ABSTRACT
Eukaryotes require 3 DNA polymerases for normal replisome operations, DNA polymerases (Pol) α, delta and epsilon. Recent biochemical and structural studies support the asymmetric use of these polymerases on the leading and lagging strands. Pol epsilon interacts with the 11-subunit CMG helicase, forming a 15-protein leading strand complex that acts processively in leading strand synthesis in vitro, but Pol epsilon is inactive on the lagging strand. The opposite results are observed for Pol delta with CMG. Pol delta is highly active on the lagging strand in vitro, but has only feeble activity with CMG on the leading strand. Pol α also functions with CMG to prime both strands, and is even capable of extending both strands with CMG present. However, extensive DNA synthesis by Pol α is sharply curtailed by the presence of either Pol epsilon or Pol delta, which limits the role of the low fidelity Pol α to the initial priming of synthesis.
Introduction
E. coli and its phages, T4 and T7, utilize multiple copies of an identical DNA polymerase for leading and lagging strand synthesis.Citation1-3 The use of identical copies of a DNA polymerase generalizes to replication fork operations in archaeal cells.Citation4,5 However, in eukaryotes the picture is more complicated; they contain 3 essential DNA polymerases and their funtions are still being sorted out. The first eukaryotic DNA polymerase to be identified was Pol α.Citation6 Pol α consists of 4 subunits ()Citation7-9; the largest subunit harbors the DNA polymerase activity and the 2 smallest subunits function together to make small RNA primers.Citation8,10 The presence of DNA polymerase and primase activities in one protein complex initially suggested Pol α might replicate the genome without needing other DNA polymerases, although the lack of a proofreading 3′–5′ exonuclease was somewhat confounding. Indeed initial biochemical studies in the simian virus 40 (SV40) replication system found that Pol α can function with SV40 T-antigen helicase to prime and extend both the leading and lagging strands.Citation9,11 Another DNA polymerase was discovered, called Pol delta, that had a proofreading 3′–5′ exonuclease,Citation12 and in vitro studies of SV40 replication demonstrated that Pol delta takes over primed sites made by Pol α and replicates both strands of the SV40 genome.Citation9,11 Pol delta consists of 4 subunits in human and 3 subunits in yeast (). Pol delta, like the bacterial Pol III replicase, requires 2 accessory factors.Citation13-15 Studies in E. coli identified these factors as a circular clamp (PCNA in eukaryotes), and an ATP driven clamp loader complex (RFC in eukaryotes).Citation16,17 These accessory factors are now known to be essential to replication in all cells from bacteria to archaea and eukaryotes.Citation3,4,9 Genetic studies in budding yeast demonstrated that a third DNA polymerase, Pol epsilon, is essential for cellular replication; it consists of 4 subunits ().Citation18 Pol epsilon, like Pol delta, utilizes the RFC clamp loader and PCNA clamp.Citation19
Figure 1. Replication fork proteins of Saccharomyces cerevisiae. The Table to the left lists the names of subunits, complexes, their molecular weight and biochemical function. SDS polyacrylamide gels of recombinant pure proteins are shown to the right of the Table.
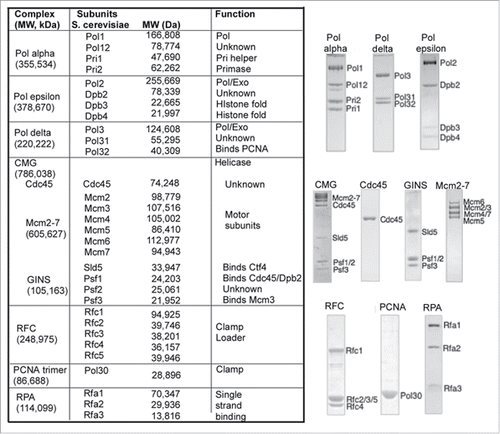
The 3 DNA polymerases, Pol α, Pol delta and Pol epsilon are members of the B-family of DNA polymerases. The largest subunit is the DNA polymerase, and in both Pol epsilon and Pol delta the large subunit also contains a 3′–5′ proofreading exonuclease, while Pol α lacks a proofreading exonuclease.Citation7,9 The 2nd largest subunit of these polymerases, referred to as the B subunit, is essential for cell viability but the role of the B subunits are not understood for any of the 3 DNA polymerases. The smallest subunits of Pol α, Pri1-Pri2, contain the primase activity.Citation7,8 The small Dpb3-Dpb4 subunits of Pol epsilon are non-essential and form a histone like heterodimer fold.Citation20 The function of the third subunit (Pol32) of Pol delta is unknown, but it contains a PCNA binding motif and both Pol31 and Pol32 are also subunits of Pol zeta, a translesion repair polymerase.Citation21,22 This review provides a brief overview of the current understanding of how these eukaryotic DNA polymerases function together at the replication fork.
