ABSTRACT
RNF20/Bre1 mediated H2B ubiquitination (H2Bub) has various physiologic functions. Recently, we found that H2Bub participates in meiotic recombination by promoting chromatin relaxation during meiosis. We then analyzed the phylogenetic relationships among the E3 ligase for H2Bub, its E2 Rad6 and their partner WW domain-containing adaptor with a coiled-coil (WAC) or Lge1, and found that the molecular mechanism underlying H2Bub is evolutionarily conserved from yeast to mammals. However, RNF20 has diverse physiologic functions in different organisms, which might be caused by the evolutionary divergency of their domain/motif architectures. In the current extra view, we not only elucidate the evolutionarily conserved molecular mechanism underlying H2Bub, but also discuss the diverse physiologic functions of RNF20 during meiosis.
Introduction
Meiosis is a specialized form of cell division of sexually reproducing organisms. After a single round of DNA replication following with 2 successive cell divisions (meiosis I and meiosis II), a single diploid germ cell gives rise to 4 haploid gametes, and the chromosome number of haploid gametes is reduced to half.Citation1 The prophase of meiosis I can be divided into 5 stages according to the different behavior of chromosomes, which are termed as leptotene, zygotene, pachytene, diplotene, diakinesis. There are some unique events of prophase I such as homologous chromosome synapsis, crossover formation, meiotic double-strand break (DSB) repairing and meiotic recombination. The homologous chromosomes segregate away from each other at the anaphase I, whereas sister chromatids segregate from each other in meiosis II. Errors in any of these events lead to failure of the gametogenesis.
The correct formation of some chromatin structures and the precise behavior of the chromosomes are essential to meiosis.Citation2 Nucleosome is the basic unit of chromosomes, it is composed of octamers of the core histone proteins and approximately 146 base pairs of DNA. A nucleosome contains 2 H2A/H2B dimers and one H3/H4 tetramer. The histone tails could be modified by different types of posttranslational modifications, including methylation,Citation3 acetylation,Citation4 phosphorylation,Citation5 SUMOylationCitation6 and ubiquitination.Citation7–9 These histone modifications play very important roles in gene transcription, gene silencing, chromosome organization and DNA repair.Citation10
Ubiquitin is a 76 residues polypeptide or small protein that is highly conserved from yeast to humans. A single ubiquitin molecule is attached to one lysine residue of the substrate protein, this is referred to as monoubiquitination. Attachment a chain of ubiquitin molecules to a specific lysine residue of substrate protein is termed as polyubiquitination. Monoubiquitination mainly contributes to DNA repair, gene expression and receptor endocytosis. Polyubiquitination is essential to protein degradation and signal transduction. Ubiquitination is mainly performed by 3 enzymes, ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2) and ubiquitin ligases (E3). Ubiquitin can be removed from target protein by deubiquitin enzymes (DUBs), which are called deubiquitination.Citation11–13 Monoubiquitination of histone H2B at the 123th Lysine residue in yeast (H2BK123ub) or the 120th lysine residue in mammals (H2BK120ub) is catalyzed by the E3 ubiquitin ligase Bre1 (RNF20/RNF40 complex in mammals). H2Bub takes part in some of the fundamental biologic processes, including cell cycle, tumogenesis, apoptosis and stem cell differentiation.Citation14 In addition, H2Bub was found to be involved in DNA damage response, chromatin segregation, DNA replication, transcription and RNA processing.Citation14 Recently, we found that Rnf20-mediated H2B ubiquitination is required for meiotic recombination. A germ cell-specific knockout of Rnf20 results in complete male infertility. The Rnf20 deficient spermatocytes arrest at the pachytene stage because of impaired programmed DSB repair. Further investigations reveal that the deletion of Rnf20 inhibits the recruitement of DSB repair factors to the proper positions on the chromatin mainly by affecting chromatin relaxation.Citation10 Here, we analyzed the phylogenetic relationships among the related E2, E3 and their partner WW domain-containing adaptor with a coiled-coil (WAC) or Lge1, and found that the molecular mechanism underlying H2Bub is evolutionarily conserved from yeast to mammals. However, RNF20 has diverse physiologic functions during meiosis, which might be caused by the evolutionary diversity of their domain/motif architectures.
