,58 Structural analysis revealed that AHD might allow AF9 to exchange between binding partners in response to changes in local concentrations and post-transcriptional modifications, suggesting that ENL/AF9 may be essential to dynamic transcriptional control.Citation20
On the other hand, PcG repress transcription.Citation27,46 Two core complexes of PcG, PRC1 and PRC2, are thought to mainly work cooperatively; PRC2 methylates histone (H3-K27me3) and this methylation recruits PRC1 onto chromatin leading to the ubiquitination of histone H2A to block initiation and elongation of RNA Polymerase II-mediated transcription.Citation4,6,9,35,36,40,41,47,53
PcG in DNA double strand break (DSB) repair and genome integrity
Several recent reports have shown that PcG proteins are also involved in DSB repair. Factors of PRC2, EZH2 and SUZ12 are recruited at Laser-induced DSB sites and Fok1-nuclease induced DSB sites.Citation5 The H3-K27me3 increased at the sites of DSB,Citation8 and ubiquitination of H2A (H2A-K119ub) at DSB sites depends on both PRC1 and PRC2,Citation18 suggesting that PRC2 is involved in the recruitment of PRC1 at DSB sites. Whereas other groups reported that H3-K27me3 at the sites of DSB did not increase, and it is consistent with other finding that PRC1 did not require PRC2 activity to be recruited to DSB sites.Citation5,16 Therefore, the relationship between PRC1 and PRC2 in DSB repair remains elusive.
PRC1 is also involved in DSB repair. The factors PRC1, BMI1 and RING1B, are recruited at DSB sites, and the knockdown of BMI1 showed the sensitivity to IR and impaired DSB repair.Citation7,8,10,12,16,31,54 BMI1 and RING1B are required for Non-Homologous End Joining (NHEJ) and Homologous Recombination (HR) in DSB repair.Citation1,7,10,12 In response to DSBs, PRC1 ubiquitinates K119/K120-H2A and K119/K120-H2AX,Citation12,16,31,54 and this ubiquitination is required for the foci formation of γH2AX, and phosphorylation of ATM, MDC1, 53BP1 and BRCA1 at DSB sites.Citation16,31,54 These results suggested that PRC1 functions upstream of ATM and is involved in initiation of ATM-signaling pathway.
On the other hand, it has been reported that the recruitment of PRC1 at DSB sites depends on ATM-signaling and BMI1 functions downstream of RNF8.Citation12 PRC1 is also recruited by CBX4, an E3 sumo ligase in the PcG complex, that is involved in the DSB repair pathway.Citation17 CBX4 binds PAR and is recruited at Laser-induced DSB sites by PARP1. Another report also suggested that PARP1 recruits PRC1 at DSB sites.Citation8 Therefore, it is likely that the recruitment of BMI1 at DSB sites also depends on PARP1 activity. Further studies should answer the question of how PcG is recruited at DSB sites and what the function of PcG is in DSB repair.
Transcriptional silencing by ATM and PcG in response to DSBs
The relationship between transcription and DNA repair has been most studied in transcription-coupled DNA repair (TCR); UV damage on the transcribed DNA template is removed by nucleotide excision repair (NER) in a transcription-coupled manner.Citation3,25,29,45 While UV damage leads to stalling of RNA Polymerase II and recruitment of DNA repair proteins and induces TCR, DNA double-strand breaks (DSBs) lead to ATM or DNA-PKcs-dependent transcriptional repressionCitation18,19,32,37,52,59 (). In TCR, Polymerase II stalled by DNA damage in gene body results in backtracking and/or removal by proteasome.Citation13 In DNA-PKcs-dependent transcriptional repression, Polymerase II at DSB site on the gene body is suggested to be removed in a proteasome-dependent manner,Citation32 while in ATM-dependent transcriptional repression, Polymerase II suggested to be stalled and remains on the gene body in response to DSB near transcription sites.Citation37
Figure 1. (A) Link between DNA damage and transcription, and pathway of DSB-induced transcriptional repression. (Left) UV damage-stalled RNA Polymerase II recruits NER proteins and induces transcription coupled repair (TCR). (Right) DSBs activate ATM and/or DNA-PKcs and promote DSB-induced transcriptional repression and DSB repair. Under the ATM-signaling, PBAF, RNF8/RNF168 and ENL/PRC1 were reportedly involved in DSB-induced transcriptional repression. (B) Models of collaboration between ENL/AF9 and PRC1 in DSB-induced transcriptional repression. I) During transcriptional activation, ENL/AF9 in SEC promotes transcriptional elongation. II) When DSB(s) is introduced near transcriptional sites, ATM phosphorylates ENL/AF9 at evolutionarily well-conserved SQ-sites. This phosphorylation of ENL/AF9 recruits PRC1 at transcription-sites through BMI1 to promote ubiquitination of H2A and transcriptional repression. Thus, ENL/AF9 in SEC would be key factors for the switch of transcriptional elongation to rapid repression, in response to cellular signaling. Dot-line indicates that it is not clear how these factors repress transcription.
