ABSTRACT
The control of genomic maintenance during S phase is crucial in eukaryotes. It involves the establishment of sister chromatid cohesion, ensuring faithful chromosome segregation, as well as proper DNA replication and repair to preserve genetic information. In animals, nuclear periphery proteins - including inner nuclear membrane proteins and nuclear pore-associated components - are key factors which regulate DNA integrity. Corresponding functional homologues are not so well known in plants which may have developed specific mechanisms due to their sessile life. We have already characterized the Gamma-tubulin Complex Protein 3-interacting proteins (GIPs) as essential regulators of centromeric cohesion at the nuclear periphery. GIPs were also shown to interact with TSA1, first described as a partner of the epigenetic regulator MGOUN3 (MGO3)/BRUSHY1 (BRU1)/TONSOKU (TSK) involved in genomic maintenance. Here, using genetic analyses, we show that the mgo3gip1 mutants display an impaired and pleiotropic development including fasciation. We also provide evidence for the contribution of both MGO3 and GIP1 to the regulation of centromeric cohesion in Arabidopsis.
Introduction
A tight regulation of DNA replication and mitosis in cycling cells ensures both the maintenance of genetic information integrity and an equational distribution of sister chromatids in daughter cells. Thus, accurate, efficient chromosome segregation is safeguarded by the maintenance of centromeric cohesion until anaphase. During S phase, DNA replication must deal with centromeric regions containing repetitive sequences and so, the cohesion between duplicated centromeres needs to be preserved. To properly monitor centromeric DNA replication, it was recently demonstrated that the slower dynamics of the replication forks did not activate the ATM- and Rad3-related (ATR) kinase involved in S phase DNA damage checkpoint.Citation1 In addition, the establishment of cohesion during S phase involves a chromosome transmission fidelity protein 7 (CTF7) which acetylates the structural maintenance of chromosome 3 (SMC3), a central component of the cohesion complex.Citation2 This step may also imply CTF18 and be coupled with the passage of the DNA replication fork.Citation2 This suggests that a mechanical link exists between cohesion establishment and DNA replication.
The regulation of DNA replication and sister chromatid cohesion remains poorly understood in plants. The ctf7 mutants exhibit defects in both DNA repair and cell division.Citation3 Sister chromatid cohesion is impaired in the mutant of minichromosome maintenance helicase-binding protein E2F-target gene 1 (ETG1). Such a defect even extends the impact to centromeric regions when CTF18 is simultaneously affected.Citation4
Recently, we demonstrated that GIPs, initially found as regulators of the recruitment of microtubule-nucleation complexes, were also key players in the regulation of the nuclear architecture.Citation5,6 GIPs located on both sides of the Nuclear Envelope (NE) play a critical role in the maintenance of centromeric cohesion in the nuclei of cycling cells studied in Arabidopsis root meristems.Citation7,8 Interestingly, GIPs were identified as partners of TSA16 which was first described as TONSOKU (TSK)-associating protein1.Citation9 TONSOKU (TSK) is specifically expressed during S phase.Citation10 Allelic mutations in the epigenetic regulator TSKCitation11 also named BRUSHY1 (BRU1)Citation12-14 and MGOUN3 (MGO3)Citation15 led to a deregulation of various biologic processes: DNA damage response during DNA replication, cell cycle progression, heterochromatin organization, chromosomal subdomain architecture as well as meristem organization and flowering transition in Arabidopsis. TSA1 was also described as a partner of the COP9 signalosome subunit 1 (CSN1).Citation16 Both TSA1 and CSN1 may be involved in seedling development. Contrary to gip and mgo3/bru1/tsk mutants, the tsa1 knocked-down mutant does not show strong growth alteration when growing under white light.Citation16 Therefore, we investigated the genetic interactions between GIPs and MGO3/BRU1/TSK in Arabidopsis. We established gip1mgo3 and gip2mgo3 lines, using the previously characterized gip1, gip2 and mgo3 mutants,Citation5,15 and compared the growth phenotypes of these mutants. We observed that gip1mgo3 presented severe growth phenotypes as described previously for gip1gip2.Citation5 The defects of centromeric chromatin organization observed in gip1mgo3 root nuclei indicate that MGO3 contributes together with GIP1 to the maintenance of centromeric cohesion in Arabidopsis. Therefore, our findings shed new light on the contribution of MGO3, in addition to GIP1, to a crosstalk between DNA replication and the establishment of sister chromatid cohesion, controlled at the nuclear periphery.