Evolutionarily conserved molecular mechanisms underlying H2B ubiquitination
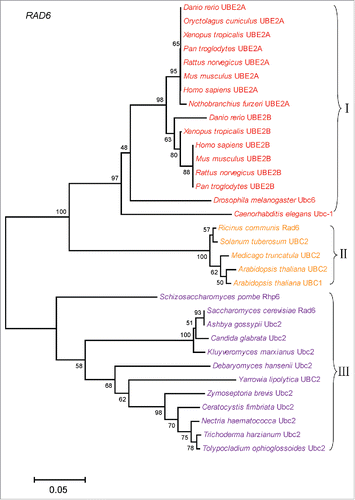
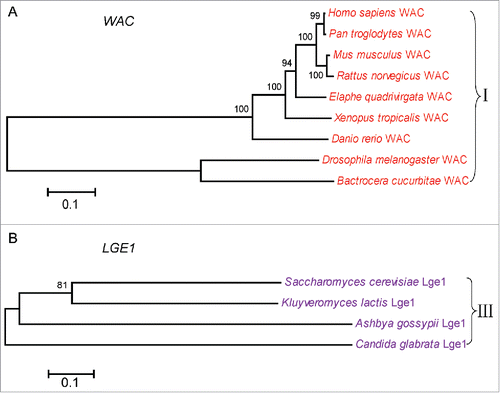
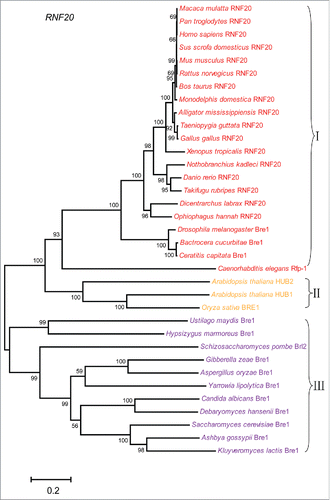
In yeast, Rad6, Bre1 and Lge1 form a complex that transfer ubiquitin to histone H2B,Citation15 among them, Rad6 works as an E2 ubiquitin-conjugating enzyme, Bre1 works as an E3 ligase in H2B ubiquitination. Lge1 interact with Bre1, and recruit it to the promoter and coding regions of actively transcribed genes in yeast.Citation15,16 RNF20 and RNF40 are the yeast homologues of Bre1 in mammalian cells, they usually form a stable heterodimeric E3 ligase, and are thought to recruit the E2 ubiquitin-conjugating enzymes UBE2B onto chromatin and transfer ubiquitin to histone H2B.Citation17 It has been reported that a novel WW domain-containing adaptor with a coiled-coil (WAC) works as a functional partner for RNF20/RNF40 in mammalian cells. WAC is not essential for H2B ubiquitination, but it facilitates the interaction between RNF20/RNF40 complex and UBE2B, thus promoting H2B ubiquitination.Citation18 To better understand the molecular mechanism underlying H2B ubiquitination in different organisms, we analyzed the phylogenetic relationship of RNF20, Rad6 together with WAC and Lge1, respectively. We downloaded the protein sequences from UniProt (http://www.uniprot.org/), Ensembl (http://asia.ensembl.org/index.html) and NCBI (https://www.ncbi.nlm.nih.gov/protein/). The longest transcript sequence was chosen as homologues sequence in 48 organisms. We constructed multi-species phylogenetic tree of 4 proteins, RNF20, WAC, Lge1 and Rad6, by using MEGA 6.0 with the Neighbor-Joining (NJ) method, and bootstrap analysis were performed using 1000 replications with the p-distance model.
