ABSTRACT
Mutations in SMARCAL1, which encodes a DNA annealing helicase with roles in DNA replication fork restart, DNA repair, and gene expression modulation, cause Schimke immuno-osseous dysplasia (SIOD), an autosomal recessive disease characterized by skeletal dysplasia, renal disease, T-cell immunodeficiency, and arteriosclerosis. The clinical features of SIOD arise from pathological changes in gene expression; however, the underlying mechanism for these gene expression alterations remains unclear. We hypothesized that changes of the epigenome alter gene expression in SIOD. To test this, we performed a genetic screen for interaction between Marcal1, the Drosophila melanogaster ortholog of SMARCAL1, and the genes of the trithorax group (trxG) and Polycomb group (PcG), which encode epigenetic regulators. SMARCAL1 and Marcal1 genetically interacted with trxG and PcG members. A homozygous null mutation of Marcal1 suppressed the wing-to-haltere transformation, ectopic Ultrabithorax (Ubx) expression, and ectopic Ubx minigene expression caused by PcG deficiency. The suppression of ectopic Ubx expression correlated with reduced chromatin accessibility of the Ubx promoter. To our knowledge, this is the first in vivo evidence for deficiency of a SMARCAL1 ortholog altering the chromatin structure of a gene.
Introduction
Mutations in SWI/SNF-related matrix-associated actin-dependent regulator of chromatin, subfamily A-like 1 (SMARCAL1) cause Schimke immuno-osseous dysplasia (SIOD, OMIM 242900).Citation1 SIOD is an autosomal recessive multisystemic disease characterized by disproportionate short stature due to skeletal dysplasia, renal disease due to focal segmental glomerulosclerosis, T-cell immunodeficiency, and arteriosclerosis.Citation2-7 Additional features of the disease include hypothyroidism, bone marrow failure, and abnormal dentition.Citation3-5,8
SMARCAL1 mutations exhibit age-dependent penetrance, variable expressivity, and poor genotype-phenotype correlation.Citation9-13 Furthermore, deficiency of the SMARCAL1 orthologs in the mouse and fly are insufficient to cause disease in the absence of other genetic or environmental insults.Citation14 These observations suggest that the molecular mechanism underlying SIOD is sensitive to genetic, epigenetic, environmental, and stochastic influences.
SMARCAL1 encodes an ATP-dependent DNA annealing helicase that maintains genomic integrity through its roles in DNA replication fork restart and DNA repair.Citation15-19 The clinical features of SIOD appear to arise from pathological gene expression changes due to SMARCAL1 deficiency.Citation7,20,21 Interleukin 7 receptor α (IL7R) deficiency contributes to the T-cell immunodeficiency, elastin (ELN) deficiency to the arteriosclerosis, and Wnt and Notch overexpression to the renal disease.Citation7,20,21 The underlying mechanism for these gene expression alterations remains unclear.
One modulator of gene expression is chromatin structure.Citation22 Transcriptionally active chromatin generally has an open conformation, whereas transcriptionally silent chromatin has a closed or compact conformation. The trithorax group (trxG) and Polycomb group (PcG) complexes regulate chromatin structure through post-translational histone modifications and chromatin remodeling to activate or silence gene expression, respectively.Citation23,24 The trxG and PcG members were discovered in Drosophila as regulators of homeotic gene expression during development. The precise spatial and temporal expression of homeotic genes defines segmentation and patterning of organisms during development,Citation25 and failure of this expression leads to homeotic transformation, that is, the transformation of one body part into another. For example, loss of PcG function causes a homeotic transformation of the mesothoracic legs (T2), and often the metathoracic legs (T3), toward the prothoracic legs (T1).Citation26-29 This transformation manifests as the development of sex combs, a characteristic of the prothoracic legs in males. Loss-of-function trxG alleles also lead to homeotic transformations; Citation30 several loss-of-function trxG alleles suppress the extra sex combs induced by loss-of-function Polycomb (Pc) mutations.Citation31
A well-studied homeotic gene regulated by trxG and PcG is Ultrabithorax (Ubx).Citation32,33 During development, Ubx expression specifies the metathorax (T3),Citation34 which is characterized by halteres, a balance organ in the adult fly. Loss of Ubx expression transforms the metathorax (T3) toward the mesothorax (T2), and manifests as a haltere-to-wing homeotic transformation.Citation35 In contrast, gain of Ubx expression in the developing wing transforms the mesothorax (T2) toward the metathorax (T3), and manifests as a wing-to-haltere homeotic transformation.Citation26,27,36,37 The trxG and PcG proteins mediate Ubx epigenetic memory of active and silent transcriptional states through cis-regulatory elements known as Polycomb/trithorax response elements (PREs).Citation24,38 The trxG and PcG protein complexes are constitutively bound to PREs in both active and silent Ubx states; however, the trxG histone methyl transferase ash1 is bound to a region approximately 1 kb downstream of the transcription start site only in the active state.Citation39 Although extensive trimethylation of H3K27, H3K9, and H3K20 is observed in the silent state, these repressive histone modifications are absent in the promoter and 5′ coding region of Ubx in the active state; Citation39 thus, trxG and PcG proteins modify the chromatin structure of Ubx to promote the activation or repression of Ubx.
Replication fork stalling-associated DNA damage causes epimutations and alterations in gene expression.Citation40-42 Because SMARCAL1 has roles in DNA replication fork restart and DNA repair, we hypothesized that epimutations cause the pathologic gene expression changes identified in SIOD patient tissues. Suggesting that epimutations underlie these pathological gene expression changes is the recent observation of increased methylation of the IL7R promoter in SMARCAL1-deficient T cells; this is a known cause of reduced IL7R expression.Citation20,43
To test further whether deficiency of SMARCAL1 orthologs alters chromatin structure and gene expression, we performed a genetic screen in Drosophila melanogaster for the interaction of SMARCAL1 orthologs with mutants of trxG and/or PcG members. Finding a genetic interaction, we then observed that a homozygous null mutation of the Drosophila melanogaster ortholog Marcal1 suppressed ectopic Ubx expression caused by loss of PcG function and that this suppression correlated with compaction of the Ubx promoter as measured by loss of micrococcal nuclease sensitivity.
