ABSTRACT
In this study, we explore the plasticity during evolution of proteins of the higher plant nuclear envelope (NE) from the most ancestral plant species to advanced angiosperms. The higher plant NE contains a functional Linker of Nucleoskeleton and Cytoskeleton (LINC) complex based on conserved Sad1-Unc84 (SUN) domain proteins and plant specific Klarsicht/Anc1/Syne homology (KASH) domain proteins. Recent evidence suggests the presence of a plant lamina underneath the inner membrane and various coiled-coil proteins have been hypothesized to be associated with it including Crowded Nuclei (CRWN; also termed LINC and NMCP), Nuclear Envelope Associated Protein (NEAP) protein families as well as the CRWN binding protein KAKU4. SUN domain proteins appear throughout with a key role for mid-SUN proteins suggested. Evolution of KASH domain proteins has resulted in increasing complexity, with some appearing in all species considered, while other KASH proteins are progressively gained during evolution. Failure to identify CRWN homologs in unicellular organisms included in the study and their presence in plants leads us to speculate that convergent evolution may have occurred in the formation of the lamina with each kingdom having new proteins such as the Lamin B receptor (LBR) and Lamin-Emerin-Man1 (LEM) domain proteins (animals) or NEAPs and KAKU4 (plants). Our data support a model in which increasing complexity at the nuclear envelope occurred through the plant lineage and suggest a key role for mid-SUN proteins as an early and essential component of the nuclear envelope.
Introduction
The nuclear envelope is a key component of eukaryotic cells and may be considered to be composed of 3 elements, the nuclear membrane, nuclear pore complexes and the nuclear lamina.Citation13,Citation19 These structural components are essential for many processes including nuclear morphology, nuclear migration, chromatin organization and regulation of gene expression.Citation15 Significant progress has been made in describing novel plant nuclear envelope proteins.Citation29,Citation35,Citation43 In Arabidopsis thaliana (A. thaliana), these include components of the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex for which functional data is slowly being revealed. Arabidopsis contains proteins of the inner nuclear envelope of the SUN domain family including Cter-Sad1-Unc84 (Cter-SUN)Citation16,Citation17,Citation28 as well as mid-SUN domain proteins in which a SUN-domain homologous to that of the C-ter SUNs is located centrally within the protein.Citation18 It also contains proteins of the outer nuclear envelope, of the KASH domain protein family including WPP Domain Interacting Proteins [WIPs], SUN interacting Nuclear Envelope Proteins [SINEs] and Arabidopsis thaliana Toll Interleukin Receptor domain KASH protein [TIK].Citation41,Citation18,Citation40 In addition, plant proteins proposed to form the nuclear lamina - Crowded Nuclei (CRWNs;Citation9,Citation37) and CRWN-interacting proteins such as KAKU4Citation14) as well as Nuclear Envelope Associated Proteins (NEAPs), which may be associated with the lamina,Citation30 have been described in A. thaliana and shown to localize to the nuclear periphery.
Sequence data now available permits comparison of components of the nuclear envelope between algae, mosses, gymnosperms and angiosperms with the components of A. thaliana. Functional analysis of these genes is challenging because they belong to small gene families as a consequence of gene and whole-genome duplication (WGD) creating duplicate genes and thus gene redundancy.Citation12,Citation33 Whole-genome duplication (WGD) is recognized as an important event for genome evolution in animals, plants and fungi and to drive key new features, with resulting increased complexity and speciation.Citation33 Following WGD, massive gene loss can occur restoring the diploid state for most duplicated loci while few duplicated genes remain and may provide new evolutionary innovation including structures (e.g. floral organs) and adaptations.Citation21 Previous analyses of plant genomes have shown that seed plants share an ancient WGD event, zeta.Citation20 A second WGD, epsilon, has been detected shortly before the diversification of angiosperms. These two WGDs are suggested to play a role in the origin and rapid diversification of the angiosperms.Citation20 Finally, the gamma WGD occurred after eudicot/monocot diversification, followed by several partial or complete duplication events.
In this study, we have selected 20 representative species based on the revised classification of eukaryotes,Citation1 their available genome sequences and gene expression description, in order to explore the evolution of components of the plant nuclear envelope. Availability of genome sequence data for the core eudicot Amborella trichopoda provides an opportunity to explore the nuclear envelope of a primitive angiosperm. Amborella, a New Caledonian shrub, has been suggested as the sole surviving sister species of all other angiosperms and is unique in sequenced plant genomes in showing no evidence of recent, lineage-specific genome duplications.Citation32 The Amborella genome therefore offers an opportunity to explore the composition of an ancestral plant nuclear envelope and the effect of genomic changes after polyploidy in other angiosperms.Citation32 Finally, RNAseq data for each species were used to describe expression levels within species to establish that the genes are active and not pseudogenes and to demonstrate gene activity and when possible tissue specific expression patterns.
The aim of the work presented in this paper was to explore the evolution of nuclear envelope proteins in unicellular algae and multicellular plants and to provide evidence for the composition of the simplest functional plant LINC complex. This study provides valuable information for mutant and other functional studies by identifying potential redundancy and specialization in nuclear envelope and lamina-like components.