Results
Phenotypic growth characterization of gip1mgo3 mutants
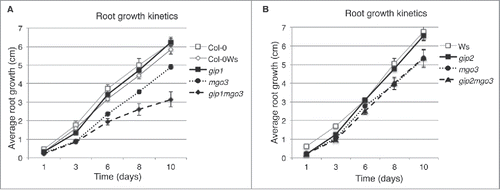
We already showed that GIP deficiency in gip1gip2 led to pleiotropic growth phenotypes and chromosomal instabilityCitation5 and that TSA1 interacted with both GIP1 and GIP2.Citation6 TSA1 was also shown to interact with MGO3/BRU1/TSK.Citation9 As BRU1 is involved in genomic maintenance,Citation12 we investigated the relationships between MGO3/BRU1/TSK and GIPs. Among the several allelic mutants described in the literature, we used herein mgo3–2, further described as mgo3.Citation15 The mgo3 mutant has a deletion of the 5′ coding region corresponding to a tetratricopeptide repeat-like domain (TPR-like) (Fig. S1).Citation15 First, we generated homozygous lines resulting from crosses between gip1 or gip2 and mgo3 mutants, and characterized the growth of the different mutant seedlings. According to the genetic backgrounds of the mutants, Columbia (Col-0) for gip1, Wassilewskija (Ws) for mgo3 and gip2, and Col-0Ws for gip1mgo3 and gip1gip2, the wild type (WT) plants remained indistinguishable from each other (Fig. S2A). The mgo3 mutant showed a reduced primary root growth compared with WT, gip1 or gip2 (Fig. S2A). While gip2mgo3 showed a similar root growth phenotype as mgo3, the mean root growth of gip1mgo3 was slower (, Fig. S2A). This was essentially due to the presence of high root length variability in gip1mgo3. Therefore, this prompted us to further split the gip1mgo3 phenotypes into 2 subsets - type a (39.13% ± 4.09; n = 1219) resembling mgo3 and type b (60.87% ± 4.09) which showed more severe and pleiotropic developmental phenotypes (Fig. S2A), like those observed in gip1gip2.Citation5 At the level of their shoots, gip2mgo3 were similar to mgo3 but differed from gip1mgo3 type a plantlets as these presented longer cotyledon petioles (Fig. S2B). Contrary to gip1gip2,Citation5 the gip1mgo3 mutants remained fertile but exhibited fasciated inflorescences and stems with a much stronger phenotype compared with mgo3 (Fig. S2C).Citation15 Such flattened shoots, as observed in mgo3/bru1/tsk, fasciata 1, 2 and tebichi mutants, were functionally linked either to defects in chromatin assembly, DNA damage repair, DNA replication, or to meristem organization.Citation11,12,15,17-19 Thus, the increased fasciation observed in gip1mgo3 suggests that GIP1 may contribute, together with MGO3, to these processes.