Results
Human SMARCAL1 and Drosophila Marcal1 genetically interact with trithorax group (trxG) and Polycomb group (PcG) members
To determine whether human SMARCAL1 or Drosophila Marcal1 genetically interact with the trxG or PcG members, we overexpressed human SMARCAL1 and Drosophila Marcal1 using the GAL4-UAS system in Drosophila melanogaster. Overexpression of human SMARCAL1 using the wing-specific GAL4 driver MS1906-GAL4 or Drosophila Marcal1 using the ubiquitously expressed GAL4 driver tubulin-GAL4 leads to ectopic wing veins that can be scored for suppression or enhancement.Citation14 Several members of the trxG and PcG suppressed or enhanced the ectopic wing veins induced by human SMARCAL1 and Drosophila Marcal1 overexpression ().
Table 1. Suppressors and enhancers of the ectopic wing vein phenotype induced by the overexpression of Drosophila Marcal1 and human SMARCAL1.
To characterize these interactions better, we studied the effect of the heterozygous or homozygous mutation of Marcal1 (i.e., Marcal1del/+ or Marcal1del/del) on the well-characterized phenotypes of several trxG and PcG alleles. The trxG member brahma (brm) encodes the ATP-dependent helicase of the brahma chromatin-remodeling complex;Citation44 expression of the dominant-negative allele brmK804R in the eye causes rough and reduced eyes.Citation45,46 Homozygous mutation of Marcal1 enhanced this phenotype (). The PcG members Polycomb (Pc), Posterior sex combs (Psc), Sex combs on midleg (Scm), and polyhomeotic proximal (ph-p), and encode members of the polycomb repressive complex 1.Citation23,47 The loss-of-function Polycomb allele Pc1 in heterozygosity led to ectopic wing veins, which were suppressed by the heterozygous mutation of Marcal1 (). Additionally, manifesting as extra sex combs, the mutant alleles Pc1, Psc1, and ScmD1 in heterozygosity and ph-p410 in hemizygosity caused a homeotic transformation of the mesothoracic legs (T2), and often the metathoracic legs (T3), toward the prothoracic legs (T1).Citation26-29 Heterozygous mutation of Marcal1 suppressed and homozygous mutation of Marcal1 further suppressed this homeotic transformation (); heterozygous or homozygous mutation of Marcal1 alone did not lead to a homeotic transformation of the mesothoracic legs (T2) toward the prothoracic legs (T1) (unpublished data). These observations suggest that Marcal1 genetically interacts with several members of the trxG and PcG.
Figure 1. The heterozygous and homozygous mutation of Marcal1 genetically interact with trithorax group (trxG) and Polycomb group (PcG) members and suppress the ectopic expression of Ultrabithorax induced by PcG loss-of-function mutations. (A) Homozygous mutation of Marcal1 enhances the small and rough eye phenotype induced by the eye-specific expression of the dominant negative allele brmK804R. (B) Heterozygous mutation of Marcal1 suppresses the ectopic wing veins (arrows) induced by the Pc1 allele in heterozygosity. The boxed regions in the overview image correspond to the higher magnification images on the right. (C) The male-specific sex combs are a dense row of bristles on the prothoracic legs and allow for successful mating. Loss-of-function PcG mutant alleles ph-p410 in hemizygosity and Pc1, Psc1, and ScmD1 in heterozygosity cause a homeotic transformation of the mesothoracic legs (T2), and often the metathoracic legs (T3), to the prothoracic legs (T1). Extra sex combs that are characteristic of the prothoracic legs are therefore observed on the mesothoracic and metathoracic legs due to loss of silencing of homeotic genes by the PcG proteins. Heterozygous mutation of Marcal1 suppresses and homozygous mutation of Marcal1 further suppresses the extra sex comb phenotype of the mesothoracic legs of these PcG mutant alleles. Data are presented as bar graphs of the percentage of mesothoracic legs with 1-3 sex comb bristles (light gray), 4-6 sex comb bristles (medium gray), or ≥7 sex comb bristles (dark gray). Diamonds denote genotypes that were not assessed due to their lethality. Statistical significance of the data was assessed using the Student's t-test. n.s., not significant; *** p < 0.001. (D) Homozygous mutation of Marcal1 suppresses the partial homeotic transformation of the wing to haltere induced by the heterozygous mutation of ph-p410 and Pc1. (E) Immunostaining of wing imaginal discs for Ultrabithorax from ph-p410/+; Pc1/+ (left panel) and ph-p410/+; Marcal1del/del; Pc1/+ (right panel) larvae. Homozygous mutation of Marcal1 suppresses the ectopic expression of Ultrabithorax (α-Ubx) induced by the heterozygous mutation of ph-p410 and Pc1. Scale bars: 100 μm. (F) Immunostaining of PBX-PRED-IDE-Ubx-lacZ wing imaginal discs for β-galactosidase (α-β-gal) in the background of ph-p410/+; Pc1/+ (left panel) and ph-p410/+; Marcal1del/del; Pc1/+ (right panel) larvae. Homozygous mutation of Marcal1 suppresses PBX-PRED-IDE-Ubx-lacZ reporter gene expression induced by the heterozygous mutation of ph-p410 and Pc1. Scale bars: 100 μm. (G) Photograph of an agarose gel stained with ethidium bromide (EtBr) following electrophoretic separation of nucleosome-protected DNA samples from Marcal11+/+ and Marcal1del/del adult flies after SphI restriction digestion and partial digestion with micrococcal nuclease (MNase) (left panel). For Southern blot analysis of the Ultrabithorax (Ubx) promoter (right panel), Ubx promoter-specific DNA was detected using a probe against the transcription start site of Ubx (lower panel). Note that Marcal1del/del decreases MNase sensitivity of the Ubx promoter. Abbreviations: bp, base pairs; DAPI, 4′, 6-diamidino-2-phenylindole; EtBr, ethidium bromide; ey>brmK804R, eyeless-GAL4, UAS-brmK804R; MNase, micrococcal nuclease; MW, molecular weight; Pc, Polycomb; PcG, polycomb group; ph-p, polyhomeotic proximal; Psc, posterior sex combs; Scm, Sex comb on midleg; trxG, trithorax group; Ubx, Ultrabithorax.