Material and methods
Homologous LINC complex and lamin-like protein detection
A Perl script was developed and applied to proteomic data to identify KASH domain proteins. The program tests the presence of the trans-membrane (TM) domain and 4 specific amino acids at the C-terminus, which are characteristic for KASH proteins. The position of the TM domain is variable and the script searches this TM domain up to 40 amino acids away from the KASH-specific C-terminal motifs detected in A. thaliana (either VIPT, VVPT, AVPT, PLPT, TVPT, LVPT or PPPS.Citation41,Citation18,Citation42). The identification of the TM domain is based on the Kyte-Doolittle method.Citation25 Only proteins, which possess a TM domain and the 4 KASH specific amino acids in the C-terminus of the protein, were selected.
For all proteins of interest a Basic Local Alignment Search Tool protein (BLASTp) was used with default parameters as well as HMMER (Hidden Markov Model-based sequence alignment tool; http//hmmer.org). The best hits were retained and used for phylogenetic analysis.Citation1990 The proteome of each species was used as reference for the BLASTp (), and the protein sequences of the LINC complex as well as the putative lamina of A. thaliana were used as queries (Supplementary Table 1). BLASTp results are given as supplementary Table 2 (mid-SUN), 3 (Cter-SUN), 4 (WIP), 5 (SINE), CRWN (6), NEAP (7) and KAKU4 (8). Reciprocal BLASTp was used to verify the relevance of all identified orthologs.
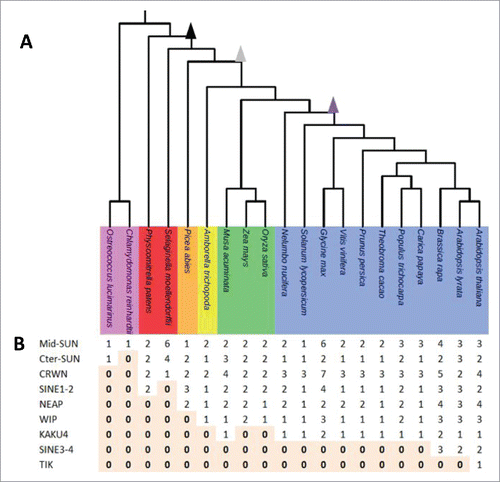
Figure 1. Distribution of components of plant nuclear envelope in the plant kingdom. (A) Selected plant lineages used in this study from left to right: Unicells Algae (pink), Moss and Club Moss (red), Gymnosperm (orange), Basal Angisoperms (yellow), Monocots (green) and Eudicots (blue). Zeta epsilon and gamma WGDs are indicated as arrow heads respectively in black, gray and purple. (B) Distribution of the 9 protein families (rows) in the 20 species (columns). Absence (0) of a given protein is highlighted in light orange.
Phylogenetic reconstruction
Selected sequences were first aligned with MUSCLE, a multiple sequence alignment tool,Citation11 using default parameters. The alignment was then refined using GblocksCitation34 Fast-Tree was then applied with default parameters, for the construction of the phylogenetic tree.Citation31 Fast-Tree infers approximately-maximum-likelihood phylogenetic trees from alignments. Finally, phylogenetic trees were drawn using the Interactive Tree Of Life ITOL.Citation26
RNA sequencing data and analysis
Data used for the RNA-seq analysis was obtained from the NCBI (http://www.ncbi.nlm.nih.gov/geo/browse/) or from the Amborella Genome Database, respectively (http://amborella.huck.psu.edu/). Five different tissues (leaves, roots, flowers, flower buds, and seeds/siliques) as well as total seedling were chosen for the analysis of the expression patterns of the genes of interest (Supplementary Table 1). The expression was analyzed for 10 species (Supplementary Table 9). Reads from RNA-Seq libraries were mapped onto the candidate gene sequences allowing no mismatches using TOPHAT v 2.0.14Citation22 with standard settings and maximum of multihits set at 1, minimum intron length set at 15 bp, and maximum intron length set as 6,000 bp. Reads were added together for each gene using HTseq-count with the overlap resolution mode set as intersection-non empty and with no strand-specific protocol.Citation3 Transcription levels in Reads Per Kilobase of transcript per Million mapped reads (RPKM) were normalized to AtSAND (At2g28390;Citation7 and Supplementary Table 10). SAND was chosen due to its constant gene expression levels across different tissues at developmental stages in Arabidopsis thaliana.Citation7 For each species, the SAND homolog with the closed sequence identity to AtSAND was chosen. Furthermore, absolute SAND expression levels (in RPKM) in different species were comparable to expression of Arabidopsis AtSAND.
Results and discussion
In order to gain an insight into the evolutionary development of known plant nuclear envelope proteins, we reconstructed the phylogenetic distribution of the LINC complex (SUN and KASH) and plant lamina (CRWN, NEAP and KAKU4) components by exploring 20 representative species including unicellular photosynthetic algae, lycophytes, mosses, gymnosperms and angiosperms for which genome sequences and gene expression data are available (; Supplementary Table 11).