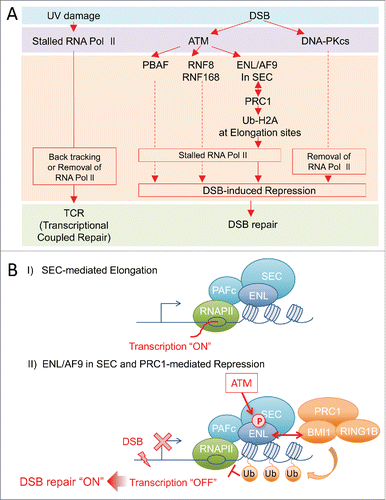
Shanbhag et al. first showed that ATM promotes ubiquitination of K119/120-H2A which prevents chromatin decondensation, leading to transcriptional silencing in response to DNA double-strand breaks (DSBs) near sites of transcriptionCitation37 (). While E3-ubiquitin-ligase RNF8 and RNF168 were suggested to be partially involved in the ubiquitination of K119/120 at H2A to silence transcription under ATM-signaling, a recent study has revealed that RNF8 is inactive toward nucleosomal H2A, and that RNF168 catalyzes ubiquitination of K13/15-H2A but not of K119/120-H2A.Citation26 Furthermore, The chromatin remodeling activity and ATM-mediated phosphorylation of PBAF complex were reported to be required for transcriptional silencing and ubiquitination of K119/120-H2A in response to DSBsCitation18 (). In addition, PRC1 and PRC2 were also showed to be involved in this DSB-induced transcriptional silencing. However, how PBAF induce ubiquitination of K119/120-H2A for DSB-induced transcriptional silencing remained unclear.
Collaboration between ENL/AF9 and PcG in transcriptional repression under ATM-signaling
The mechanism of transcriptional repression by PRC1 in response to DSBs was revealed through the study of the interaction between PRC1 and ENL/AF9Citation52 (). BMI1 and RING1B, a E3-ubiquitin ligase of PRC1, were identified as ENL-interacting proteins. In transcriptional activation, ENL in SEC binds transcriptional activation sites and promotes transcription, but PRC1 does not, indicating that these factors have different functions in transcriptional activation (). In response to DSBs, ENL/AF9 in SEC was phosphorylated by ATM, the master kinase of the DSB response,Citation39 and this phosphorylation increases the interaction between ENL in SEC and BMI1/RING1B in PRC1. The phosphorylated ENL in SEC recruits PRC1 at transcriptional elongation sites leading to the ubiquitination of H2A and results in transcriptional repression (). Therefore, this mechanism enables PRC1 to halt, rapidly and transiently, mobile Pol II at transcriptional elongation sites in response to cellular emergency signaling, such as ATM-signaling ().