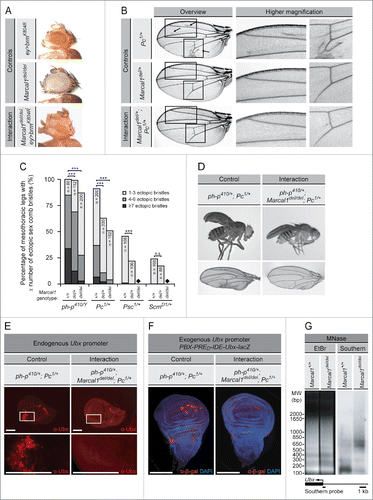
Marcal1 deficiency suppresses the derepression of Ultrabithorax (Ubx) expression associated with deficiency of Polycomb activity
The loss of PcG function not only causes homeotic transformation of the mesothoracic legs but also a wing-to-haltere transformation via derepression of Ubx expression in the developing wing.Citation26,27,36,37 Homozygous mutation of Marcal1 suppressed the partial wing-to-haltere transformation induced by the loss-of-function alleles Pc1 and ph-p410 in heterozygosity (), as well as the ectopic expression of Ubx in the wing imaginal disc (). This epistatic interaction between the homozygous mutation of Marcal1 and the PcG alleles Pc1 and ph-p410 in heterozygosity was not limited to the endogenous Ubx gene located at 89D, but also occurred with a Ubx promoter-lacZ transgene (PBX-PRED-IDE-Ubx-lacZ) inserted at 46D ().Citation38
To determine if the homozygous mutation of Marcal1 caused these changes in Ubx expression by altering binding of Marcal1 to the Ubx promoter, we performed chromatin immunoprecipitation (ChIP) and DNA adenine methyltransferase identification (DamID) analyses of the Ubx promoter in embryos and Kc157 cells, respectively; however, neither method detected Marcal1 binding (data not shown). To determine therefore if the homozygous mutation of Marcal1 effected these changes in Ubx expression indirectly through alteration of chromatin structure, we performed a micrococcal nuclease (MNase)-Southern blot assay of the endogenous Ubx promoter. Because MNase is an endo-exonuclease that induces double-strand DNA breaks within nucleosome linker regions, MNase digestion of chromatin followed by Southern blot analysis assesses the chromatin accessibility of a region of interest.Citation48,49 Electrophoretic separation and ethidium bromide visualization of nucleosome-protected DNA samples from Marcal1+/+ and Marcal1del/del adult flies after partial digestion with MNase demonstrated a nucleosomal DNA ladder with mono-, di-, tri-, and tetranucleosomal fragments (). Upon Southern blot hybridization using a probe against the Ubx promoter, we observed decreased detection of low molecular weight nucleosomal DNA fragments and increased detection of high molecular weight nucleosomal DNA fragments in Marcal1del/del flies (). We conclude, therefore, that the homozygous mutation of Marcal1 reduced MNase digestion of the Ubx promoter consistent with compaction or closure of the Ubx promoter.
Discussion
We have demonstrated that SMARCAL1 and Marcal1 genetically interact with trxG and PcG members, and that the homozygous mutation of Marcal1 suppresses the derepression of Ubx expression associated with the deficiency of Polycomb activity. The altered Ubx gene expression due to the homozygous mutation of Marcal1 correlates with the expected phenotypic change as well as reduced chromatin accessibility of the Ubx promoter. To our knowledge, this is the first report of the deficiency of a SMARCAL1 ortholog associating with altered chromatin accessibility of a promoter.
We have previously observed that SMARCAL1 deficiency pathologically increases or decreases gene expression to contribute to the vascular, immune, and kidney diseases of SIOD;Citation7,20,21 however, the mechanism by which SMARCAL1 deficiency alters the expression of these genes is unknown. We considered 4 nonexclusive hypotheses for SMARCAL1 deficiency altering gene expression including 1) an effect of SMARCAL1 on transcription, 2) an effect of SMARCAL1 on gene promoter structure, 3) an effect of SMARCAL1 deficiency through unrepaired DNA lesions impeding transcription, and 4) an effect of SMARCAL1 deficiency on gene expression through replication stress-induced alterations of chromatin structure.
Regarding the first potential mechanism, SMARCAL1 might alter gene expression as part of the RNA polymerase complex. We previously observed that Marcal1 preferentially binds promoters and transcriptionally active chromatin and genetically interacts with genes encoding components of the transcription factor machinery.Citation14 However, evidence against this model includes 1) the failure of SMARCAL1 orthologs to co-purify or co-immunoprecipitate with RNA polymerase IICitation50-52 and 2) the failure of RNA polymerase II to co-purify or co-immunoprecipitate with SMARCAL1 or Marcal1.Citation14
Regarding the second potential mechanism, SMARCAL1 deficiency might influence gene expression by altering the promoter structure of a gene or of a regulator of that gene. Indeed, Sharma et al. have demonstrated that bovine SMARCAL1 is able to negatively regulate the transcription of MYC by altering the conformation of its promoter in an ATP-dependent manner.Citation53 Alternatively, SMARCAL1 deficiency might indirectly alter expression of a gene of interest by effects on upstream regulators, such as transcription factors or microRNAs. Indeed, increased expression of microRNA regulators of ELN in an SIOD aorta appear to contribute to the elastin deficiency observed in the SIOD aorta.Citation54 Further studies are, however, required to define the nature and distribution of the chromatin conformational changes and the mechanism(s) by which SMARCAL1 is recruited to promoters.