Phylogenetic analysis of inner nuclear membrane proteins
Cter-SUN proteins
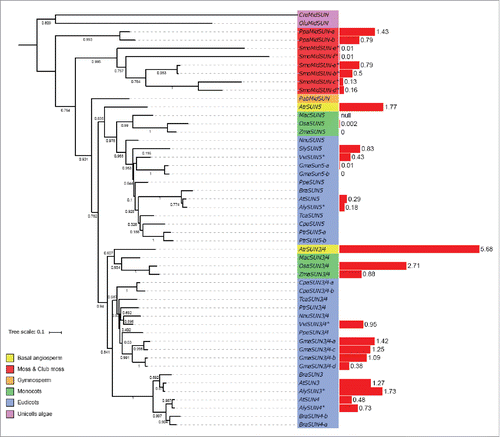
The SUNs are divided into 2 subfamilies according to the position of the SUN domain: the mid-SUN with a central SUN domain and the Cter-SUNs having a SUN domain at the C-terminus. The potential origin of the 2 classes of SUN domain proteins remains obscure and is discussed in Graumann et al., 2015. The SUNs are key members of the LINC complex and expressed in all tissues.Citation27,Citation18 Blastp and HMMER analysis revealed 3three Cter-SUN proteins across all the 19 species studied, other than Chlamydomonas reinhardtii, where no Cter-SUN protein was detected (). Monocots and eudicots form 2 paraphyletic groups, with the Vitis vinifera homolog showing greater similarity to the monocot Cter-SUN sequence. The Brassicaceae form a monophyletic group and the duplication of the Cter-SUN gene seems to have occurred late in evolution because duplicated Cter-SUNs remain grouped within a given species ().
Figure 2. Phylogenetic tree of Cter-SUN proteins and gene expression levels. Left: maximum likelihood tree of Cter-SUN protein homologues constructed from an alignment. Bootstrap values are presented. The color of the label shows the lineage of the plant. The gene label is constructed with the 3 letters from the species name (supplementary Table 4) and the gene name of the A. thaliana homologues. Right: red bar represents the value of the transcription level in seedlings expressed in RPKM, except for species indicated by *, the RNA-seq data was obtained from leaf tissue (Supplementary Table 2).
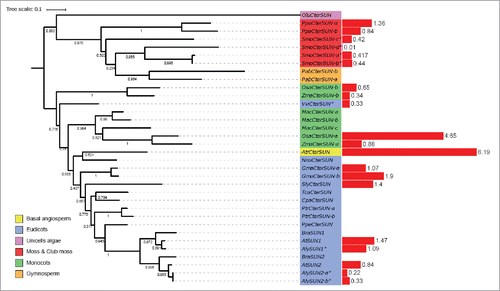
Expression data for the Cter-SUNs shows a similar transcript level for all the tissue analyzed in different species (Supplementary Fig. 1). In some cases, one of the 2 Cter-SUNs is more strongly expressed in the seedling (e.g.,: AtSUN1 more strongly expressed than AtSUN2; OsaSUN-a more strongly expressed than OsaSUN-b) ((WIPs)). A. trichopoda encodes only one Cter-SUN that is highly expressed in all tissues. The simplest functional LINC complex may therefore be based on a single Cter-SUN, and strengthens the suggestion that duplication of the Cter-SUN gene occurred after speciation.
One or 2 Cter-SUN proteins were identified in most plants and the moss, although 4 close homologues were identified for the club moss Selageinella molendorfii. In A. thaliana, SUN1 and SUN2 share almost the same activity and localization.Citation17 This is in contrast to mammals, where 5 Cter-SUN orthologues have clearly differentiated functions. It appears that the gene duplication resulting in these orthologues occurred earlier in the evolution of mammals. One likely consequence is the lack of specificity of function of plant Cter-SUN homologues; for example, a disruption of a single SUN gene results in an infertility phenotype in animals,Citation8 but in A. thaliana, a single Cter-SUN deletion does not affect meiosis or fertility whereas the double mutant atsun1 atsun2 impacts fertility and cell division.Citation36 This suggests a significant redundancy in Cter-SUN function in plants and that double knock-out or knock-down mutants are required for recognizable phenotypes to be obtained.
mid-SUN proteins
All the species considered contain at least a mid-SUN protein and overall 50 mid-SUN homologues were identified during our bioinformatic screen. The mid-SUN angiosperm homologues are clustered in 2 groups, SUN3/SUN4 and SUN5. In each mid-SUN homologous group, the basal angiosperm, monocots and eudicots form monophyletic groups. This suggests that mid-SUN (3, 4) gene duplication occurred after speciation between angiosperms and gymnosperms (). In all tissue analyzed the SUN3/SUN4 group tends to be more ubiquitously and highly expressed than the SUN5 group, this is also true for the A. trichopoda homologues (Supplementary Fig. 1). It has been suggested that AtSUN5 has a meiotic functionCitation18 and this is also true for maize with ZmaSUN5,Citation27 although while the double mutants of SUN3, SUN4 and SUN5 are viable, a sun3 sun4 sun5 triple mutant is lethal.Citation18 A. trichopoda has 2 mid-SUN proteins, one SUN3/SUN4 homolog and a SUN5 homolog. This suggests that the simplest LINC complex has 2 mid-SUNs each with a specific or partially overlapping function.
In summary, the majority of the 20 species possess at least one mid-SUN and one Cter-SUN protein except for Chlamydomonas reinhardtii, which has only one mid-SUN protein. Interestingly, in common with Cter SUNs, the club moss Selaginella has the highest number of mid-Sun protein homologues (6). These results are in good agreement with previous studies that have highlighted the conservation of both Cter- and mid-SUN proteins in most eukaryotesCitation27,Citation18 and suggest the that the LINC complex was present in the Last Evolutionary Common Ancestor (LECA;Citation23). Mid-SUN homologues and Cter-SUN proteins were detected in the unicellular algae examined, suggesting that SUN emergence pre-dates the evolution of multicellularity. The evolutionary relationship between Cter-SUN and mid-SUN proteins has yet to be described. This study suggests that SUN domain proteins may also be among the earliest evolving components of the plant nuclear envelope and that mid-SUN proteins may have significance nuclear function in the absence of Cter-SUN.