Centromere organization is impaired in gip1mgo3 mutants
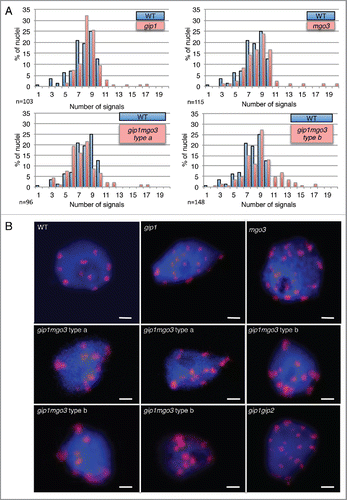
In meristematic root cells, nuclei are enlarged in both gip1mgo3 type b and gip1gip2 (), while interphasic cortical microtubule organization is not significantly altered in these mutants nor in gip1mgo3 type a, gip1 and mgo3 mutants (, Fig. S3). More specifically, while chromocentres which mainly correspond to pericentromeric heterochromatin at the nuclear periphery,Citation20 reach the number of 10 in Arabidopsis WTs (either ecotypes Ws, Col-0 or Col-0Ws), far more than 10 heterochromatin signals are present in about 20% of gip1mgo3 type b plantlets compared with gip1mgo3 type a, gip1 or mgo3 (less than 5%). Thus, the gip1mgo3 type b phenotype was similar to that described previously for gip1gip2 ().Citation7 For a more accurate description of the centromere defect, we evaluated the number of centromeric signals in 4C flow sorted nuclei, using the FISH centromere-specific pAL probe. While in WT the number of pAL signals did not exceed 10, we observed more than 10 signals in 6 to 7% of the nuclei in the gip1 and mgo3 mutants as well as in the gip1mgo3 type a mutants (). This percentage increased up to 19% in the gip1mgo3 type b mutants. A similarly increased number of pAL signals was observed for the less affected gip1gip2 type1 mutants.Citation7 These data argue for a synergistic contribution of MGO3 and GIP1 to centromeric cohesion. In addition, we observed an irregular distribution of pAL signals in the nuclei of both gip1mgo3 type a (8.6%; n = 93) and b (22%; n = 77) mutants compared with WT (). The centromeric histone H3 variant (CENH3) signals, detected using immunolabelling, confirmed these results (Fig. S4). This indicates that MGO3, together with GIP1, may contribute to the spatial centromeric chromatin organization.
Figure 2. Analysis of root meristematic nuclei from gip1, mgo3, gip1mgo3 compared with gip1gip2. (A) Detection of chromatin by DAPI staining (blue) and microtubules by immuno-labeling with antibodies directed against α-tubulin (red) performed on whole mount meristems of the different seedlings (gip1, mgo3, gip1mgo3 and gip1gip2). Images were captured by confocal microscopy and correspond to Z-stack projections of focal planes. Bars = 10 µm. (B) Meristematic nuclei representative for different WT backgrounds (Ws Col-0), Col-0Ws) were compared with gip1, mgo3, gip1mgo3 type a and b and gip1gip2. Images were captured by confocal microscopy and correspond to Z-stack projections of focal planes. Bars = 2 µm. For Z-stacks, slides were acquired in 0.35 µm intervals.
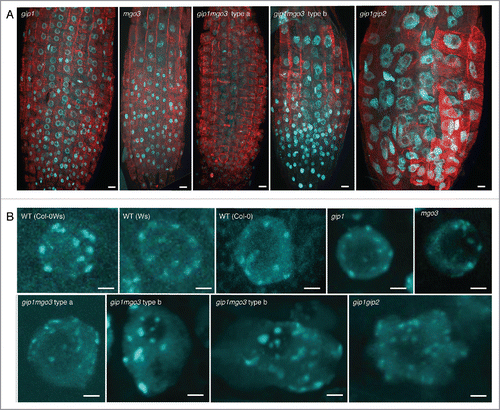
Figure 3. The gip1, mgo3 and gip1mgo3 mutants exhibit centromeric cohesion defects. (A) Number of pAL signals in 4C flow-sorted nuclei from WT, gip1, mgo3 and 2 seedling phenotypes (types a and b) of gip1mgo3 mutants. A Student t-test was used to calculate the confidence of values for signals >10. P < 0.05 (B) FISH detection of centromeric pAL signals in 4C nuclei from WT, gip1, mgo3, gip1mgo3 type a and b compared with gip1gip2 mutants. The image stacks of nuclei were collected with a Z-step size of 0.34 µm. Bars = 2 µm.
Discussion
In the present study, we have analyzed the genetic interactions between MGO3 and GIP1 in the control of centromeric chromatin. Previously, we showed the involvement of both GIP1 and GIP2 at the nuclear interfaceCitation7 and herein, the contribution of GIP1 together with MGO3 to the maintenance of centromeric cohesion.