Regarding the third potential mechanism, SMARCAL1 deficiency might alter gene expression through unrepaired DNA lesions impairing RNA polymerase II progression. Such DNA lesions are repaired by transcription-coupled nucleotide excision repair (TC-NER).Citation55 Although SMARCAL1 is recruited to sites of DNA damage and has roles in DNA repair,Citation15-19,56 evidence against this model includes 1) the insensitivity of SMARCAL1-deficient dermal fibroblasts to ultraviolet light-induced transcriptional inhibition, which induces DNA damage that is repaired by global-genome nucleotide excision repair (GG-NER) and TC-NER and 2) the insensitivity of SMARCAL1-deficient dermal fibroblasts to illudin S, which induces DNA damage that is repaired exclusively by TC-NER.Citation57
Regarding the fourth potential mechanism, SMARCAL1 deficiency might alter gene expression through replication stress-induced alterations on chromatin structure. The function of SMARCAL1 as a DNA replication stress response protein required for replication fork restart has been clearly established.Citation15-19 There is also increasing evidence that replication stress leads to both transient and permanent alterations in chromatin structure that change gene expression.Citation40-42 SMARCAL1 deficiency might, therefore, indirectly alter gene expression through replication stress-induced alterations of chromatin structure.
Sources of replication stress include DNA lesions, DNA secondary structures, misincorporation of ribonucleotides, collision of the replication and transcription machinery, and depletion of factors required for efficient DNA replication.Citation58 Consequently, DNA damage from replication stress is unevenly distributed throughout the human genome and also differently distributed in the genomes of different species. This latter observation could explain the poor modeling of SIOD in other organisms.
This model of replication stress-induced epimutations is consistent with prior findings of DNA topological structural changes in the context of the deficiency of SMARCAL1 orthologs including 1) SMARCAL1-deficient cells having increased S1 nuclease sensitivity when pulsed with potassium permanganate,Citation59 2) SMARCAL1-deficient T cells showing increased methylation of the IL7R promoter that corresponds with the reduced expression of IL7R,Citation20 and herein, 3) Drosophila Marcal1 deficiency suppressing derepression of the Ubx gene due to PcG mutations via reduced accessibility of the Ubx promoter.
This study provides evidence for the deficiency of SMARCAL1 orthologs altering chromatin structure and gene expression. Observations in SIOD patient tissues have shown that a trait of interest can arise from either increased or decreased expression of key genes in tissues of SIOD patients.Citation7,20,21 Some gene expression changes appear to arise from effects of SMARCAL1 activity at promoters,Citation53 whereas others might arise from epigenetic changes induced by replication stress. Future genome-wide studies to assess the relationship between chromatin structure as well as epigenetic marks and gene expression will provide valuable insight into the mechanism by which SMARCAL1 deficiency alters gene expression.
Materials and methods
Drosophila melanogaster lines
The Drosophila Marcal1 overexpression transgenic line UAST-Marcal1/CyO; tubulin-GAL4/TM3, Sb1 and the human SMARCAL1 overexpression transgenic line MS1096-GAL4; UAST-SMARCAL1 have been previously described.Citation14 The C96-GAL4 UAS-Hrs/MKRS transgenic line was a kind gift from Dr. Hugo Bellen (Baylor College of Medicine, Houston, TX, USA). The UAS-brmK804R transgenic line was a kind gift from Dr. Jessica Treisman (New York University, New York, NY, USA).Citation45 All overexpression lines utilized the GAL4-UAS system in which tissue-specific expression of the yeast transcriptional activator GAL4 drives expression of a gene of interest through the upstream activating sequence (UAS).Citation60
The amorphic allele Marcal1del was generated by imprecise P element excision and has been previously described.Citation14 The loss-of-function Polycomb allele Pc1, Posterior sex combs allele Psc1, Sex combs on midleg allele ScmD1, and polyhomeotic proximal allele ph-p410 were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN, USA). The Polycomb allele Pc6 was obtained from the Kyoto Stock Center (Kyoto, Japan). The Enhancer of zeste alleles E(z)32 and E(z)61 were a kind gift from Dr. Thomas Grigliatti (University of British Columbia, Vancouver, BC, Canada). The Imitation SWI alleles Iswi1 and Iswi2 were a kind gift from Dr. John Tamkun (University of California, Santa Cruz, CA, USA). The transgenic reporter line PBX-PRED-IDE-Ubx-lacZ was a kind gift from Dr. Jürg Müller (Max Planck Institute of Biochemistry, Martinsried, Germany).Citation38 All other Drosophila stocks used in this study were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN, USA).
Anti-Marcal1 antibody production
A polyclonal antibody to Marcal1 was generated in rabbit by injecting the N-terminus of Marcal1 (amino acids 1-400). This domain does not display homology to other Drosophila proteins by BLASTp. The Marcal1 peptide was produced in E. coli using the pET28a expression system (Novagen, Madison, WI, USA). The antiserum was further purified by affinity purification. The polyclonal antibody is specific by Western and immunohistochemistry analyses (unpublished data).