KASH protein homologues
KASH proteins are diverse in sequence and structureCitation40 but possess a conserved C-terminal region with a TM domain and a conserved motif of 4 amino acids at the extreme C-terminus. For the detection of the KASH domain protein homologues, 2 strategies were used. The first was a BLASTp analysis based on known A. thaliana KASH domain proteins (Supplementary Table 1). This analysis permitted detection of KASH protein homologues in all the organisms studied, except for the unicellular algae where no KASH protein was detected (Supplementary Table 12). Using this method, 32 SINE homologues were found, whereas WIP and potential TIK proteins [or TIK-like proteins] (Supplementary Table 12) were much less common and were found mainly in eudicots. An exception is Brassicaceae, where several potential WIP (3 in A. lyrata, 4 in Brassica rapa) and TIK (2 in A. lyrata and 1 in B. rapa) homologues were identified, these were also detected in Glycine max (2 WIP, 1 TIK), Prunus persica (1 TIK), Carica papaya (1 WIP), Musa acuminata (1 WIP), A. trichopoda (1 TIK) and the gymnosperm Picea abies (1 TIK) (Supplementary Table 12). To expand the data collected by Blastp, a script was developed to detect proteins with the TM domain and C-terminal motif.
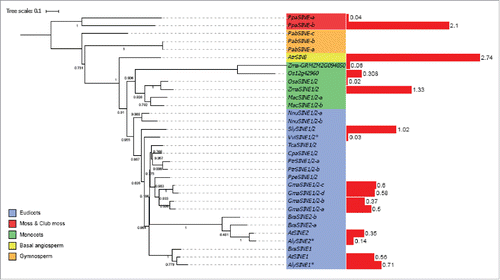
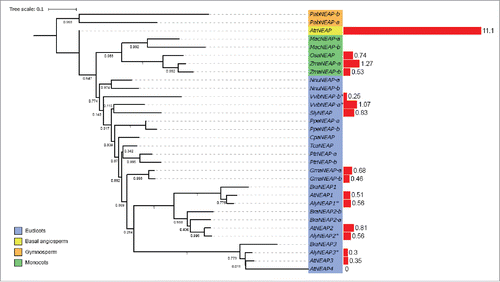
All the identified plant KASH domain proteins have been divided into 3 groups: SINEs, WIPs and TIK.Citation40 Six KASH protein clusters were revealed (Supplementary Fig. 2). One includes WIP proteins detected in the monocotyledons and the basal angiosperms (Supplementary Table 12), as well as 7 new putative WIP proteins to those detected previously by BLASTp. For SINE proteins, 3 clusters were detected, for SINE1/2, SINE3 and SINE4 adding respectively 2, 6 and 12 SINE proteins to those already identified. The high number of proteins in the SINE3 and SINE4 cluster found only by the script was due to weak conservation of these proteins. One much smaller cluster includes the TIK-like proteins. Only four putative homologues were added but these were shown subsequently to lack either the TIR domain or the C-terminal TM domain, therefore suggesting that the TIK proteinCitation18 may be unique to A. thaliana. An additional cluster (other) had low sequence similarity and was not included subsequently. The three WIP proteins in A. thaliana show previously described propertiesCitation38,Citation39 of a cytoplasmic domain at the N-terminus, with AtWIP1 and AtWIP2 having 3 coiled-coil domains but AtWIP3 only one. The C-terminal region is well conserved and the coiled-coil domains align, with all proteins detected as homologues having a C-terminal predicted TM domain and KASH motif, except AlyWIP1, which lacks homology at the C-terminal region but is well conserved at the N-terminus.
All SINEs have a typical KASH TM domain and C-terminal amino acid motif.Citation42 The SINE gene family comprises 4 genes in A. thaliana, with similarity between AtSINE1 and AtSINE2, characterized by an Armadillo repeat domain near the N-terminus; and between AtSINE3 and AtSINE4. The Perl script added only 2 sequences in the SINE1/SINE2 group while it added 14 proteins to the SINE3/SINE4 cluster. In this case the Blastp approach was less efficient than the Perl script because of the absence of well-conserved domains in the N-terminus. After removal of sequences with the lowest similarity or without the conserved domain, SINE1/SINE2 proteins are present in all species except the unicellular algae and club moss, while SINE3/SINE4 were absent (). In summary, the SINE1/SINE2 cluster and WIP proteins are detected in basal angiosperms whereas the TIK protein is detected only in A. thaliana.
Phylogenetic analysis of the outer nuclear membrane proteins
WIP proteins
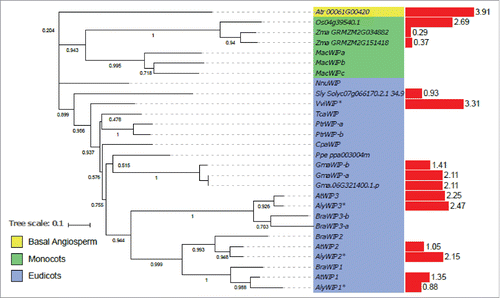
The WIP protein family was the first KASH family detected in A. thalianaCitation41). WIP proteins were not detected in unicellular algae, moss, club moss or gymnosperms; suggesting that they are angiosperm specific proteins. One WIP homolog was detected for A. trichopoda. The monocots form a monophyletic group, with one protein for rice, 2 and 3 for maize and Musa acuminata suggesting gene duplication (). The eudicots form a paraphyletic group because the WIP homolog of Nelumbo nucifera differs from, and is positioned outside, the WIPs of eudicots. The Brassicaceae on the other hand, form a monophyletic group (). This suggests that an ancestral duplication in the Brassicaceae ancestor gave rise to WIP1/WIP2 and WIP3, and then WIP1 and WIP2 resulted from a more recent gene duplication. All three genes are expressed in all the tissues analyzed. In A. thaliana AtWIP3 transcripts are more abundant than AtWIP1 and AtWIP2 in all tissues. This may be due to redundancy in AtWIP1 and AtWIP2 function, and in A. trichopoda, the WIP homolog is highly expressed (Supplementary Fig. 1).