MGO3/BRU1/TSK plays a role in stalling replication forksCitation12 as described for its homolog in humans, Tonsoku-Like (TONSL),Citation21,22 acting as a reader of new epigenetic marks linked to new histone incorporation at the post-replicative state.Citation23 Beside their role in centromeric cohesion, GIPs must also be involved in the loading/maintenance of CENH3 at centromeres.Citation7 The current challenge is to untangle the dynamics of these processes and to characterize the chaperones involved in CENH3 loading.Citation8 In animals, a dual role was described for the histone chaperone nucleosome assembly protein 1 (NAP1) controlling sister chromatid separation independently of its function in nucleosome assembly.Citation24 It is worth noting that human TONSL was shown to be an H3–1/H4 chaperone needed for nucleosome assembly after the restart of stalled replication forks,Citation25 but its role in the control of centromeric replication has not been investigated so far.
In interphase cells, GIPs were found on both sides of the NE. TSA1 and GIP1 exhibited a similar spotty appearance at the NE periphery,Citation6,11 while BRU1–EGFP fusion proteins, detected in the nucleus, were partially excluded from the nucleolus.Citation13 Nuclear localization of GIPs at the centromeres may contribute, together with MGO3, to the control of centromeric cohesion during S phase as previously suggested.Citation26 Interestingly, the minimal domain of interaction of TSK was partly overlapping with that of GIP1 in the C-terminal region of TSA1.Citation6,9 These data argue for a spatiotemporal regulation of GIP1, TSA1 and MGO3 interactions to properly ensure centromere cohesion. In addition, in tsk mutants, a stabilization of TSA1 was observed without any changes in the TSA1 transcript level,Citation16 contrary to mgo3 in which we showed that TSA1 was upregulated (3 to 4 times, Fig. S5). This corroborates that a tight regulation exists between TSA1 and MGO3.
In plants so far, only the etg1ctf18 mutants which are affected in DNA replication and sister chromatid cohesion, respectively,Citation4 have been described to impair centromeric cohesion, but to a lesser extent than for gip1gip2 or gip1mgo3. Our study highlights MGO3 and GIP1 as key actors whose dynamic interplay may allow the coordination of DNA replicationCitation12 and centromeric cohesion at the NE periphery, although the underlying mechanisms remain to be elucidated.
In addition, as observed in ctf7,Citation3 post-replicative repair is affected in the etg1 mutants and leads to the formation of endogenous Double Strand Breaks as well as the activation of the DNA damage response.Citation4 Similarly, mutations in the structural maintenance of chromosome 5/6 complex (SMC5/6) lead to defects in sister chromosome cohesion as well as impaired DNA repair in Arabidopsis.Citation27 All these data argue for a tight control between DNA replication, DNA repair and sister chromatid cohesion. As postreplicative DNA repair is impaired in bru1,Citation12 we need to further investigate this aspect in relation with GIPs and the nuclear periphery.
Altogether, our data reveal a particular network at the nuclear periphery involved in centromeric cohesion establishment/maintenance. This sustains proper chromosome segregation which is crucial for cell division and plant development. A further characterization of the dynamic interplay between GIP1 and MGO3 may help to understand the functional crosstalks taking place at the nuclear envelope periphery in plants. Since both GIP and MGO3 are conserved in humans as MOZART1Citation28 and TONSL, respectively, this may allow the investigation of their roles in defects of nuclear architectureCitation29 and chromatid cohesion in humansCitation30 linked to cancer progression.
Materials and methods
Plants and growth
gip1, gip2, gip1gip2 and mgo3 mutants have been described previously.Citation5,15 The Arabidopsis lines were grown in vitro on Murashige and Skoog medium (SERVA Electrophoresis) at 20°C with a 16h photoperiod (70 µmol m-2 s-1 fluorescent lighting). Homozygous T-DNA insertion lines of gip1 and gip2 were crossed with mgo3–2 to produce the gip1mgo3 and gip2mgo3 mutants. All the investigations were performed on homozygous F3 lines. For genotyping, we used the same primers as described previously.Citation5,15
Whole mount root tip immunostaining
7-day-old Arabidopsis seedlings were fixed as described.Citation5 We used the primary monoclonal antibody anti-α-tubulin (clone DM1A; Sigma-Aldrich, 1/5000) and the Alexa 568–conjugated goat anti-mouse IgG secondary antibody (1:300) (Molecular Probes). Root tips were mounted in antifade Vectashield (Vector Laboratories), with DAPI (2 µg/ml).