This mechanism would have benefits for smoothly restarting paused RNA Polymerase II-mediated elongation after the end of the cellular signaling. We showed that ENL phosphorylation induces a charge alteration and/or conformational change in SEC and enables its interaction with BMI1 in PRC1 without influencing other binding partners of SEC. After a decrease in kinase activity of ATM by completion of DNA repair, dephosphorylation of ENL in SEC may release PRC1 at elongation sites. This would lead to de-ubiquitination of ubiquitinated H2A on the template around paused RNA Polymerase II by USP16, as previously reported,Citation37 and thus, RNA Polymerase II on the template could restart smoothly at the elongation sites with SEC. Therefore, this SEC-mediated transcriptional control may ensure a rapid “off and on” state of the transcriptional machinery in cellular signaling.
Mutation of ENL or BAF180 that led to inability to repress transcription in response to DSBs resulted in cellular sensitivity to IR, suggesting that a defect in transcriptional repression inhibits DSB repairCitation18,52 (). Additionally, the recruitment of KU70 proteins at DSB sites was reduced in the ENL mutation.Citation52 Therefore, it is suggested that ATM-controlled transcriptional repression mediated by SEC and PRC1 enables DSB repair proteins to access to DSB sites and promotes DSB repair.Citation18,52 It seems that ATM may maintain genome stability by regulating SEC-mediated elongation and PRC1-mediated repression to promote DSB repair instead of transcription for genome stability.
ENL/AF9 in SEC is a key factor controlling both elongation and repression
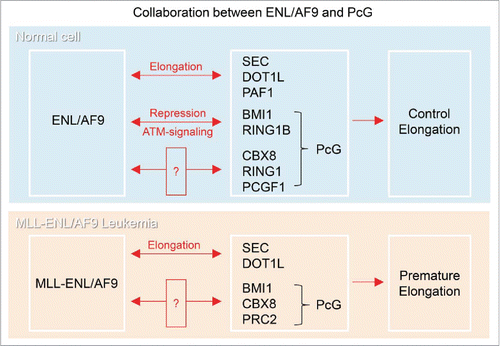
Whereas ENL/AF9 has been reported to interact with transcriptional elongation factors, such as PAF1, Dot1L and other SEC factors to promote transcriptional elongation, recently we found that ENL/AF9 interacts with transcriptional repressive factors, BMI1 and RING1B in PcG to repress transcription, as described above ().Citation2,14,21,23,28,33,52,57 In addition, ENL/AF9 has been reported to interact with other PcG proteins, CBX8, RING1 and PCGF1Citation11,15,28,33 (, top). While the meaning of these interactions were not clear, CBX8 interacts with ENL/AF9 through their AHD domain in vivo and in vitro, and ENL/AF9 and CBX8 were found co-localized in the nuclei.Citation11,15,28 From structural analysis, it has been proposed that AHD of ENL/AF9 might allow ENL/AF9 to exchange between binding partners in response to post-transcriptional modifications for control of transcription.Citation20 Therefore, ENL/AF9 could change binding partners between factors in transcriptional activation and repression in response to cellular signaling, and this mechanism of changing partner may enable SEC to switch transcription rapidly between elongation and repression.
Figure 2. (Top) Collaboration between ENL/AF9 and PcG. ENL/AF9 was reported to interact directly with SEC, DOT1L and PAFc and promote transcriptional elongation. ENL also interacts with PcG proteins, CBX8, RING1 and PCGF1, but the cellular function of the interaction remains unknown. We found that ENL interacts with the E3-ubiquitin ligase complex in Polycomb Repressive Complex 1 (PRC1), BMI1 and RING1B, and these work together to repress transcription under ATM-signaling. Therefore, ENL functions in both transcriptional elongation and repression depending on the circumstances. (Bottom) Collaboration of MLL-ENL/AF9 with PcG in Leukemia. PcG is involved in the tumorigenesis in MLL-ENL/AF9 rearranged Leukemia. “?” show the interaction, of which meaning is not clear or controversial.