Genetic studies indicate pol epsilon and pol delta act on different strands
Considering that DNA consists of 2 strands, one may expect that only 2 polymerase molecules would be needed for DNA replication. The fact that there are 3 different essential DNA polymerases suggested that Pol α, with its intrinsic priming activity, probably functions as the primase while Pol epsilon and Pol delta perform bulk leading and lagging strand synthesis.Citation7-9 Supporting this view is the fact that Pol α has no proofreading exonuclease and thus has lower fidelity compared to Pols epsilon and delta, which each contain a 3′–5′ proofreading exonuclease.Citation7-9 Recent studies have identified mutations in the exonuclease active sites of Pol epsilon and Pol delta that are associated with colorectal cancer, consistent with a central role of these polymerases in cellular replication.Citation23,24 It has long been thought that Pol epsilon and Pol delta function on different strands (i.e. leading vs lagging), and this presumption was supported by early studies on exonuclease mutants of Pols epsilon and delta.Citation25 However, these early studies could not assign the DNA polymerases to one strand or the other.
The first strong evidence that identified the strand upon which a DNA polymerase functions came from genetic studies using a Pol epsilon active site mutant that leaves a mutation signature on the strand that it acts upon.Citation26,27 These studies provided convincing evidence for Pol epsilon on the leading strand in the budding yeast (Saccharomyces cerevisiae).Citation27 Similar studies using active sites mutants of Pol delta indicated that Pol delta performs lagging strand synthesis.Citation28 Further genetic studies employed mutations in the active sites of Pols epsilon and delta that frequently misincorporate rNTPs, with the same conclusions.Citation29 Experiments in fission yeast (Schizosaccharomyces pombe) also supported the assignments of Pol epsilon and Pol delta on the leading and lagging strands, respectively.Citation30 Additional support was provided by DNA-protein cross-linking studies demonstrating that Pol epsilon cross-links to the leading strand and Pol delta cross-links to the lagging strand.Citation31 Despite these observations, there remain caveats to the experiments, and a recent report concluded that Pol delta acts as the major replicase on both strands, similar to replication of the SV40 genome.Citation32 In addition, the catalytic domain of Pol2, the polymerase subunit of Pol epsilon, can be deleted and cells survive, although the cells show severe defects in S phase progression.Citation33 It was presumed that Pol delta takes over in these mutant cells, possibly similar to studies showing that Pol I takes over chromosome replication in E. coli when the replicative Pol III is deleted.Citation34 In contrast to the deletion of the catalytic domain of Pol2, active site point mutants of Pol2 were lethal, indicating that Pol epsilon is normally used for replication.Citation35 Considering that the issue of polymerase assignment to particular strands is not fully resolved, additional information from a different line of investigation may help clarify the issue. Recent experiments have reconstituted the eukaryotic replsome from pure proteins in vitro, and the biochemical studies have shed new light onto this question.
The eukaryotic cellular helicase
Biochemical reconstitution studies of eukaryotic cellular replication have lagged far behind the bacterial in vitro replication field. This has in part been due to the difficulty in identification of the eukaryotic cellular helicase. Bacterial, archaeal and many phage and viral systems utilize a homohexameric helicase to unwind DNA at the replication fork.Citation36,37 The eukaryotic replicative helicase was believed for many years to be the heterohexameric Mcm2-7 complex.Citation38 Indeed, it has long been known that a double hexamer of the Mcm2-7 complex is loaded onto an origin in a G1 phase specific licensing step. The SV40 large T-antigen helicase is also assembled as a double hexamer at the viral origin,Citation39 and thus Mcm2-7 was expected to perform as a helicase in similar fashion to SV40 large T-antigen. In addition, the archaeal Mcm homohexamer is an active helicase, supporting the hypothesis that Mcm2-7 may be the eukaryotic cellular helicae.Citation40 While the isolated Mcm2-7 complex is nearly devoid of helicase activity, a Mcm4,6,7 subassembly is a robust 3′–5′ helicase.Citation41 But more recent studies discovered that the true cellular helicase requires 5 additional proteins.Citation42
Identification and purification of the active eukarytotic helicase was accomplished in the Drosophila system.Citation42,43 The active helicase is a complex of 11 different subunits, Mcm2-7, Cdc45 protein, and the heterotetramer GINS (go-ichi-ni-san). This helicase complex, referred to as CMG (Cdc45, Mcm2-7, GINS), displays robust helicase activity.Citation42,43 The Cdc45 and GINS accessory subunits do not interact with ATP, and thus are proposed to provide helicase activity by holding the Mcm2-7 complex in an active conformation for helicase activity. Numerous cell biology and biochemical studies have shown that the Cdc45 and GINS are chaperoned to the Mcm2-7 complex in a multistep reaction at an origin that involves several initiation factors and 2 cell cycle kinases (reviewed in Citation44). Hence, activation of origins of replication appear to be intricately intertwined with activation of the CMG helicase, and possibly even defined by this process.