Immunocytochemistry
7-day-old Arabidopsis seedlings were placed for 20 min on ice in 4% paraformaldehyde in PEM buffer (50 mM PIPES, 5 mM EGTA, 5 mM MgSO4, pH 6.9) and processed as described.Citation7 Primary anti-CENH3 polyclonal antibodies (Novus Biologicals; 1/500) were used in overnight incubation at 4°C; signals were detected using Alexa 568 fluor dye-conjugated secondary antibodies (1:300, Life Technologies) and counterstained with 2µg/ml DAPI.
Flow sorting of nuclei
The nuclei of 7-day-old plantlets were isolated and flow-sorted according to their endoploidy level after formaldehyde fixation using a FACS Aria (BD Biosciences), as described previously.Citation31
FISH
The pAL plasmidCitation32 was used for the detection of the 180 bp centromeric tandem repeats. Probes were labeled with Orange 552 dUTP (Enzo Life Sciences (Els) Ag,) using the nick translation DNA labeling system (Enzo Life Sciences (Els) Ag). Slides were processed as describedCitation7 previously and incubated with 20 µl of pAL probe per slide. After denaturation at 80°C for 2 min, hybridization was performed overnight at 37°C. After successive washes at 42°C in 2xSSC, in 50% formamide in 2xSSC and then 2xSCC, the material was mounted in Vectashield (Vector Laboratories) containing DAPI (2 µg/ml). The different frequencies of sister chromatid cohesion were tested for significance, using the 2-sided Fisher exact test.
Confocal microscopy
Confocal images were acquired with a Zeiss LSM 780 microscope equipped with 20x/0.8 or 63x/1.4 oil objectives. pAL centromeric signals as well as DAPI were observed using a laser beam with the excitation wavelengths of 405 and 488 nm, respectively.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KNCL_1276142_Supplementary_Files.zip
Download Zip (22.2 MB)Acknowledgments
Microscopy was performed at the Strasbourg-Esplanade cellular imaging facility (CNRS, Université de Strasbourg, Région Alsace, Fondation ARC pour la Recherche sur le cancer and Ligue contre le cancer). We thank V. Schaeffly, C. Tritsch, S. Mangold for their technical help. We are also very grateful to M. Delarue for providing the mgo3–2 mutants and helpful discussions, and L. Blech for the corrections of the English manuscript.
Funding
This work was supported by the Center National de la Recherche Scientifique (CNRS), the Ministère de l'Enseignement Supérieur et de la Recherche (MESR) and by the PHC Procope 30746TB.