SEC containing p-TEFb is involved in rapid transcriptional induction, including induction of heat shock genes, serum-inducible genes and certain developmentally controlled genes, by phosphorylation of the RNA polymerase II CTD. This leads to release of paused RNA polymerase II from the pause sites and switches transcriptional elongation to the “on” state.Citation23,42,58 During elongation, the phosphorylation of ENL/AF9 in SEC by ATM recruits PRC1 at transcriptional elongation sites through ENL/AF9, and this leads SEC-mediated rapid transcriptional pausing, the “off” state.Citation52 Therefore, SEC may switch transcription rapidly “on and off” state by controlling the phosphorylation of RNA polymerase II and the recruitment of PRC1. ENL/AF9 in SEC would be key to controlling this repression mechanism, in response to cellular signaling.
DSBs are thought to arise not only by DNA damage (UV, IR and environmental materials) but by proliferation and senescence (arising from the misregulation of DNA replication, transcription and chromosomal segregation). In addition to DSBs, other DNA damage and DNA replication stress induce activation of ATR-signaling and/or DNA-PKcs-signaling. ATR and DNA-PKcs phosphorylate at the same SQ sites as ATM,Citation39 thus the phosphorylation of ENL/AF9 may arise from ATR and DNA-PKcs leading to increase in the interaction with PRC1 and promotion of repression, in response to other cellular events in addition to DSBs. Furthermore, ENL/AF9 would has other modifications that leads to change transcriptional binding partners for rapid “on and off” state mediated by SEC. There may be more other mechanisms to discover about the control of transcription by SEC.
Collaboration between MLL-ENL/AF9 oncoproteins and PcG in leukemia
Several frequent translocation partners in MLL have been found in SEC.Citation21,23,58 The SEC-mediated transcriptional elongation is disrupted in MLL-SEC rearranged leukemia, suggesting that misregulation of this elongation stage of transcription often causes leukemia.Citation14,21-23,44,55,58 MLL-ENL/AF9 oncoproteins are recruited to the normal target genes of MLL, leading to the rest of the SEC recruitment and, therefore, to premature activation of transcriptional elongationCitation21,28,34,51 ().
Surprisingly, unexpected collaborations between MLL-ENL/AF9 Leukemia and PcG have been reported (). MLL-AF9 collaborates with BMI1, a factor in Polycomb repressive complex 1 (PRC1), and overcomes senescence for the development of leukemic stem cells.Citation43 CBX8, a protein in PRC1, interacts with MLL-fused oncoprotein, MLL-ENL/AF9, and this interaction causes leukemia by promoting abnormally activated transcription.Citation24,48,56 It is not clear whether the interaction between CBX8 and MLL-ENL/AF9 recruits Tip60 at transcription sitesCitation48 or inhibits PRC1 function.Citation24 Polycomb repressive complex 2 (PRC2) is also required for MLL-AF9 leukemiaCitation30,38,49,50 (). It is poorly understood how these factors, MLL-ENL/AF9 and PcG, collaborate during tumorigenesis. This unexpected collaboration may arise from inappropriate MLL-ENL/AF9 and PcG activity, such as mutational activity and alteration of expression levels, which predisposes to cancer. Alternatively, uncovered novel functional collaboration between ENL/AF9 and PcG in transcriptional regulation would cause tumorigenesis in MLL-ENL/AF9 Leukemia.
Conclusion and future perspectives
ATM controls transcriptional switches from SEC-mediated elongation to PRC1-mediated repression by regulated collaboration between ENL/AF9 in SEC and E3-ubiquitin ligase of PRC1.Citation52 ATM may also control other factors involved in transcriptional repression, such as other subunits of PRC1, PRC2, RNF8, RNF168 and PBAF, in response to DSBs.Citation18,Citation37,52 It remains unclear how RNF8, RNF168 and PBAF regulate ubiquitination of H2A for repression, and the relationship between PRC1 and PRC2 in the repression. Further studies on how these factors regulate DSB-induced transcriptional repression under ATM-signaling remain important.
We are only just beginning to understand the mechanism of functional collaboration between ENL/AF9 and PcG for SEC-mediated rapid control of transcription, “on and off” state, in response to cellular signaling. Therefore, further investigation into the mechanisms of collaboration between ENL/AF9 and PcG will provide new insights into the system of transcriptional control by SEC, and possibly into ENL/AF9-MLL rearranged Leukemia.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Shirley McCready for editing the text.