Identification of the cellular helicase provided the missing link necessary to reconstitute the eukaryotic replication fork in vitro. However, CMG is present at very low concentrations in the cell and amounts needed for intensive biochemical studies required expression by recombinant means. Recombinant Drosophila CMG has been expressed in the baculovirus system,Citation43 and CMG of budding yeast and human have also been expressed by recombinant means.Citation45,46 In all cases, the isolated recombinant CMG displays ATP dependent 3′–5′ DNA helicase activity.Citation43,45,46 The 3′–5′ polarity of unwinding places CMG on the leading strand of a replication fork. Studies of bacterial and viral homohexameric helicases suggest that they encircle single-strand DNA and use ATP to track along it while excluding the other strand of DNA from inside the ring.Citation36,37,47 In this “steric exclusion” model of unwinding, the helicase unwinds dsDNA as it tracks along the strand that it encircles, acting as a wedge to separate the strand that it sterically excludes (i.e., the strand that it does not encircle).
Reconstitution of the leading strand replisome
Pull-outs of CMG subunits from extracts of budding yeast, followed by mass spectrometry analysis has identified several additional factors that travel with CMG, referred to as the replisome progression complex (RPC).Citation48,49 In addition to CMG, the RPC contains Pol α, Ctf4, Mcm10, The Mrc1-Tof1-Csm3 heterotrimer involved in checkpoint control, the FACT complex for nucleosome handling and Topoisomerase I. The RPC is presumed to function at a moving replication fork, along with Pol epsilon, Pol delta, RFC, PCNA and the RPA single-strand binding protein.
The first biochemical reconstitution of a functional eukaryotic leading strand replication fork entailed use of 27 different recombinant polypeptides ( and Citation45,46), and reconstitution of a coupled leading-lagging strand replisome in vitro utilized 31 different proteins ().Citation50 Initiation at a yeast origin has also been accomplished by recombinant proteins in the absence of Pol delta, RFC and PCNA.Citation51 Studies of the 3 DNA polymerases in the reconstituted replication fork system,Citation45,50 and studies of individual DNA polymerases,Citation52,53 has provided insight into the mechanisms by which DNA polymerases are assigned to their respective DNA strands at a replication fork.
Figure 2. Example of leading-lagging strand replication in vitro. (A) The substrate used in the in vitro assays is a 3 kb forked DNA that has no dG residues on the leading strand and no dC residues on the lagging strand. Therefore 32P-dCTP specifically labels the leading strand product and 32P-dGTP labels lagging strand products. (B) Leading strand reaction time course using 32P-dCTP and either Pols α + epsilon, or Pols α + epsilon + delta. (C) While a low level of lagging strand products are observed with only Pols α + epsilon, significantly more lagging strand fragments are formed by addition of Pol delta. (D) Quantitation of DNA synthesis shows similar extents of leading and lagging strand replication. The panels of the figure are adapted with permission fromCitation50. Specifically, panels a, b are from Supplemental Figure 6a,b, panel c is from Figure 5b, and panel d is from Figure 7b of Citation50.
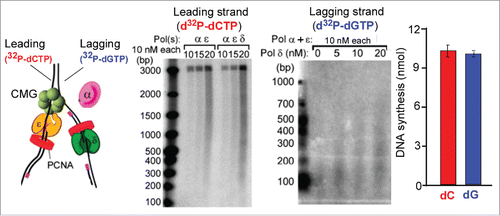
Biochemical study of pure CMG helicase, DNA polymerases, PCNA clamp, RFC clamp loader and RPA single-strand binding protein used a pre-formed DNA fork substrate.Citation45,50 The forked DNA lacks dC residues on one strand and dG residues on the other (). This enables specific labeling of leading or lagging strand synthesis depending on whether 32P-dCTP or 32P-dGTP is used (). Processive leading strand synthesis was observed using CMG and Pol epsilon, along with RFC, PCNA and RPA.Citation45 Substituting Pol delta for Pol epsilon resulted in comparatively little synthesis, indicating that CMG stabilizes Pol epsilon but not Pol delta. Hence, asymmetric polymerase use on the leading strand is recapitulated in vitro. Pol epsilon function on the leading strand is consistent with the bulk of genetic studies. However, it is important to note that future studies that include other proteins that travel with forks could alter these conclusions.