According to the phylogenetic analysis (), the RNF20/Bre1 proteins can be divided into 3 groups: animals, plants, and fungi. Fungi and worm only have one H2Bub E3 ligase, Bre1 or Rfp-1. Arabidopsis thaliana genome encodes 2 E3 ligases, HUB1 and HUB2, which may form a tetramer with 2 copies of the each enzyme.Citation19,20 Mammalian genomes also encode 2 E3 ligases, RNF20 and RNF40. They will form stable heterodimer for ubiquitinating H2B histone. Therefore, the E3 ligases of plants and mammals are more complex compared with that of the other species. We also constructed multiple species phylogenetic tree of Rad6. As shown in , the phylogenetic relationship of Rad6 is consistent with RNF20, and 3 distinct branches can be found during evolution. The fungi genomes only have a single copy of this E2 enzyme, while mammalian genomes encode 2 copies of this E2, UBE2A and UBE2B, suggesting a more complex functional requirement for this ubiquitin-conjugating enzyme. UBE2A is the isoform of UBE2B, they share 96% identity at protein level. Next we want to know whether their functional partners have similar evolutionary pattern. We constructed multiple species phylogenetic tree of WAC and Lge1 (), respectively. And we found that there were no WAC homologues in yeast, while no Lge1 homologues in animals. WAC has WW domain and coiled-coil domain. In mammalian cells, the coiled-coil domain of WAC interacts with the coiled-coil domain of RNF20/RNF40 complex. WAC stabilizes RNF20/RNF40 and facilitates the association between RNF20/RNF40 and hRAD6, thus promoting H2B ubiquitination. And WAC regulates gene transcription by promoting RNF20/RNF40 E3 ligase activity for H2B ubiquitination at active transcription sites.Citation18 Lge1 also have coiled-coil domain, it regulates transcriptional elongation by promoting H2B ubiquitination.Citation15 The sporulation efficiency of lge1 mutant strain is very low, which is similar to that of bre1 mutant strain.Citation16,21 So, although we don't know whether WAC is evolved from Lge1, both of them may play similar roles in H2B ubiquitination, in other words, they might be functional analogs to each other. Together, our phylogenetic analysis reveals that the molecular mechanisms underlying H2B ubiquitination is evolutionarily conserved.
Figure 1. Multiple species phylogenetic tree of RNF20. Phylogenetic tree of RNF20 proteins was constructed by MEGA 6.0 with the Neighbor-Joining (NJ) method. We performed 1000 bootstrap replicates. And I, II, III represent different groups, with I representing animals, II representing plants, III representing fungi.
Conserved functions of H2Bub during meiosis
Once histone H2B is modified by ubiquitin, nucleosome-nucleosome stacking interactions will be disrupted, the modification will lead to chromatin relaxation. Eventually, chromatin structures form an open and biochemically accessible conformation in which the transcriptional factors are easier to interact with promoter region.Citation22,23 H2Bub is often enriched in the promoter region of actively transcriptional genes in both yeast and mammalian somatic cells.Citation18,24-26 Previously, it was found that the knockout of Rad6 mammalian homolog, HR6B, results in male infertility due to defect in spermatogenesis. Detailed analysis of the knockout mice revealed that the synaptonemal complex and meiotic recombination were affected in the spermatocytes, thus strongly implicate the role of H2Bub in chromatin remodeling.Citation27 In addition, it has been reported that H2Bub is required for the recruitment of a DSB-creating complex at hotspots during meiosis in budding yeast.Citation28 Our previous study proved that the decreased level of H2Bub lead to more condensed chromatin structure in yeast. And it has been reported that Rad6-Bre1-mediated histone H2B ubiquitination modulates the formation of double-strand breaks during meiosis. So it is possible that condensed chromatin excludes the binding of DSB formation factors, thus reducing DSBs formation in yeast mutants. Recently, we confirmed that RNF20-mediated H2Bub might also be involved in programmed DSB formation in mouse, but its major function is to regulate meiotic recombination by promoting chromatin relaxation and facilitating the recruitment of some early DNA repair factors to the chromatin, and this mechanism is evolutionarily conserved from yeast to mammals.