Funding
This work was funded by the Grants-in-Aid for Scientific Research and from the Ministry of Education, Culture, Sports, Science and Technology, Japan, to A.Y. (24310037 and 22131005) and to A.U. (22131006).
Reference
- Bartocci C, Diedrich JK, Ouzounov I, Li J, Piunti A, Pasini D, Yates JR, 3rd, Lazzerini Denchi E. Isolation of chromatin from dysfunctional telomeres reveals an important role for Ring1b in NHEJ-mediated chromosome fusions. Cell Rep 2014; 7:1320-32; PMID:24813883; http://dx.doi.org/10.1016/j.celrep.2014.04.002
- Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KS, Allis CD, Roeder RG. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc Natl Acad Sci U S A 2011; 108:15751-15756; PMID:21896721; http://dx.doi.org/10.1073/pnas.1111498108
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 1985; 40:359-69; PMID:3838150; http://dx.doi.org/10.1016/0092-8674(85)90150-3
- Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J 2006; 25:2465-74; PMID: 16710298; http://dx.doi.org/10.1038/sj.emboj.7601144
- Campbell S, Ismail IH, Young LC, Poirier GG, Hendzel MJ. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell cycle (Georgetown, Tex) 2013; 12:2675-83; PMID:23907130; http://dx.doi.org/10.4161/cc.25795
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell 2005; 20:845-54; PMID:16359901; http://dx.doi.org/10.1016/j.molcel.2005.12.002
- Chagraoui J, Hebert J, Girard S, Sauvageau G. An anticlastogenic function for the Polycomb Group gene Bmi1. Proc Natl Acad Sci U S A 2011; 108:5284-9; PMID: 21402923; http://dx.doi.org/10.1073/pnas.1014263108
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A 2010; 107:18475-80; PMID:20937877; http://dx.doi.org/10.1073/pnas.1012946107
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 2004; 7:663-76; PMID:15525528; http://dx.doi.org/10.1016/j.devcel.2004.10.005
- Facchino S, Abdouh M, Chatoo W, Bernier G. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J Neurosci 2010; 30:10096-111; PMID: 20668194; http://dx.doi.org/10.1523/JNEUROSCI.1634-10.2010
- Garcia-Cuellar MP, Zilles O, Schreiner SA, Birke M, Winkler TH, Slany RK. The ENL moiety of the childhood leukemia-associated MLL-ENL oncoprotein recruits human Polycomb 3. Oncogene 2001; 20:411-9; PMID: 11313972; http://dx.doi.org/10.1038/sj.onc.1204108
- Ginjala V, Nacerddine K, Kulkarni A, Oza J, Hill SJ, Yao M, Citterio E, van Lohuizen M, Ganesan S. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol 2011; 31:1972-1982; PMID:21383063; http://dx.doi.org/10.1128/MCB.00981-10
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol 2008; 9:958-70; PMID:19023283; http://dx.doi.org/10.1038/nrm2549
- He N, Chan CK, Sobhian B, Chou S, Xue Y, Liu M, Alber T, Benkirane M, Zhou Q. Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc Natl Acad Sci U S A 2011; 108:E636-45; PMID: 21873227; http://dx.doi.org/10.1073/pnas.1107107108
- Hemenway CS, de Erkenez AC, Gould GC. The polycomb protein MPc3 interacts with AF9, an MLL fusion partner in t(9;11)(p22;q23) acute leukemias. Oncogene 2001; 20:3798-805; PMID:11439343; http://dx.doi.org/10.1038/sj.onc.1204478
- Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol 2010; 191:45-60; PMID :20921134; http://dx.doi.org/10.1083/jcb.201003034
- Ismail IH, Gagne JP, Caron MC, McDonald D, Xu Z, Masson JY, Poirier GG, Hendzel MJ. CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res 2012; 40:5497-510; PMID:22402492; http://dx.doi.org/10.1093/nar/gks222
- Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Kunzel J, Lobrich M, Jeggo PA, Downs JA. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol Cell 2014; 55:723-32; PMID:25066234
- Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 2007; 447:730-4; PMID:17554310; http://dx.doi.org/10.1038/nature05842
- Leach BI, Kuntimaddi A, Schmidt CR, Cierpicki T, Johnson SA, Bushweller JH. Leukemia fusion target AF9 is an intrinsically disordered transcriptional regulator that recruits multiple partners via coupled folding and binding. Structure (London, England : 1993) 2013; 21:176-83; PMID:23260655; http://dx.doi.org/10.1016/j.str.2012.11.011
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev 2011; 25:1486-98; PMID:21764852
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 2010; 37:429-37; PMID:20159561; http://dx.doi.org/10.1016/j.molcel.2010.01.026
- Luo ZJ, Lin CQ, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Bio 2012; 13:543-7; PMID:22895430; http://dx.doi.org/10.1038/nrm3417
- Maethner E, Garcia-Cuellar MP, Breitinger C, Takacova S, Divoky V, Hess JL, Slany RK. MLL-ENL inhibits polycomb repressive complex 1 to achieve efficient transformation of hematopoietic cells. Cell Rep 2013; 3:1553-66; PMID: 23623499; http://dx.doi.org/10.1016/j.celrep.2013.03.038
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 2014; 15:465-81; PMID:24954209; http://dx.doi.org/10.1038/nrm3822
- Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 2012; 150:1182-95; PMID:22980979; http://dx.doi.org/10.1016/j.cell.2012.08.005
- Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer 2010; 10:669-82; PMID:20865010; http://dx.doi.org/10.1038/nrc2931
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, et al.. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 2007; 110:4445-54; PMID:17855633; http://dx.doi.org/10.1182/blood-2007-05-090514
- Mullenders L. DNA damage mediated transcription arrest: Step back to go forward. DNA Repair 2015; 36:28-35; PMID:26422136; http://dx.doi.org/10.1016/j.dnarep.2015.09.005
- Neff T, Sinha AU, Kluk MJ, Zhu N, Khattab MH, Stein L, Xie H, Orkin SH, Armstrong SA. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci U S A 2012; 109:5028-33; PMID:22396593; http://dx.doi.org/10.1073/pnas.1202258109
- Pan MR, Peng G, Hung WC, Lin SY. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J Biol Chem 2011; 286:28599-607; PMID: 21676867; http://dx.doi.org/10.1074/jbc.M111.256297
- Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat struct Mol Biol 2012; 19:276-82; PMID:22343725; http://dx.doi.org/10.1038/nsmb.2224
- Park G, Gong Z, Chen J, Kim JE. Characterization of the DOT1L network: implications of diverse roles for DOT1L. Protein J 2010; 29:213-23; PMID:20431927; http://dx.doi.org/10.1007/s10930-010-9242-8
- Rubnitz JE, Morrissey J, Savage PA, Cleary ML. ENL, the gene fused with HRX in t(11;19) leukemias, encodes a nuclear protein with transcriptional activation potential in lymphoid and myeloid cells. Blood 1994; 84:1747-52; PMID:8080983
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell 2007; 128:735-45; PMID:17320510; http://dx.doi.org/10.1016/j.cell.2007.02.009
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 2007; 8:9-22; PMID:17173055; http://dx.doi.org/10.1038/nrg1981
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 2010; 141:970-81; PMID:20550933; http://dx.doi.org/10.1016/j.cell.2010.04.038
- Shi J, Wang E, Zuber J, Rappaport A, Taylor M, Johns C, Lowe SW, Vakoc CR. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene 2013; 32:930-8; PMID:22469984; http://dx.doi.org/10.1038/onc.2012.110
- Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 2013; 14:197-210; PMID:23847781; http://dx.doi.org/10.1038/nrm3546
- Simon JA, Kingston RE. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Bio 2009; 10:697-708; PMID:19738629; http://dx.doi.org/10.1038/nrn2731