References
- Aze A, Sannino V, Soffientini P, Bachi A, Costanzo V. Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Nat Cell Biol 2016; 18:684-91; PMID:27111843; http://dx.doi.org/10.1038/ncb3344
- Kenna MA, Skibbens RV. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol Cell Biol 2003; 23:2999-3007; PMID:12665596; http://dx.doi.org/10.1128/MCB.23.8.2999-3007.2003
- Bolanos-Villegas P, Yang X, Wang HJ, Juan CT, Chuang MH, Makaroff CA, Jauh GY. Arabidopsis CHROMOSOME TRANSMISSION FIDELITY 7 (AtCTF7/ECO1) is required for DNA repair, mitosis and meiosis. Plant J 2013; 75:927-40; PMID:23750584; http://dx.doi.org/10.1111/tpj.12261
- Takahashi N, Quimbaya M, Schubert V, Lammens T, Vandepoele K, Schubert I, Matsui M, Inze D, Berx G, De Veylder L. The MCM-binding protein ETG1 aids sister chromatid cohesion required for postreplicative homologous recombination repair. PLoS Genet 2010; 6:e1000817; PMID:20090939; http://dx.doi.org/10.1371/journal.pgen.1000817
- Janski N, Masoud K, Batzenschlager M, Herzog E, Evrard JL, Houlne G, Bourge M, Chaboute ME, Schmit AC. The GCP3-interacting proteins GIP1 and GIP2 are required for gamma-tubulin complex protein localization, spindle integrity, and chromosomal stability. Plant Cell 2012; 24:1171-87; PMID:22427335; http://dx.doi.org/10.1105/tpc.111.094904
- Batzenschlager M, Masoud K, Janski N, Houlne G, Herzog E, Evrard JL, Baumberger N, Erhardt M, Nomine Y, Kieffer B, et al. The GIP gamma-tubulin complex-associated proteins are involved in nuclear architecture in Arabidopsis thaliana. Front Plant Sci 2013; 4:480; PMID:24348487; http://dx.doi.org/10.3389/fpls.2013.00480
- Batzenschlager M, Lermontova I, Schubert V, Fuchs J, Berr A, Koini MA, Houlne G, Herzog E, Rutten T, Alioua A, et al. Arabidopsis MZT1 homologs GIP1 and GIP2 are essential for centromere architecture. Proc Natl Acad Sci U S A 2015; 112:8656-60; PMID:26124146; http://dx.doi.org/10.1073/pnas.1506351112
- Schmit AC, Herzog E, Chaboute ME. GIP/MZT1 proteins: Key players in centromere regulation. Cell Cycle 2015; 14:3665-6; PMID:26517054; http://dx.doi.org/10.1080/15384101.2015.1112614
- Suzuki T, Nakajima S, Morikami A, Nakamura K. An Arabidopsis protein with a novel calcium-binding repeat sequence interacts with TONSOKU/MGOUN3/BRUSHY1 involved in meristem maintenance. Plant Cell Physiol 2005; 46:1452-61; PMID:15964904; http://dx.doi.org/10.1093/pcp/pci155
- Suzuki T, Nakajima S, Inagaki S, Hirano-Nakakita M, Matsuoka K, Demura T, Fukuda H, Morikami A, Nakamura K. TONSOKU is expressed in S phase of the cell cycle and its defect delays cell cycle progression in Arabidopsis. Plant Cell Physiol 2005; 46:736-42; PMID:15746155; http://dx.doi.org/10.1093/pcp/pci082
- Suzuki T, Inagaki S, Nakajima S, Akashi T, Ohto MA, Kobayashi M, Seki M, Shinozaki K, Kato T, Tabata S, et al. A novel Arabidopsis gene TONSOKU is required for proper cell arrangement in root and shoot apical meristems. Plant J 2004; 38:673-84; PMID:15125773; http://dx.doi.org/10.1111/j.1365-313X.2004.02074.x
- Takeda S, Tadele Z, Hofmann I, Probst AV, Angelis KJ, Kaya H, Araki T, Mengiste T, Mittelsten Scheid O, Shibahara K, et al. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev 2004; 18:782-93; PMID:15082530; http://dx.doi.org/10.1101/gad.295404
- Ohno Y, Narangajavana J, Yamamoto A, Hattori T, Kagaya Y, Paszkowski J, Gruissem W, Hennig L, Takeda S. Ectopic gene expression and organogenesis in Arabidopsis mutants missing BRU1 required for genome maintenance. Genetics 2011; 189:83-95; PMID:21705754; http://dx.doi.org/10.1534/genetics.111.130062
- Ohno Y, Nishimura T, Hattori T, Takeda S. BRU1 maintains configuration of the euchromatic subchromosomal domain in the nucleus of arabidopsis. Plant Mol Biol Rep 2013; 32:19-27; http://dx.doi.org/10.1007/s11105-013-0596-x