SINE proteins
SINEs in A. thalianaCitation42 comprise 2 groups, SINE1/SINE2 and SINE3/SINE4. AtSINE1 is more expressed in guard cells, and its armadillo domain forms F-actin-associated fibers involved in nuclear positioning while AtSINE2 is suggested to be involved in the immunity response of leaves.Citation42 No expression and activity data was available for AtSINE3 and AtSINE4.
SINE1/SINE2 proteins were not found in unicellular algae and in club moss, but in contrast to WIPs, 2 and 3 SINE homologues were found in moss and gymnosperms, respectively (). The angiosperms form a monophyletic group and one SINE1/SINE2 homolog was detected for A. trichopoda and positioned at the base of the angiosperm group (). The phylogenetic analysis of SINE3 and SINE4 is not possible due to the low similarity between sequences and a lack of conserved domains. Although SINE3 and SINE4 are detected in the Brassicaceae group, the other sequences are divergent. In the monocots, 2 protein homologues were detected for Musa acuminata, Oryza sativa and Zea mays. However, the phylogeny suggests the presence of recent gene duplication in Musa acuminata (). In contrast, the gene duplication between the 2 other monocots seems to have occurred before their speciation. All the eudicots possess at least one SINE1/SINE2 homolog. Four homologues that group together were found in Glycine max, suggesting a recent gene duplication. As for WIPs, Brassicaceae proteins cluster together, and one group of homologues is detected for each of SINE1 and SINE2. The organization between the 2 groups suggests a gene duplication to form SINE1 and SINE2.
In A. thaliana, AtSINE1 and AtSINE2 are expressed at the same level in all tissues, but at a higher level than AtSINE3 and AtSINE4. However, SINE1/SINE2 homologues in most other species show the lowest level of expression of all KASH proteins for all tissues analyzed expect for maize, rice and A. trichopoda (Supplementary Fig. 1). In these species WIP and SINE expression is at the same level for all tissues. In A. thaliana, AtWIPs are more highly expressed than AtSINE.
Phylogenetic analysis of the putative nuclear lamina and nuclear-envelope associated proteins
Proteins of the lamin family are restricted to animals.Citation5 However, to date, 3 protein families have been suggested to be components of the putative lamina in A. thaliana, CRWN,Citation9,Citation37 KAKU4Citation14 and a novel nuclear envelope associated protein family, NEAPCitation30.
CRWN proteins
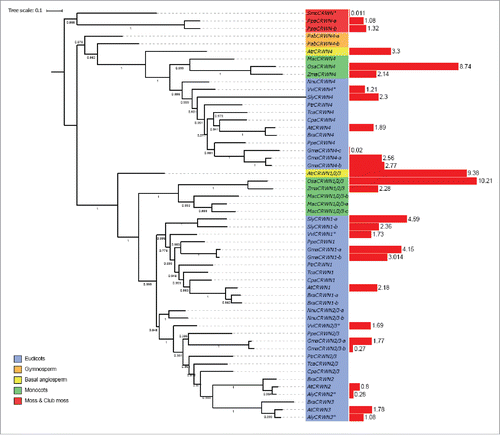
The CRWN gene family is imported into the nucleus through an NLS and extensive coiled-coil domains reminiscent of the animal lamins are hypothesized to allow polymerisation of the protein to form the plant lamina. Fifty CRWN proteins were detected by BLASTp and pHMMER in all multicellular plants but are absent from unicellular algae. In most species 2 homologues were detected for each species. Two clusters of CRWN proteins were defined in a previous publicationCitation6 and were also identified here as 2 main phylogenetic groups: CRWN1/CRWN2/CRWN3 and CRWN4. The clusters of CRWN4 homologues constitute monophyletic groups and only one protein was found for all species except for Glycine max (). Gymnosperm homologues seem to have only the CRWN4 lineage while A. lyrata has lost the CRWN4 lineage meaning that some functional redundancy exists between the 2 monophyletic groups.
For the second group made up of the homologues of the 3 other CRWN proteins, the same organization was found and only one homolog in A. trichopoda was detected (). In the monocot group, only Musa acuminata possesses 3 homologues, the other monocots possessing only one (). In the eudicot group, 2 clusters can be distinguished: one for the homologues of AtCRWN1 and the other for AtCRWN2/AtCRWN3. This reveals a gene duplication, which occurred after the speciation creating monocots and eudicots. The other duplication, which gave rise to CRWN2 and CRWN3, occurred after Brassicaceae speciation and formed a monophyletic group. The genes belonging to the cluster CRWN1/CRWN2/CRWN3 show higher expression in comparison to CRWN4. Other than in the Brassicaceae, CRWN2 is less expressed than CRWN1 and CRWN3 for all the tissues analyzed. Surprisingly, no lamin-like proteins were detected in the chlorophyte unicellular algae. Previous studies have shown the presence of other lamin-like proteins in unicells like NE81 and NUP1.Citation10,Citation24 It is likely that several proteins have evolved in different systems to fulfil a similar role and this would reward further study.