Protein interaction studies reveal that Pol epsilon binds directly to CMG, forming a 15-protein “CMGE” complex.Citation54 Recent single-particle EM 3D reconstruction studies of CMGE reveal that Pol epsilon sits on the GINS-Cdc45 accessory factors, and on the C-terminal side of the Mcm2-7 motor subunits.Citation55 Keeping in mind that CMG encircles the leading strand, the interaction of Pol epsilon with CMG may confine Pol epsilon function to the leading strand. In this view, the asymmetric use of Pol epsilon over Pol delta in leading strand synthesis is facilitated by a direct connection between CMG and Pol epsilon.
Lagging strand synthesis
Pol delta has been studied for many years, and characterization of its biochemical properties indicated that it is well suited to the actions required on the lagging strand. For instance, Pol delta is highly efficient in switching with Pol α on a primed site.Citation56 The Pol delta-PCNA complex is uniquely capable of functioning with Fen1 nuclease in the removal of RNA primers to produce a ligatable nick for sealing Okazaki fragments, while Pol epsilon-PCNA does not fulfill this function.Citation53 Pol delta-PCNA also contains a limited strand displacement activity that provides an efficient substrate for Fen1 nuclease.Citation52 In addition, the strand displacement activity of Pol delta-PCNA has recently been shown to be processive during the 1–2 seconds required for action with Fen1 during Okazaki fragment repair.Citation57 On a primed RPA coated single-strand DNA, Pol delta-PCNA is processive but quickly self-ejects from the PCNA clamp shortly after completing replication (i.e. it only stays with completed DNA the few seconds needed to function with Fen1).Citation58 This self-ejection feature of Pol delta-PCNA may underlie the observation that Pol delta-PCNA has only feeble activity on the leading strand with CMG.Citation45 Specifically, Pol delta-PCNA extends DNA faster than CMG unwinds DNA, and thus Pol delta-PCNA will “bump” into CMG which may trigger the self-ejection of Pol delta from PCNA resulting in the observed distributive synthesis with CMG.Citation45 Biochemical studies also show that Pol delta rapidly switches with Pol epsilon on PCNA-primed ssDNA, a lagging strand mimic.Citation45 Extrapolation of these actions to a moving replication fork suggests that even if Pol epsilon were to assemble with PCNA at a lagging strand primed site, Pol delta would soon switch with it and take over.
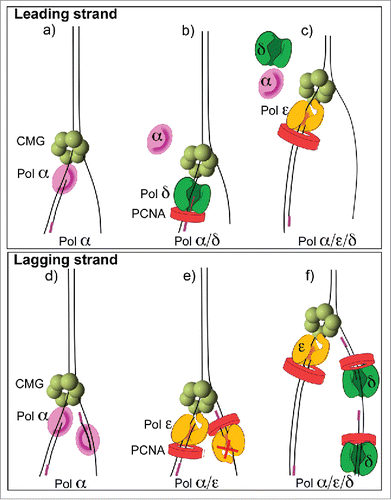
Reconstitution of an efficient leading-lagging strand replication system required the addition of all 3 DNA polymerases, along with CMG, RFC, PCNA and RPA.Citation50 In the presence of all 3 DNA polymerases, the leading and lagging strands were synthesized in equal amounts, and the Okazaki fragments were the length expected from in vivo studies (). Experiments using individual polymerases, or combinations of 2 DNA polymerases with CMG, provided insight into the diferent functions of Pols α, epsilon and delta on the leading and lagging strands (summarized in ). One unexpected finding was that Pol epsilon could not extend lagging strand primers into Okazaki fragments, even in the complete absence of Pol delta. Hence, CMG-Pol epsilon is not capable of acting on the lagging strand. Furthermore, in the absence of both Pol epsilon and Pol delta, Pol α is capable of priming and extending both the leading and lagging strands with CMG (explained further in the next section). However, the presence of Pol epsilon results in a Pol epsilon switch with Pol α, which stimulates the leading strand but inhibits the lagging strand (). The mechanism that prevents Pol epsilon function on the lagging strand is not yet understood. One possibility is that Pol epsilon is inefficient in extending short primed sites made by Pol α, as suggested from studies of isolated Pol epsilon.Citation59 In summary, Pol epsilon and Pol delta are only efficient in synthesis when they are on the “right” strand. Specifically, Pol epsilon is most active on the leading strand and Pol delta is most active on the lagging strand (). The in vitro results indicate that asymmetric activity of Pol epsilon and Pol delta on the leading and lagging strands is an inherent property of the core helicase/polymerase components of the replisome.