- Guyomarc'h S, Benhamed M, Lemonnier G, Renou JP, Zhou DX, Delarue M. MGOUN3: evidence for chromatin-mediated regulation of FLC expression. J Exp Bot 2006; 57:2111-9; PMID:16728410; http://dx.doi.org/10.1093/jxb/erj169
- Li W, Zang B, Liu C, Lu L, Wei N, Cao K, Deng XW, Wang X. TSA1 interacts with CSN1/CSN and may be functionally involved in Arabidopsis seedling development in darkness. J Genet Genomics 2011; 38:539-46; PMID:22133685; http://dx.doi.org/10.1016/j.jgg.2011.08.007
- Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell 2001; 104:131-42; PMID:11163246; http://dx.doi.org/10.1016/S0092-8674(01)00197-0
- Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K, Abe K, Ichikawa H, Valentine L, et al. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J 2006; 25:5579-90; PMID:17110925; http://dx.doi.org/10.1038/sj.emboj.7601434
- Inagaki S, Suzuki T, Ohto MA, Urawa H, Horiuchi T, Nakamura K, Morikami A. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell 2006; 18:879-92; PMID:16517762; http://dx.doi.org/10.1105/tpc.105.036798
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci U S A 2002; 99:14584-9; PMID:12384572; http://dx.doi.org/10.1073/pnas.212325299
- Duro E, Lundin C, Ask K, Sanchez-Pulido L, MacArtney TJ, Toth R, Ponting CP, Groth A, Helleday T, Rouse J. Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol Cell 2010; 40:632-44; PMID:21055984; http://dx.doi.org/10.1016/j.molcel.2010.10.023
- O'Donnell L, Panier S, Wildenhain J, Tkach JM, Al-Hakim A, Landry MC, Escribano-Diaz C, Szilard RK, Young JT, Munro M, et al. The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol Cell 2010; 40:619-31; PMID:21055983; http://dx.doi.org/10.1016/j.molcel.2010.10.024
- Saredi G, Huang H, Hammond CM, Alabert C, Bekker-Jensen S, Forne I, Reverón-Gómez N, Foster BM, Mlejnkova L, Bartke T, et al. H4K20me0 marks post-replicative chromatin and recruits the TONSL–MMS22L DNA repair complex. Nature 2016; 534:714-8; PMID:27338793; http://dx.doi.org/10.1038/nature18312
- Moshkin YM, Doyen CM, Kan TW, Chalkley GE, Sap K, Bezstarosti K, Demmers JA, Ozgur Z, van Ijcken WF, Verrijzer CP. Histone chaperone NAP1 mediates sister chromatid resolution by counteracting protein phosphatase 2A. PLoS Genet 2013; 9:e1003719; PMID:24086141; http://dx.doi.org/10.1371/journal.pgen.1003719
- Campos EI, Smits AH, Kang YH, Landry S, Escobar TM, Nayak S, Ueberheide BM, Durocher D, Vermeulen M, Hurwitz J, et al. Analysis of the histone H3.1 interactome: a suitable chaperone for the right event. Mol Cell 2015; 60:697-709; PMID:26527279; http://dx.doi.org/10.1016/j.molcel.2015.08.005
- Chaboute ME, Berr A. GIP contributions to the regulation of centromere at the interface between the nuclear envelope and the nucleoplasm. Front Plant Sci 2016; 7:118; PMID:26904080; http://dx.doi.org/10.3389/fpls.2016.00118
- Watanabe K, Pacher M, Dukowic S, Schubert V, Puchta H, Schubert I. The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 2009; 21:2688-99; PMID:19737979; http://dx.doi.org/10.1105/tpc.108.060525
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 2010; 328:593-9; PMID:20360068; http://dx.doi.org/10.1126/science.1181348
- Bell ES, Lammerding J. Causes and consequences of nuclear envelope alterations in tumour progression. Eur J Cell Biol 2016; 95:449-464; PMID:27397692
- Barber TD, McManus K, Yuen KW, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci U S A 2008; 105:3443-8; PMID:18299561; http://dx.doi.org/10.1073/pnas.0712384105
- Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysak MA, Fuchs J, Schubert I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 2004; 113:258-69; PMID:15480725; http://dx.doi.org/10.1007/s00412-004-0316-2
- Martinez-Zapater JM, Estelle MA, Somerville CR. A highly repeated DNA sequence in Arabidopsis thaliana. Mol Gen Genet 1986; 204:417-23; http://dx.doi.org/10.1007/BF00331018