Drosophila genetic studies
The Drosophila Marcal1 and human SMARCAL1 overexpression screen was carried out at 28°C and has been previously described.Citation14,21 Drosophila have 5 longitudinal veins (L1, L2, L3, L4, and L5) as well as an anterior and posterior cross vein (ACV and PCV); overexpression of Marcal1 and SMARCAL1 leads to an ectopic vein parallel and anterior to L2, an ectopic vein extending laterally from the PCV, a partially missing or completely absent ACV or PCV, and distal bending or splitting of longitudinal veins L2, L4, and L5.Citation14,21 We screened for trxG and PcG alleles that lead to the suppression or enhancement of the ectopic wing veins induced by the overexpression of Marcal1 and SMARCAL1. Ectopic wing veins observed in the F1 progeny of the crosses were scored as previously published.Citation14,21 The reference wing vein phenotype was determined by crossing Marcal1 overexpression flies to w1118 mutants of 3 genetic backgrounds; the scores from these crosses were averaged to provide a reference score. To determine whether there was any non-specific interaction between the various mutant alleles and the GAL4-UAS system, all mutant lines were crossed to the C96-GAL4, UAS-Hrs/MKRS transgenic line and the degree of wing margin scalloping in the desired F1 progeny was scored. Any mutant alleles that interacted with the GAL4-UAS system (i.e., those that enhanced or suppressed the wing margin scalloping phenotype of the C96-GAL4, UAS-Hrs/MKRS transgenic line) were excluded; all mutant alleles presented here had no detectable non-specific interactions with the GAL4-UAS system. Ten or more wings were analyzed for each cross and scored by 2 independent readers. Scores for each cross were compared to the reference scores to determine whether the wing vein phenotype was suppressed or enhanced. Where there was a discrepancy between the first 2 reads, a third read was completed by C. F. B.
All other crosses to assess the effect of Marcal1del/+ and Marcal1del/del on the well-characterized phenotypes of trxG and PcG alleles were performed at 25°C. Representative images of eyes or wings were acquired using an MZ16 Stereomicroscope (Leica Microsystems, Buffalo Grove, IL, USA).
Immunostaining of wing imaginal discs
Wing imaginal discs were dissected in 1× phosphate-buffered saline (PBS) on ice. Following fixation with 4% paraformaldehyde for 4 minutes, the wing imaginal discs were washed 2× with washing buffer (0.1% Tween 20, 1× PBS) for 15 minutes, blocked with blocking buffer (20% bovine serum albumin, 2% normal goat serum, 0.1% Tween 20, 1× PBS) for 1 hour, and then incubated with mouse anti-Ultrabithorax (1:200, FP3.38, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA, USA) or mouse anti-β-galactosidase (1:200, Promega, Madison, WI, USA) primary antibody diluted in blocking buffer overnight at 4°C. Samples were then washed 6× with washing buffer for 15 minutes and incubated with a fluorophore-conjugated secondary antibody for 4 hours at room temperature. Samples were washed 6× with washing buffer for 15 minutes and the nuclei stained with 4′, 6-diamidino-2-phenylindole (DAPI, 1:1,000, Fluka, St. Louis, MO, USA). Representative images of wing imaginal discs were acquired using an Axiovert 200 inverted microscope, an AxioCam MR microscope camera, and the AxioVision software version 4.0 (Carl Zeiss Microscopy, Thornwood, NY, USA).
Chromatin immunoprecipitation (ChIP)
Formaldehyde cross-linking of chromatin from Drosophila embryos was performed as previously described.Citation61 Briefly, Marcal1+/+ and Marcal1del/del embryos were dechorionated with 3% NaOCl in Embryo Wash Buffer (0.4% NaCl, 0.03% Triton X-100) and extensively washed with Embryo Wash Buffer. The samples were then cross-linked with Cross-linking Solution (1.8% formaldehyde, 1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, 50 mM HEPES, pH 8.0) in the presence of n-heptane for 15 minutes; the cross-linking reaction was stopped by centrifuging the samples at 16,100 × g for 1 minute and resuspending the samples in Stop Solution (0.125 M glycine, 1× PBS, 0.01% Triton X-100). The samples were then sequentially washed in Wash Solution A (10 mM HEPES pH 7.6, 10 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 0.25% Triton X-100) and Wash Solution B (10 mM HEPES pH 7.6, 200 mM NaCl, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 0.01% Triton X-100). Samples were resuspended in 1 ml Lysis Buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1) and sonicated. Chromatin immunoprecipitations of formaldehyde cross-linked chromatin from embryos were performed using the ChIP Assay Kit (Upstate/Millipore, Billerica, MA, USA) as previously described. 30 μl of Protein A agarose beads were incubated with 20 μl of rabbit anti-Marcal1 antibody prior to incubation with sonicated lysates. The Ubx promoter and the upstream bxd cis-regulatory domain of Ubx were amplified by PCR using primers that have been previously described.Citation62
DNA adenine methyltransferase identification (DamID)
DamID analysis of Marcal1 binding was performed as previously described.Citation14,63
Micrococcal nuclease-Southern blot assay
Nuclei purification and chromatin digestion with micrococcal nuclease (MNase) were performed as previously described,Citation64 with the following modifications. Nuclei were isolated from Marcal1+/+ and Marcal1del/del flies using NucBuster Extraction Reagent 1 from the NucBuster Protein Extraction Kit (TB338, Novagen, Madison, WI, USA). Adult flies were homogenized using a Dounce homogenizer (Wheaton, Millville, NJ, USA) and the homogenate was filtered using glass wool. The filtrate was vortexed for 15 seconds, incubated at 4°C for 5 minutes, vortexed for 15 seconds, and centrifuged at 16,100 × g for 5 minutes to pellet the nuclei. The nuclei were resuspended in RSB buffer (10 mM NaCl, 3 mM MgCl2, 10 mM Tris-Cl, pH 7.5, 0.5% NP-40, protease inhibitor) and adjusted to 1 mM CaCl2 for MNase digestion; each sample was digested with 9 U of MNase (N3755, Sigma-Aldrich, St. Louis, MO, USA) at 37°C for 5 minutes. The reaction was terminated by the addition of EDTA to 5 mM. The subsequent RNase A digestion, proteinase K digestion, and DNA extraction were performed as previously described.Citation64
Southern blot analysis was performed as previously described,Citation14 with the following modifications. Ten micrograms of nucleosome-protected DNA were digested with the SphI restriction enzyme overnight; the SphI restriction enzyme cuts within intron 1 of Ubx and approximately 6.8 kb upstream of the transcription start site of Ubx (). The DNA was re-isolated by ethanol precipitation, separated by agarose gel electrophoresis, and then transferred to a nitrocellulose membrane. To detect Ubx promoter-specific nucleosome-protected DNA, a probe to the Ubx promoter was generated by PCR amplification using the 5′-GATTGGGAACTGAAACTGTAGTC-3′ forward primer and 5′-CCGCTGATAATGTGGATAATAAAAC-3′ reverse primer and radioactively labeled with32 P using the Amersham Rediprimer II DNA Labeling System (RPN1633, GE Healthcare, Pittsburgh, PA, USA).