NEAP proteins
The NEAP proteins are characterized by a TM region at the C-terminus, a functional NLS and extensive coiled-coil domains.Citation30 NEAP1, NEAP2, and NEAP3 were identified in gymnosperms and angiosperms and 28 proteins were detected while NEAPs are absent from the more ancestral species moss, club moss and unicellular algae (Supplementary Table 11). The monocots form a monophyletic group with 2 potential specific gene duplications for Musa acuminata and Zea mays (). As for monocots, the eudicots form a monophyletic group (), and the gene duplication seems specific to species. So the 3 NEAP genes in Brassicaceae appear to result from a duplication event during the speciation of Brassicaceae. The single NEAP gene in A. trichopoda is expressed at very high level. Lack of expression of AtNEAP4 and absence of protein homologues in other species imply that it is a pseudogene. The other NEAP genes are expressed in seedlings and in other tissues but at a low level (Supplementary Fig. 1).
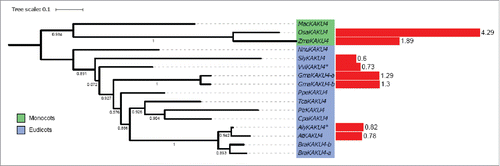
KAKU4 proteins
KAKU4 homologues are only detected in angiosperms. Only one KAKU4 homolog is detected in each species except for Glycine max and Brassica rapa. KAKU4 is therefore a recent addition specific to angiosperms and as it interacts with CRWN1 and CRWN4, it could link CRWN proteins to other components at the nuclear periphery. Analysis of KAKU4 phylogeny reveals 2 monophyletic groups, for the monocot and eudicot homologues (). The protein was not detected in basal angiosperms, gymnosperms, moss, club moss and unicellular algae. Either KAKU4 is a protein with specific function in angiosperms, or was not detected due to a high variability between species. KAKU4 homologues are expressed to comparable levels in all tissues (Supplementary Fig. 1).
Discussion
The results presented reveal functional conservation of the proteins of the plant nuclear envelope with those of other kingdoms, but surprising diversity in protein sequence. summarizes the occurrence of each of the components in the study, together with their function. SUN domain proteins, lamina component CRWN and the KASH domain proteins SINE1-2 involved in actin binding are present before the Zeta WGD (though C-ter SUNs and CRWN are absent from the Chlamydomonas); putative NE- anchored lamina components NEAPs are first found after the Zeta WGD. Binding of RanGAP to the NE by the KASH proteins designated WIP originates with the gamma WGD of the angiosperms; the mechanism of anchorage of RanGAP in gymnosperms and mosses therefore warrants further study. CRWN interacting KAKU4 and KASH protein TIK appear to be of later origin and were only detected in the Brassicas, suggesting specialist functions.
Table 1. protein classes and their origins and function as derived in this study.
Data from A. trichopoda suggests a minimal angiosperm LINC complex, with 2 KASH domain proteins (one WIP and one SINE), 3 SUN domain proteins (one Cter-SUN and 2 mid-SUNs) and putative lamina constituents (2 CRWNs) together with one NEAP. The moss P. patens has 4 SUNs, 2 KASH and 2 putative lamina constituents. The gymnosperm P. abies has 3 SUNs, 3 KASH (SINEs) and putative lamina components (2 CRWN) together with 2 NEAPs (). Evolution of SUN, KASH and lamina constituents appears to have accompanied WGD and partial genome duplication events and to have resulted in a range of homologues during angiosperm speciation. The results presented also indicate a plant nuclear envelope which has developed significant complexity and redundancy through gene duplication explaining the need for multiple knockout mutants; for instance the double mutant atsun1 atsun2Citation41 or the quintuple mutant wifi (atwip1 atwip2 atwip3 atwit1 atwit2)Citation44 before strong phenotypes are observed.
The data also suggests evolution from an ancestral LINC complex, in which SUN domain proteins are multifunctional, to a more multifaceted LINC complex containing an increasing number of KASH and lamin-like proteins. A key role for mid-SUN proteins is suggested. This is commensurate with the demonstration that SUNs play a fundamental role in chromatin interaction with the nuclear envelope during its reformation in plant mitosis15 and in telomere attachment in meiosis.36 KASH domain proteins appear to be evolving, with SINEs preceding WIPs, with TIK only identified in Arabidopsis. It is suggested that increasing specialization accompanies the acquisition of additional KASH homologues and that specific functions of the later evolving proteins (for instance RanGTP anchorage and nuclear movement in the pollen tube) are undertaken by other nuclear envelope components in their absence. A similar pattern of evolution of KASH proteins is suggested in ophisthokonts;Citation42 commenting on novel plant KASH proteins noted that while some are highly conserved (e.g. Nesprin 1 and 2, ANC-1 and MSP-300) others are restricted in distribution (e.g., Klarsicht homologues to insects and KDP-1 to nematodes) suggesting origins after SUN domain proteins and rapid evolution linked to diversifying function. Finally, higher plants have evolved a lamina-like structure based, like animal cells, on coiled-coil proteins. This appears to have arisen with the CRWN proteins present in mosses and clubmosses (lycophytes) and with KAKU4 arising later. These data are consistent with previous reports suggesting that SUN domain proteins were the first nuclear envelope proteins linking chromatin to the nuclear envelope,Citation5 predating the evolution of lamins. Indeed lamins are prone to rapid evolution as they interact with fewer partners than components of the LINC complex.Citation23 One of the striking results of our analysis is the absence of CRWN in unicellular species despite the fact that the lamina is involved in basic function such as nuclear morphology and chromatin organization which are both important for the regulation of gene expression. Although we cannot exclude if lamins and CRWN are subjected to fast evolution leading to unsuccessful recovery of homologs by Blast and HMMMER analyses, it is tempting to speculate that convergent evolution occurred in animals and plants, with increasing functionality and complexity through the introduction of LBR and LEM proteins in animal and KAKU4 in plants. Similar observations were recently presented for the PRC1 polycomb group complex which is a conserved function but with poor sequence homology despite the presence of the conserved RING-domain, again suggesting convergent evolution between plant and animals. Interestingly, the PRC1 complex is involved in the regulation of gene expression through the binding of trimethylated Histone H3 at lysine 27 (H3K27me3) a well-known repressive epigenetic mark also enriched in Lamina-Associated Domains (LADs).Citation4 Exploring the possible connection between the nuclear envelope components and the PRC1 repressive complex will lead to a better understanding of the functions of the nuclear envelope in the regulation of chromatin organization and gene expression.