Figure 3. Asymmetric use of DNA polymerases at a replication fork. The diagrams refer to conclusions from in vitro replisome reconstitution reactions demonstrating that Pol epsilon is the dominant enzyme on the leading strand and Pol delta is the dominant enzyme on the lagging strand Citation38,43. (A) Pol α functions with CMG on the leading strand, (B) Pol delta switches with Pol α on the leading strand but lowers synthesis, (C) Pol epsilon takes over from both Pols α and delta to provide the most synthesis on the leading strand. (D) Pol α can function on the lagging strand in the absence of other polyemrases, (E) Pol epsilon takes over from Pol α on both strands, but is only active on the leading strand, (F) Pol delta extends Okazaki fragments in the presence of Pol epsilon and Pol α.
Priming of DNA synthesis
One may question why Pol α contains a DNA polymerase activity, considering that the cell has 2 other DNA polymerases for the leading and lagging strands. Conceptually, only the RNA priming activity of Pol α should be required for the replication fork. The relevance of DNA synthesis by Pol α has not garnered much attention. Archaea do not have a 4-subunit Pol α and instead contain only the heterodimer primase (i.e., Pri1-Pri2).Citation4,5 Archaea differ from eukaryotes in having circular chromosomes and perhaps the DNA polymerase of Pol α is important to convert the product of telomerase to double-strand DNA (i.e. telomere second strand synthesis). Regardless of the polymerase function, all eukaryotes studied thus far have a 4-subunit Pol α. In vitro studies show that Pol α requires CMG for priming activity in the presence of the RPA single-strand binding protein.Citation50 The dependence of priming activity on the interaction of primase with its cognate helicase is a common theme in bacterial and phage systems.Citation1,2,3 Studies in the SV40 system also demonstrate that Pol α requires interaction with the SV40 T-antigen helicase for priming activity on plasmid DNA containing the SV40 origin.Citation60
As described above, Pol α can prime and extend both the leading and lagging strands in combination with CMG helicase,Citation50 but the presence of Pol epsilon or Pol delta stops Pol α activity.Citation50 This remains the case even when the Pol epsilon or Pol delta is functioning on the “wrong” strand. For example, Pol α lagging strand products are inhibited by Pol epsilon, and conversely, leading strand extension by Pol α is inhibited by Pol delta.Citation50 These observations suggest that in the cell, where all 3 DNA polymerases are present simultaneously, that Pol α DNA polymerase activity will be sharply curtailed by DNA polymerases epsilon and delta. However, the priming function of Pol α is not curtailed.
Conclusion
Recent studies demonstrate that a functional eukaryotic replisome can be reconstituted from pure proteins, thus laying a foundation for future mechanistic studies. The initial studies reveal asymmetric functional properties of Pol epsilon and Pol delta on the leading and lagging strands, respectively, yet much remains to be understood. The current stage of development has only used naked DNA of limited sequence context. In the cell, the replisome will encounter a vast combinatorial landscape of nucleotide sequence, various types of DNA lesions, transcribing RNA polymerase and many other protein blocks. Additionally, eukaryotic DNA is packaged into nucleosomes, some of which condense DNA into highly packed heterochromatic regions. There also exist cohesion rings that hold the 2 daughter chromosomes together, possibly a consequence of a replisome that replicates through the giant cohesion rings. The replisome copies every nucleostide of the entire genome, and thus must have mechanisms to cope with and traverse all impediments to replication. How the replisome deals with the many biological impediments it encounters is completely unknown. Furthermore, the replisome is regulated by a variety of posttranslational modifications. For example, induction of the DNA damage checkpoint response results in phosphorylation of Mcm subunits, and an initial study indicates that CMG helicase activity is altered in response to DNA damage induced phosphorylation.Citation61 DNA damage also leads to ubiquitinylation of PCNA, and this is associated with translesion bypass performed by a variety of specialized translesion DNA polymerases that function with ubiquitinated PCNA.Citation62 The Mcm10 subunit of the RPC is an essential protein, yet little is known about its function, and the Ctf4 homotrimer is known to cross-link Pol α to CMG, but the functional consequence of this interaction is still unknown.Citation55,63 Interestingly, the Mcm2 subunit binds a H3-H4 heterotetramer.Citation64,65 Whether the Mcm2 subunit, in the context of CMG, can bind histones is not yet known, but the results with pure Mcm2 indicate that the interaction faciliates nucleosome assembly onto DNA.Citation58 The FACT nucleosome handling factor is also a member of the RPC. Thus it seems likely that the repllsome machinery is directly involved in nucleosome handling, and thus might play a direct role in the transfer of epigenetic information to the next generation.