Statistical analysis
For the genetic interaction studies assessing the effect of the heterozygous or homozygous mutation of Marcal1 on the homeotic transformation of the prothoracic legs to the mesothoracic legs by the PcG mutant alleles, statistical significance of the data was evaluated by the 2-tailed Student's t-test. p values were corrected for multiple comparisons using the Bonferroni method. A p value of less than 0.05 was considered to be statistically significant.
Abbreviations
| brm | = | brahma |
| ChIP | = | chromatin immunoprecipitation |
| DamID | = | DNA adenine methyltransferase identification |
| ELN | = | elastin |
| IL7R | = | interleukin 7 receptor α chain |
| MNase | = | micrococcal nuclease |
| Pc | = | Polycomb |
| PcG | = | Polycomb group |
| ph-p | = | Polyhomeotic proximal |
| PRE | = | Polycomb/trithorax response element |
| Psc | = | Posterior sex combs |
| Scm | = | Sex comb on midleg |
| SIOD | = | Schimke immuno-osseous dysplasia |
| SMARCAL1 | = | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin, subfamily A-like 1 |
| trxG | = | trithorax group |
| UAS | = | upstream activating sequence |
| Ubx | = | Ultrabithorax |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by the New Investigator Award jointly sponsored by the SickKids Foundation and the Canadian Institutes of Health Research Institute of Human Development, Child and Youth Health (XG09-025 to CFB); the Michael Smith Foundation for Health Research (CI-SCH-O1899(07-1) to CFB); The Little Giants Foundation (CFB); and the Asociacion Española de Displasias Oseas Minoritarias (CFB). MM was supported by a Four Year Doctoral Fellowship from the University of British Columbia. CFB is a scholar of the Michael Smith Foundation for Health Research and a Clinical Investigator of the Child & Family Research Institute.
References
- Boerkoel CF, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, Andre JL, Bogdanovic R, Burguet A, Cockfield S, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet 2002; 30:215-20; PMID:11799392; http://dx.doi.org/10.1038/ng821
- Schimke RN, Horton WA, King CR. Chondroitin-6-sulphaturia, defective cellular immunity, and nephrotic syndrome. Lancet 1971; 2:1088-9; PMID:4106927; http://dx.doi.org/10.1016/S0140-6736(71)90400-4
- Spranger J, Hinkel GK, Stoss H, Thoenes W, Wargowski D, Zepp F. Schimke immuno-osseous dysplasia: A newly recognized multisystem disease. J Pediatr 1991; 119:64-72; PMID:2066860; http://dx.doi.org/10.1016/S0022-3476(05)81040-6
- Ehrich JH, Burchert W, Schirg E, Krull F, Offner G, Hoyer PF, Brodehl J. Steroid resistant nephrotic syndrome associated with spondyloepiphyseal dysplasia, transient ischemic attacks and lymphopenia. Clin Nephrol 1995; 43:89-95; PMID:7736684
- Boerkoel CF, O'Neill S, Andre JL, Benke PJ, Bogdanovic R, Bulla M, Burguet A, Cockfield S, Cordeiro I, Ehrich JH, et al. Manifestations and treatment of Schimke immuno-osseous dysplasia: 14 new cases and a review of the literature. Eur J Pediatr 2000; 159:1-7; PMID:10653321; http://dx.doi.org/10.1007/s004310050001
- Clewing JM, Antalfy BC, Lücke T, Najafian B, Marwedel KM, Hori A, Powel RM, Safo Do AF, Najera L, SantaCruz K, et al. Schimke immuno-osseous dysplasia: A clinicopathological correlation. J Med Genet 2007; 44:122-30; PMID:16840568; http://dx.doi.org/10.1136/jmg.2006.044313
- Morimoto M, Yu Z, Stenzel P, Clewing JM, Najafian B, Mayfield C, Hendson G, Weinkauf JG, Gormley AK, Parham DM, et al. Reduced elastogenesis: A clue to the arteriosclerosis and emphysematous changes in Schimke immuno-osseous dysplasia? Orphanet J Rare Dis 2012; 7:70; PMID:22998683; http://dx.doi.org/10.1186/1750-1172-7-70
- Morimoto M, Kerouredan O, Gendronneau M, Shuen C, Baradaran-Heravi A, Asakura Y, Basiratnia M, Bogdanovic R, Bonneau D, Buck A, et al. Dental abnormalities in Schimke immuno-osseous dysplasia. J Dent Res 2012; 91:29S-37S; PMID:22699664; http://dx.doi.org/10.1177/0022034512450299
- Lama G, Marrone N, Majorana M, Cirillo F, Salsano ME, Rinaldi MM. Spondyloepiphyseal dysplasia tarda and nephrotic syndrome in three siblings. Pediatr Nephrol 1995; 9:19-23; PMID:7742215; http://dx.doi.org/10.1007/BF00858959
- Lou S, Lamfers P, McGuire N, Boerkoel CF. Longevity in Schimke immuno-osseous dysplasia. J Med Genet 2002; 39:922-5; PMID:12471207; http://dx.doi.org/10.1136/jmg.39.12.922
- Lücke T, Billing H, Sloan EA, Boerkoel CF, Franke D, Zimmering M, Ehrich JH, Das AM. Schimke-immuno-osseous dysplasia: New mutation with weak genotype-phenotype correlation in siblings. Am J Med Genet A 2005; 135:202-5; PMID:15880370; http://dx.doi.org/10.1002/ajmg.a.30691
- Clewing JM, Fryssira H, Goodman D, Smithson SF, Sloan EA, Lou S, Huang Y, Choi K, Lucke T, Alpay H, et al. Schimke immunoosseous dysplasia: Suggestions of genetic diversity. Hum Mutat 2007; 28:273-83; PMID:17089404; http://dx.doi.org/10.1002/humu.20432