Abbreviations
| A. thaliana | = | Arabidopsis thaliana |
| BLASTp | = | Basic Local Alignment Search Tool protein |
| CRWN | = | Crowded Nuclei (also termed LINC for Little Nuclei and NMCP for Nuclear Matrix Constituent Protein) |
| HMMER | = | Hidden Markov Model-based sequence alignment tool |
| KASH | = | Klarsicht/Anc1/Syne homology |
| LBR | = | Lamin B receptor |
| LEM | = | Lamin-Emerin-Man1 |
| LINC | = | Linker of Nucleoskeleton and Cytoskeleton |
| NEAP | = | Nuclear Envelope Associated Protein |
| RPKM | = | Reads Per Kilobase of transcript per Million mapped reads |
| SUN | = | Sad1-Unc84 |
| SINEs | = | SUN interacting Nuclear Envelope Proteins |
| TIK | = | Toll Interleukin Receptor domain KASH protein |
| TM | = | trans-membrane |
| WGD | = | whole-genome duplication |
| WIPs | = | WPP Domain Interacting Proteins |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
2016NUCLEUS0010R-s01.zip
Download Zip (1.7 MB)Funding
The work was supported by the CNRS, INSERM, Blaise Pascal and Oxford Brookes Universities. KG is a Leverhulme Trust Early Career Fellow.
References
- Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al. The revised classification of eukaryotes. J Eukaryot Microbiol 2012; 59:429-514; PMID:23020233; http://dx.doi.org/10.1111/j.1550-7408.2012.00644.x
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-410; PMID:2231712; http://dx.doi.org/10.1016/S0022-2836(05)80360-2
- Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31:166-9; PMID:25260700; http://dx.doi.org/10.1093/bioinformatics/btu638
- Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell 2013; 152:1270-84; PMID:23498936
- Cavalier-Smith T. Kingdoms protozoa and chromista and the eozoan root of the eukaryotic tree. Biol Lett 2010; 6:342-5; PMID:20031978; http://dx.doi.org/10.1098/rsbl.2009.0948
- Ciska M, Moreno Diaz de la Espina S. NMCP/LINC proteins: putative lamin analogs in plants? Plant Signal Behav 2013; 8:e26669; PMID:24128696; http://dx.doi.org/10.4161/psb.26669
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol 2005; 139:5-17; PMID:16166256; http://dx.doi.org/10.1104/pp.105.063743
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 Is Required for Telomere Attachment to Nuclear Envelope and Gametogenesis in Mice. Dev Cell 2007; 12:863-72
- Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. LITTLE NUCLEI Genes Affecting Nuclear Morphology in Arabidopsis thaliana. Plant Cell 2007; 19:2793-803; PMID:17873096; http://dx.doi.org/10.1105/tpc.107.053231
- DuBois KN, Alsford S, Holden JM, Buisson J, Swiderski M, Bart JM, Ratushny AV, Wan Y, Bastin P, Barry JD, et al. NUP-1 Is a Large Coiled-Coil Nucleoskeletal Protein in Trypanosomes with Lamin-Like Functions. PLoS Biol 2012; 10:e1001287; PMID:22479148; http://dx.doi.org/10.1371/journal.pbio.1001287
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 2004; 5:113; PMID:15318951; http://dx.doi.org/10.1186/1471-2105-5-113
- Gaut BS, Ross-Ibarra J. Selection on Major Components of Angiosperm Genomes. Science 2008; 320:484-6; PMID:18436777; http://dx.doi.org/10.1126/science.1153586
- Gerace L, Burke B Functional organization of the nuclear envelope. Annu Rev Cell Biol 1988; 4:335-74; PMID:2461721; http://dx.doi.org/10.1146/annurev.cb.04.110188.002003
- Goto C, Tamura K, Fukao Y, Shimada T, Hara-Nishimura I. The novel nuclear envelope protein KAKU4 modulates nuclear morphology in arabidopsis. Plant Cell 2014; 26:2143-55; PMID:24824484; http://dx.doi.org/10.1105/tpc.113.122168
- Graumann K, Evans DE. The plant nuclear envelope in focus. Biochem Soc Trans 2010a; 38:307-11; http://dx.doi.org/10.1042/BST0380307
- Graumann K, Evans DE. Plant SUN domain proteins: Components of putative plant LINC complexes? Plant Signal. Behav 2010b; 5:154-6; http://dx.doi.org/10.4161/psb.5.2.10458
- Graumann K, Runions J, Evans DE. Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J 2010; 61:134-44; PMID:19807882; http://dx.doi.org/10.1111/j.1365-313X.2009.04038.x
- Graumann K, Vanrobays E, Tutois S, Probst AV, Evans DE, Tatout C. Characterization of two distinct subfamilies of SUN-domain proteins in Arabidopsis and their interactions with the novel KASH-domain protein AtTIK. J Exp Bot 2014; 65:6499-512; PMID:25217773
- Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol 2010; 2:a000539; PMID:20300205
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011; 473:97-100; PMID:21478875; http://dx.doi.org/10.1038/nature09916