Abbreviations
| CMG | = | Cdc45-Mcm2-7-GINS |
| FACT | = | facilitates chromatin transcription |
| GINS | = | Go-Ichi-Ni-San |
| MCM | = | minichromosome maintenance |
| PCNA | = | proliferating cell nuclear antigen |
| Pol | = | DNA polymerase |
| RFC | = | replication factor C |
| RPA | = | replication protein A |
| RPC | = | replisome progression complex |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
The authors are grateful for funding from the US National Institutes of Health (GM115809) and the Howard Hughes Medical Institute.
References
- Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem 2001; 70:181-208; PMID:11395406; http://dx.doi.org/10.1146/annurev.biochem.70.1.181
- McHenry CS. Bacterial replicases and related polymerases. Curr Opin Chem Biol 2011; 15:587-94; PMID:21855395; http://dx.doi.org/10.1016/j.cbpa.2011.07.018
- O'Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol 2013; 5:a010108; PMID:23818497; http://dx.doi.org/10.1101/cshperspect.a010108
- Beattie TR, Bell SD. Molecular machines in archaeal DNA replication. Curr Opin Chem Biol 2011; 15:614-9; PMID:21852183; http://dx.doi.org/10.1016/j.cbpa.2011.07.017
- Kelman LM, Kelman Z. Archaeal DNA replication. Annu Rev Genet 2014; 48:71-97; PMID:25421597; http://dx.doi.org/10.1146/annurev-genet-120213-092148
- Kaguni LS, Rossignol JM, Conaway RC, Lehman IR. Isolation of an intact DNA polymerase-primase from embryos of Drosophila melanogaster. Proc Natl Acad Sci U S A 1983; 80:2221-5; PMID:6403945; http://dx.doi.org/10.1073/pnas.80.8.2221
- Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol 2005; 40:115-28; PMID:15814431; http://dx.doi.org/10.1080/10409230590935433
- Kaguni LS, Rossignol JM, Conaway RC, Banks GR, Lehman IR. Association of DNA primase with the β/gamma subunits of DNA polymerase α from Drosophila melanogaster embryos. J Biol Chem 1983; 258:9037-9; PMID:6409898
- MacNeill S. The Eukaryotic Replisome: A Guide to Protein Structure and Function. Springer Netherlands, 2012.
- Conaway RC, Lehman IR. A DNA primase activity associated with DNA polymerase α from Drosophila melanogaster embryos. Proc Natl Acad Sci U S A 1982; 79:2523-7; PMID:6806812; http://dx.doi.org/10.1073/pnas.79.8.2523
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem 1998; 67:721-51; PMID:9759502; http://dx.doi.org/10.1146/annurev.biochem.67.1.721
- Byrnes JJ, Downey KM, Black VL, So AG. A new mammalian DNA polymerase with 3′ to 5′ exonuclease activity: DNA polymerase delta. Biochemistry 1976; 15:2817-23; PMID:949478; http://dx.doi.org/10.1021/bi00658a018
- Melendy T, Stillman B. Purification of DNA polymerase delta as an essential simian virus 40 DNA replication factor. J Biol Chem 1991; 266:1942-9; PMID:1671044
- Tan CK, Castillo C, So AG, Downey KM. An auxiliary protein for DNA polymerase-delta from fetal calf thymus. J Biol Chem 1986; 261:12310-6; PMID:3745189
- Tsurimoto T, Stillman B. Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol Cell Biol 1989; 9:609-19; PMID:2565531; http://dx.doi.org/10.1128/MCB.9.2.609
- Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 1992; 69:425-37; PMID:1349852; http://dx.doi.org/10.1016/0092-8674(92)90445-I
- Stukenberg PT, Studwell-Vaughan PS, O'Donnell M. Mechanism of the sliding β-clamp of DNA polymerase III holoenzyme. J Biol Chem 1991; 266:11328-34; PMID:2040637
- Morrison A, Araki H, Clark AB, Hamatake RK, Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell 1990; 62:1143-51; PMID:2169349; http://dx.doi.org/10.1016/0092-8674(90)90391-Q
- Lee SH, Pan ZQ, Kwong AD, Burgers PM, Hurwitz J. Synthesis of DNA by DNA polymerase epsilon in vitro. J Biol Chem 1991; 266:22707-17; PMID:1682323
- Pursell ZF, Kunkel TA. DNA polymerase epsilon: a polymerase of unusual size (and complexity). Prog Nucleic Acid Res Mol Biol 2008; 82:101-45; PMID:18929140; http://dx.doi.org/10.1016/S0079-6603(08)00004-4
- Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. J Biol Chem 2012; 287:17281-7; PMID:22465957; http://dx.doi.org/10.1074/jbc.M112.351122
- Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc Natl Acad Sci U S A 2012; 109:12455-60; PMID:22711820; http://dx.doi.org/10.1073/pnas.1206052109
- Albertson TM, Ogawa M, Bugni JM, Hays LE, Chen Y, Wang Y, Treuting PM, Heddle JA, Goldsby RE, Preston BD. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci U S A 2009; 106:17101-4; PMID:19805137; http://dx.doi.org/10.1073/pnas.0907147106
- Briggs S, Tomlinson I. Germline and somatic polymerase epsilon and delta mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol 2013; 230:148-53; PMID:23447401; http://dx.doi.org/10.1002/path.4185
- Shcherbakova PV, Pavlov YI. 3′–>5′ exonucleases of DNA polymerases epsilon and delta correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics 1996; 142:717-26; PMID:8849882
- Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol 2008; 18:521-7; PMID:18824354; http://dx.doi.org/10.1016/j.tcb.2008.08.005
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 2007; 317:127-30; PMID:17615360; http://dx.doi.org/10.1126/science.1144067
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell 2008; 30:137-44; PMID:18439893; http://dx.doi.org/10.1016/j.molcel.2008.02.022
- Clausen AR, Lujan SA, Burkholder AB, Orebaugh CD, Williams JS, Clausen MF, Malc EP, Mieczkowski PA, Fargo DC, Smith DJ, et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol 2015; 22:185-91; PMID:25622295; http://dx.doi.org/10.1038/nsmb.2957
- Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet 2011; 7:e1002407; PMID:22144917; http://dx.doi.org/10.1371/journal.pgen.1002407
- Yu C, Gan H, Han J, Zhou ZX, Jia S, Chabes A, Farrugia G, Ordog T, Zhang Z. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell 2014; 56:551-63; PMID:25449133; http://dx.doi.org/10.1016/j.molcel.2014.09.017
- Johnson RE, Klassen R, Prakash L, Prakash S. A Major Role of DNA Polymerase delta in Replication of Both the Leading and Lagging DNA Strands. Mol Cell 2015; 59:163-75; PMID:26145172; http://dx.doi.org/10.1016/j.molcel.2015.05.038
- Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell 1999; 3:679-85; PMID:10360184; http://dx.doi.org/10.1016/S1097-2765(00)80361-5
- Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem 1999; 274:22283-8; PMID:10428796; http://dx.doi.org/10.1074/jbc.274.32.22283
- Niwa O, Bryan SK, Moses RE. Alternate pathways of DNA replication: DNA polymerase I-dependent replication. Proc Natl Acad Sci U S A 1981; 78:7024-7; PMID:7031666; http://dx.doi.org/10.1073/pnas.78.11.7024
- Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol 2011; 21:240-8; PMID:21282052; http://dx.doi.org/10.1016/j.sbi.2011.01.002
- Patel SS, Picha KM. Structure and function of hexameric helicases. Annu Rev Biochem 2000; 69:651-97; PMID:10966472; http://dx.doi.org/10.1146/annurev.biochem.69.1.651
- Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell 2008; 31:287-93; PMID:18657510; http://dx.doi.org/10.1016/j.molcel.2008.05.020
- Mastrangelo IA, Hough PV, Wall JS, Dodson M, Dean FB, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature 1989; 338:658-62; PMID:2539565; http://dx.doi.org/10.1038/338658a0
- Chong JP, Hayashi MK, Simon MN, Xu RM, Stillman B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc Natl Acad Sci U S A 2000; 97:1530-5; PMID:10677495; http://dx.doi.org/10.1073/pnas.030539597
- Ishimi Y. A DNA helicase activity is associated with an MCM4, −6, and −7 protein complex. J Biol Chem 1997; 272:24508-13; PMID:9305914; http://dx.doi.org/10.1074/jbc.272.39.24508
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A 2006; 103:10236-41; PMID:16798881; http://dx.doi.org/10.1073/pnas.0602400103
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell 2010; 37:247-58; PMID:20122406; http://dx.doi.org/10.1016/j.molcel.2009.12.030
- Siddiqui K, On KF, Diffley JF. Regulating DNA replication in eukarya. Cold Spring Harb Perspect Biol 2013; 5; PMID:23838438; http://dx.doi.org/10.1101/cshperspect.a012930