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 2013; 49:808-24; PMID:23473600; http://dx.doi.org/10.1016/j.molcel.2013.02.013
- Smith E, Lin CQ, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev 2011a; 25:661-72; PMID:21460034; http://dx.doi.org/10.1101/gad.2015411
- Smith LL, Yeung J, Zeisig BB, Popov N, Huijbers I, Barnes J, Wilson AJ, Taskesen E, Delwel R, Gil J, et al. Functional crosstalk between Bmi1 and MLL/Hoxa9 axis in establishment of normal hematopoietic and leukemic stem cells. Cell Stem Cell 2011b; 8:649-62; PMID:21624810; http://dx.doi.org/10.1016/j.stem.2011.05.004
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell 2010; 38:439-51; PMID:20471949; http://dx.doi.org/10.1016/j.molcel.2010.04.012
- Spivak G, Hanawalt PC. Photosensitive human syndromes. Mutat Res 2015; 776:24-30; PMID:26255937; http://dx.doi.org/10.1016/j.mrfmmm.2014.11.003
- Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol 2014; 15:340-56; PMID:24755934; http://dx.doi.org/10.1038/nrm3789
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 2007; 9:1428-35; PMID:18037880; http://dx.doi.org/10.1038/ncb1663
- Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, Hess JL. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell 2011; 20:563-75; PMID:22094252; http://dx.doi.org/10.1016/j.ccr.2011.09.008
- Tanaka S, Miyagi S, Sashida G, Chiba T, Yuan J, Mochizuki-Kashio M, Suzuki Y, Sugano S, Nakaseko C, Yokote K, et al. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood 2012; 120:1107-17; PMID:22677129; http://dx.doi.org/10.1182/blood-2011-11-394932
- Thiel AT, Feng Z, Pant DK, Chodosh LA, Hua X. The trithorax protein partner menin acts in tandem with EZH2 to suppress C/EBPalpha and differentiation in MLL-AF9 leukemia. Haematologica 2013; 98, 918-27; PMID:23349306; http://dx.doi.org/10.3324/haematol.2012.074195
- Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 1992; 71:691-700; PMID:1423624; http://dx.doi.org/10.1016/0092-8674(92)90602-9
- Ui A, Nagaura Y, Yasui A. Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair. Mol Cell 2015; 58, 468-82; PMID:25921070; http://dx.doi.org/10.1016/j.molcel.2015.03.023
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004; 431:873-8; PMID:15386022; http://dx.doi.org/10.1038/nature02985
- Wu CY, Kang HY, Yang WL, Wu J, Jeong YS, Wang J, Chan CH, Lee SW, Zhang X, Lamothe B, et al. Critical role of monoubiquitination of histone H2AX protein in histone H2AX phosphorylation and DNA damage response. J Biol Chem 2011; 286:30806-15; PMID: 21690091; http://dx.doi.org/10.1074/jbc.M111.257469
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 2010; 17, 198-212; PMID:20153263; http://dx.doi.org/10.1016/j.ccr.2009.12.040
- Zeisig BB, Arteaga MF, Thirant C, So CW. Collaboration between PcG proteins and MLL fusions in Leukemogenesis: an emerging paradigm. Cancer Cell 20:551-3; PMID:22094247; http://dx.doi.org/10.1016/j.ccr.2011.10.031
- Zeisig DT, Bittner CB, Zeisig BB, Garcia-Cuellar MP, Hess JL, Slany RK. The eleven-nineteen-leukemia protein ENL connects nuclear MLL fusion partners with chromatin. Oncogene 2005; 24:5525-32; PMID: 15856011; http://dx.doi.org/10.1038/sj.onc.1208699
- Zhou Q, Li TD, Price DH. RNA Polymerase II Elongation Control. Annu Rev Biochem 2012; 81:119-43; PMID:22404626; http://dx.doi.org/10.1146/annurev-biochem-052610-095910
- Harding SM, Boiarsky JA, and Greenberg RA. ATM Dependent silencing links nucleolar chromatin reorganization to DNA damage recognition. J Cell Rep 2005; 13:2:251–259. http://dx.doi.org/1016/j.cel.rep.2015.08.085