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 2004; 428:617-24; PMID:15004568; http://dx.doi.org/10.1038/nature02424
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14:R36; PMID:23618408; http://dx.doi.org/10.1186/gb-2013-14-4-r36
- Koreny L, Field MC. Ancient eukaryotic origin and evolutionary plasticity of nuclear lamina. Genome Biol Evol 2016; PMID:27189989
- Krüger A, Batsios P, Baumann O, Luckert E, Schwarz H, Stick R, Meyer I, Gräf R. Characterization of NE81, the first lamin-like nucleoskeleton protein in a unicellular organism. Mol Biol Cell 2012; 23:360-70; PMID:22090348
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol 1982; 157:105-32; PMID:7108955; http://dx.doi.org/10.1016/0022-2836(82)90515-0
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 2011; 39:W475-8; PMID:21470960; http://dx.doi.org/10.1093/nar/gkr201
- Murphy SP, Simmons CR, Bass HW. Structure and expression of the maize (Zea mays L.) SUN-domain protein gene family: evidence for the existence of two divergent classes of SUN proteins in plants. BMC Plant Biol 2010; 10:269; PMID:21143845; http://dx.doi.org/10.1186/1471-2229-10-269
- Oda Y, Fukuda H. Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. Plant J 2011; 66:629-41; PMID:21294795; http://dx.doi.org/10.1111/j.1365-313X.2011.04523.x
- Parry G. The plant nuclear envelope and regulation of gene expression. J Exp Bot 2015; 66:1673-85; PMID:25680795; http://dx.doi.org/10.1093/jxb/erv023
- Pawar V. Poulet A, Détourné G, Tatout C, Vanrobays E, Evans DE Graumann K. A novel family of plant nuclear envelope-associated proteins. J Exp Bot 2016; doi:10.1093/jxb/erw332.
- Price MN, Dehal PS, Arkin AP. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS One 2010; 5:e9490; PMID:20224823; http://dx.doi.org/10.1371/journal.pone.0009490
- Project AG, Albert VA, Barbazuk WB, dePamphilis CW, Der JP, Leebens-Mack J, Ma H, Palmer JD, Rounsley S, Sankoff D, et al. The Amborella Genome and the Evolution of Flowering Plants. Science 2013; 342:1241089; PMID:23403266
- Soltis PS, Soltis DE. Ancient WGD events as drivers of key innovations in angiosperms. Curr Opin Plant Biol 2016; 30:159-165; PMID:27064530; http://dx.doi.org/10.1016/j.pbi.2016.03.015
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 2007; 56:564-77; PMID:17654362; http://dx.doi.org/10.1080/10635150701472164
- Tamura K, Goto C, Hara-Nishimura I. Recent advances in understanding plant nuclear envelope proteins involved in nuclear morphology. J Exp Bot 2015; 66:1641-7; PMID:25711706; http://dx.doi.org/10.1093/jxb/erv036
- Varas J, Graumann K, Osman K, Pradillo M, Evans DE, Santos JL, Armstrong SJ. Absence of SUN1 and SUN2 proteins in Arabidopsis thaliana leads to a delay in meiotic progression and defects in synapsis and recombination. Plant J 2015; 81:329-46; PMID:25412930; http://dx.doi.org/10.1111/tpj.12730
- Wang H, Dittmer TA, Richards EJ. Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol 2013; 13:200; PMID:24308514; http://dx.doi.org/10.1186/1471-2229-13-200
- Xu XM, Meulia T, Meier I. Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol 2007; 17:1157-63; PMID:17600715; http://dx.doi.org/10.1016/j.cub.2007.05.076
- Zhao Q, Brkljacic J, Meier I. Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 2008; 20:1639-1651; PMID:18591351; http://dx.doi.org/10.1105/tpc.108.059220
- Zhou X, Meier I. Efficient plant male fertility depends on vegetative nuclear movement mediated by two families of plant outer nuclear membrane proteins. Proc Natl Acad Sci U S A 2014; 111:11900-5; PMID:25074908; http://dx.doi.org/10.1073/pnas.1323104111
- Zhou X, Graumann K, Evans DE, Meier I. Novel plant SUN–KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J Cell Biol 2012; 196:203-11; PMID:22270916; http://dx.doi.org/10.1083/jcb.201108098
- Zhou X, Graumann K, Wirthmueller L, Jones JDG, Meier I. Identification of unique SUN-interacting nuclear envelope proteins with diverse functions in plants. J Cell Biol 2014; 205:677-92; PMID:24891605; http://dx.doi.org/10.1083/jcb.201401138
- Zhou X, Graumann K, Meier I. The plant nuclear envelope as a multifunctional platform LINCed by SUN and KASH. J Exp Bot 2015a; 66:1649-59; http://dx.doi.org/10.1093/jxb/erv082
- Zhou X, Groves NR, Meier I. Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1. Nucl Austin Tex 2015b; 6:144-53