- Georgescu RE, Langston L, Yao NY, Yurieva O, Zhang D, Finkelstein J, Agarwal T, O'Donnell ME. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol 2014; 21:664-70; PMID:24997598; http://dx.doi.org/10.1038/nsmb.2851
- Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc Natl Acad Sci U S A 2012; 109:6042-7; PMID:22474384; http://dx.doi.org/10.1073/pnas.1203734109
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 2011; 146:931-41; PMID:21925316; http://dx.doi.org/10.1016/j.cell.2011.07.045
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 2006; 8:358-66; PMID:16531994; http://dx.doi.org/10.1038/ncb1382
- Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase α within the eukaryotic replisome. EMBO J 2009; 28:2992-3004; PMID:19661920; http://dx.doi.org/10.1038/emboj.2009.226
- Georgescu RE, Schauer GD, Yao NY, Langston LD, Yurieva O, Zhang D, Finkelstein J, O'Donnell ME. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. Elife 2015; 4:e04988; PMID:25871847; http://dx.doi.org/10.7554/eLife.04988
- Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015; 519:431-5; PMID:25739503; http://dx.doi.org/10.1038/nature14285
- Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem 2009; 284:4041-5; PMID:18835809; http://dx.doi.org/10.1074/jbc.R800062200
- Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev 2004; 18:2764-73; PMID:15520275; http://dx.doi.org/10.1101/gad.1252304
- Langston LD, Zhang D, Yurieva O, Georgescu RE, Finkelstein J, Yao NY, Indiani C, O'Donnell ME. CMG helicase and DNA polymerase epsilon form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci U S A 2014; 111:15390-5; PMID:25313033; http://dx.doi.org/10.1073/pnas.1418334111
- Sun J, Shi Y, Georgescu RE, Z Y, Chait BT, Li H, O'Donnell ME. The Architecture of a Eukaryotic Replisome. Nat Struct Mol Biol 2015; 22:976-82; PMID:26524492; http://dx.doi.org/10.1038/nsmb.3113
- Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase α and delta during initiation of leading and lagging strand synthesis. J Biol Chem 1991; 266:1961-8; PMID:1671046
- Stodola JL, Burgers PM. Resolving individual steps of Okazaki-fragment maturation at a millisecond timescale. Nat Struct Mol Biol 2016; 23(5):402-8; PMID:27065195
- Langston LD, O'Donnell M. DNA polymerase delta is highly processive with proliferating cell nuclear antigen and undergoes collision release upon completing DNA. J Biol Chem 2008; 283:29522-31; PMID:18635534; http://dx.doi.org/10.1074/jbc.M804488200
- Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol 2006; 13:35-43; PMID:16369485; http://dx.doi.org/10.1038/nsmb1040
- Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature 1994; 369:207-12; PMID:7910375; http://dx.doi.org/10.1038/369207a0
- Ilves I, Tamberg N, Botchan MR. Checkpoint kinase 2 (Chk2) inhibits the activity of the Cdc45/MCM2-7/GINS (CMG) replicative helicase complex. Proc Natl Acad Sci U S A 2012; 109:13163-70; PMID:22853956; http://dx.doi.org/10.1073/pnas.1211525109
- McIntyre J, McLenigan MP, Frank EG, Dai X, Yang W, Wang Y, Woodgate R. Posttranslational Regulation of Human DNA Polymerase iota. J Biol Chem 2015; 290:27332-44; PMID:26370087; http://dx.doi.org/10.1074/jbc.M115.675769
- Simon AC, Zhou JC, Perera RL, van Deursen F, Evrin C, Ivanova ME, Kilkenny ML, Renault L, Kjaer S, Matak-Vinkovic D, et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 2014; 510:293-7; PMID:24805245; http://dx.doi.org/10.1038/nature13234
- Huang H, Stromme CB, Saredi G, Hodl M, Strandsby A, Gonzalez-Aguilera C, Chen S, Groth A, Patel DJ. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat Struct Mol Biol 2015; 22:618-26; PMID:26167883; http://dx.doi.org/10.1038/nsmb.3055
- Wang H, Wang M, Yang N, Xu RM. Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2. Protein Cell 2015; 6:693-7; PMID:26186914; http://dx.doi.org/10.1007/s13238-015-0190-0f