- Dekel B, Metsuyanim S, Goldstein N, Pode-Shakked N, Kovalski Y, Cohen Y, Davidovits M, Anikster Y. Schimke immuno-osseous dysplasia: Expression of SMARCAL1 in blood and kidney provides novel insight into disease phenotype. Pediatr Res 2008; 63:398-403; PMID:18356746; http://dx.doi.org/10.1203/PDR.0b013e31816721cc
- Baradaran-Heravi A, Cho KS, Tolhuis B, Sanyal M, Morozova O, Morimoto M, Elizondo LI, Bridgewater D, Lubieniecka J, Beirnes K, et al. Penetrance of biallelic SMARCAL1 mutations is associated with environmental and genetic disturbances of gene expression. Hum Mol Genet 2012; 21:2572-87; PMID:22378147; http://dx.doi.org/10.1093/hmg/dds083
- Postow L, Woo EM, Chait BT, Funabiki H. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem 2009; 284:35951-61; PMID:19841479; http://dx.doi.org/10.1074/jbc.M109.048330
- Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev 2009; 23:2394-9; PMID:19793864; http://dx.doi.org/10.1101/gad.1836409
- Yusufzai T, Kong X, Yokomori K, Kadonaga JT. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev 2009; 23:2400-4; PMID:19793863; http://dx.doi.org/10.1101/gad.1831509
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev 2009; 23:2405-14; PMID:19793861; http://dx.doi.org/10.1101/gad.1839909
- Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev 2009; 23:2415-25; PMID:19793862; http://dx.doi.org/10.1101/gad.1832309
- Sanyal M, Morimoto M, Baradaran-Heravi A, Choi K, Kambham N, Jensen K, Dutt S, Dionis-Petersen KY, Liu LX, Felix K, et al. Lack of IL7Ralpha expression in T cells is a hallmark of T-cell immunodeficiency in Schimke immuno-osseous dysplasia (SIOD). Clin Immunol 2015; 161:355-65; PMID:26499378; http://dx.doi.org/10.1016/j.clim.2015.10.005
- Morimoto M, Myung C, Beirnes K, Choi K, Asakura Y, Bokenkamp A, Bonneau D, Brugnara M, Charrow J, Colin E, et al. Increased Wnt and Notch signaling: A clue to the renal disease in Schimke immuno-osseous dysplasia? Orphanet J Rare Dis 2016; 11:149; PMID:27816064; http://dx.doi.org/10.1186/s13023-016-0519-7
- Bell O, Tiwari VK, Thoma NH, Schubeler D. Determinants and dynamics of genome accessibility. Nat Rev Genet 2011; 12:554-64; PMID:21747402; http://dx.doi.org/10.1038/nrg3017
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell 2007; 128:735-45; PMID:17320510; http://dx.doi.org/10.1016/j.cell.2007.02.009
- Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol 2014; 15:340-56; PMID:24755934; http://dx.doi.org/10.1038/nrm3789
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet 2005; 6:893-904; PMID:16341070; http://dx.doi.org/10.1038/nrg1726
- Saget O, Randsholt NB. Transposon-induced rearrangements in the duplicated locus ph of Drosophila melanogaster can create new chimeric genes functionally identical to the wild type. Gene 1994; 149:227-35; PMID:7958995; http://dx.doi.org/10.1016/0378-1119(94)90154-6
- Tiong SY, Russell MA. Clonal analysis of segmental and compartmental homoeotic transformations in polycomb mutants of Drosophila melanogaster. Dev Biol 1990; 141:306-18; PMID:1976555; http://dx.doi.org/10.1016/0012-1606(90)90387-X
- Tearle RG, Nusslein-Volhard C. Tubingen mutants and stock list. Drosoph Inf Serv 1987; 66:209-69
- Ruhf ML, Braun A, Papoulas O, Tamkun JW, Randsholt N, Meister M. The domino gene of Drosophila encodes novel members of the SWI2/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development 2001; 128:1429-41; PMID:11262242
- Ingham PW, Whittle R. Trithorax: A new homeotic mutation of Drosophila melanogaster causing transformations of abdominal and thoracic imaginal segments. Mol Gen Genet 1980; 179:607-14; http://dx.doi.org/10.1007/BF00271751
- Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci U S A 1988; 85:8136-40; PMID:3141923; http://dx.doi.org/10.1073/pnas.85.21.8136
- Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J 1994; 13:2553-64; PMID:7912192
- Tillib S, Petruk S, Sedkov Y, Kuzin A, Fujioka M, Goto T, Mazo A. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol Cell Biol 1999; 19:5189-202; PMID:10373568; http://dx.doi.org/10.1128/MCB.19.7.5189
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature 1978; 276:565-70; PMID:103000; http://dx.doi.org/10.1038/276565a0
- Lewis EB. Genes and developmental pathways. Am Zool 1963; 3:33-56; http://dx.doi.org/10.1093/icb/3.1.33
- van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell 2005; 17:695-707; PMID:15749019; http://dx.doi.org/10.1016/j.molcel.2005.02.013
- White RAH, Akam ME. Contrabithorax mutations cause inappropriate expression of Ultrabithorax products in Drosophila. Nature 1985; 318:567-70; http://dx.doi.org/10.1038/318567a0
- Fritsch C, Brown JL, Kassis JA, Muller J. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 1999; 126:3905-13; PMID:10433918
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev 2006; 20:2041-54; PMID:16882982; http://dx.doi.org/10.1101/gad.388706
- Schiavone D, Guilbaud G, Murat P, Papadopoulou C, Sarkies P, Prioleau MN, Balasubramanian S, Sale JE. Determinants of G quadruplex-induced epigenetic instability in REV1-deficient cells. EMBO J 2014; 33:2507-20; PMID:25190518; http://dx.doi.org/10.15252/embj.201488398
- Papadopoulou C, Guilbaud G, Schiavone D, Sale JE. Nucleotide pool depletion induces G-quadruplex-dependent perturbation of gene expression. Cell Rep 2015; 13:2491-503; PMID:26686635; http://dx.doi.org/10.1016/j.celrep.2015.11.039
- Khurana S, Oberdoerffer P. Replication stress: A lifetime of epigenetic change. Genes (Basel) 2015; 6:858-77; PMID:26378584
- Kim HR, Hwang KA, Kim KC, Kang I. Down-regulation of IL-7Ralpha expression in human T cells via DNA methylation. J Immunol 2007; 178:5473-9; PMID:17442928; http://dx.doi.org/10.4049/jimmunol.178.9.5473
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 1992; 68:561-72; PMID:1346755; http://dx.doi.org/10.1016/0092-8674(92)90191-E
- Elfring LK, Daniel C, Papoulas O, Deuring R, Sarte M, Moseley S, Beek SJ, Waldrip WR, Daubresse G, DePace A, et al. Genetic analysis of brahma: The Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 1998; 148:251-65; PMID:9475737
- Papoulas O, Daubresse G, Armstrong JA, Jin J, Scott MP, Tamkun JW. The HMG-domain protein BAP111 is important for the function of the BRM chromatin-remodeling complex in vivo. Proc Natl Acad Sci U S A 2001; 98:5728-33; PMID:11331758; http://dx.doi.org/10.1073/pnas.091533398
- Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 2013; 20:1147-55; PMID:24096405; http://dx.doi.org/10.1038/nsmb.2669
- Keene MA, Elgin SC. Micrococcal nuclease as a probe of DNA sequence organization and chromatin structure. Cell 1981; 27:57-64; PMID:6799212; http://dx.doi.org/10.1016/0092-8674(81)90360-3
- Levy A, Noll M. Chromatin fine structure of active and repressed genes. Nature 1981; 289:198-203; PMID:6779207; http://dx.doi.org/10.1038/289198a0
- Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci U S A 2008; 105:8580-4; PMID:18562274; http://dx.doi.org/10.1073/pnas.0804424105
- Jeronimo C, Langelier MF, Zeghouf M, Cojocaru M, Bergeron D, Baali D, Forget D, Mnaimneh S, Davierwala AP, Pootoolal J, et al. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol Cell Biol 2004; 24:7043-58; PMID:15282305; http://dx.doi.org/10.1128/MCB.24.16.7043-7058.2004
- Robert F, Blanchette M, Maes O, Chabot B, Coulombe B. A human RNA polymerase II-containing complex associated with factors necessary for spliceosome assembly. J Biol Chem 2002; 277:9302-6; PMID:11773074; http://dx.doi.org/10.1074/jbc.M110516200
- Sharma T, Bansal R, Haokip DT, Goel I, Muthuswami R. SMARCAL1 negatively regulates c-Myc transcription by altering the conformation of the promoter region. Sci Rep 2015; 5:17910; PMID:26648259; http://dx.doi.org/10.1038/srep17910
- Morimoto M, Wang KJ, Yu Z, Gormley AK, Parham D, Bogdanovic R, Lucke T, Mayfield C, Weksberg R, Hendson G, et al. Transcriptional and posttranscriptional mechanisms contribute to the dysregulation of elastogenesis in Schimke immuno-osseous dysplasia. Pediatr Res 2015; 78:609-17; PMID:26309238; http://dx.doi.org/10.1038/pr.2015.156
- Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: Molecular mechanisms and biological effects. Cell Res 2008; 18:73-84; PMID:18166977; http://dx.doi.org/10.1038/cr.2008.6
- Keka IS, Mohiuddin , Maede Y, Rahman MM, Sakuma T, Honma M, Yamamoto T, Takeda S, Sasanuma H. Smarcal1 promotes double-strand-break repair by nonhomologous end-joining. Nucleic Acids Res 2015; 43:6359-72; PMID:26089390; http://dx.doi.org/10.1093/nar/gkv621
- Baradaran-Heravi A, Raams A, Lubieniecka J, Cho KS, DeHaai KA, Basiratnia M, Mari PO, Xue Y, Rauth M, Olney AH, et al. SMARCAL1 deficiency predisposes to non-Hodgkin lymphoma and hypersensitivity to genotoxic agents in vivo. Am J Med Genet A 2012; 158A:2204-13; PMID:22888040; http://dx.doi.org/10.1002/ajmg.a.35532
- Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol 2014; 16:2-9; PMID:24366029; http://dx.doi.org/10.1038/ncb2897
- Baradaran-Heravi A, Morimoto M, Tolhuis B, Shaw C, Sanyal M, Raams A, Bokenkamp A, Cho KS, Myung C, Leung D, et al. Schimke immuno-osseous dysplasia: A Mendelian complex trait arising from impaired DNA maintenance and global alterations in gene expression. 60th Annual American Society of Human Genetics Meeting, November 2–6, 2010, Washington, DC, United States.
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993; 118:401-15; PMID:8223268
- Cavalli G, Orlando V, Paro R. Mapping DNA target sites of chromatin-associated proteins by formaldehyde cross-linking in Drosophila embryos. In: Bickmore WA, ed. Chromosome Structural Analysis: A Practical Approach. Oxford: Oxford University Press, 1999:20-37
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell 2004; 14:637-46; PMID:15175158; http://dx.doi.org/10.1016/j.molcel.2004.05.009
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet 2006; 38:694-9; PMID:16628213; http://dx.doi.org/10.1038/ng1792
- Ner SS, Harrington MJ, Grigliatti TA. A role for the Drosophila SU(VAR)3-9 protein in chromatin organization at the histone gene cluster and in suppression of position-effect variegation. Genetics 2002; 162:1763-74; PMID:12524347