Diverse physiologic functions of RNF20/Bre1 during meiosis
Although the molecular mechanism and functions of H2Bub are evolutionarily conserved from yeast to mammals, the physiologic functions of their E3 ligases are distinct to each other. In yeast, Bre1-mediated H2B ubiquitination regulates the formation of DSBs during meiosis. The DSBs reduced significantly in the bre1 mutant strain. The H2B K123R mutant strain mimics the phenotype of the bre1 mutant strain, and the sporulation efficiency seriously decreased in this mutant strain.Citation28,29 HUB1 and HUB2 are the yeast homologues of Bre1 in Arabidopsis thaliana. In Arabidopsis thaliana, histone H2B can be monoubiquitinated at lysine 143 (H2BK143) by the E2 enzymes UBC1, UBC2 and the E3 ligase HUB1, HUB2, which are the homologues of Rad6 and RNF20, respectively. HUB1/HUB2 regulates gene expression by interacting with the transcript elongation factor (FACT).Citation30 Deletion of FACT results in abnormal flower structure and severely reduced seed production.Citation30 In addition, HUB1/HUB2 mediated H2B monoubiquitination modulates flowing time,Citation30,31 and seed dormancy by modulating chromatin remodeling.Citation20 In Drosophila melanogaster, dBre1 regulates BMP signaling and the H3K4 trimethylation levels, which are involved in germline stem cell (GSC) maintenance, self-renewal and differentiation. Deletion of dBre1 decreased H3K4 trimethylation levels and lead to the loss of GSC.Citation32 Therefore, dBre1 play a key role in cell fate determination during fly oogenesis. All these results are not exactly match the functional role of RNF20 during mammalian meiosis, because germ cell-specific knockout Rnf20 in mice results in spermatocytes arresting at the pachytene stage due to over condensed chromatin structure.Citation10 The functional disparity of RNF20 family proteins maybe come from their structural divergence during evolution.
To understand the functional disparity of RNF20 family proteins during meiosis, we compared their domain architectures in several well-studied species by using SMART (http://smart.embl-heidelberg.de/). Except the conserved RING domain, all of them have coiled-coil motifs, but not any other motif/domain. With the evolution of the genome from yeast to mammals, the coiled-coil motifs of RNF20 family proteins turn to more and more complex ( right). The coiled-coil motifs of RNF20 family proteins are longer and more complicated in mouse and Drosophila, while they are relative shorter and simpler in A. thaliana and yeast. Usually, the coiled-coil motif is involved in protein-protein interaction,Citation33,34 the complication of the coiled-coil motifs indicates RNF20 family proteins have more partners/substrates. Indeed, some other new RNF20 substrates have been identified recently. For example, RNF20 is found to be co-localized and interacted with transcription factor activator protein 2a (AP-2a). And RNF20 promotes its polyubiquitination and subsequent degradation in ubiquitin–proteasome-dependent manner.Citation35 It also could promote tumor suppressor Ebp1 degradation through mediating its polyubiquitination.Citation36 In addition, RNF20 facilitates spindle assembly during mitosis by interacting with and ubiquitinating another protein, Eg5.Citation37 So the evolutionary divergency of RNF20's coiled-coil motifs might result in their diverse physiological functions during meiosis and other biological processes.
Conclusion and perspective
The molecular mechanism underlying H2Bub is evolutionarily conserved from yeast to mammals, and the functions of H2Bub during meiosis is also conserved, but the functional roles of the major E3 ligases for H2Bub are diverse from each other, most likely due to the divergence at their coiled-coil motifs during evolution. Except promoting chromatin relaxation, H2Bub also participates in DNA damage response, chromatin segregation, DNA replication, transcription, and RNA processing. Moreover, H2Bub has crosstalk with multiple epigenetic modifications. Bre1-mediated H2Bub is required for methylation of H3 at lysine 4 and lysine 79 in yeast.Citation38 RNF20/RNF40 mediated H2Bub also promotes the formation of H3K4me3, which regulates transcriptional elongation and increases elongation rate of tissue-specific genes.Citation39 Whether these functions are used in meiosis or not still need further investigation. On the other hand, histone is not the only substrate of RNF20. It could promote mono- or poly-ubiquitination of some proteins, thus changing their fates ultimately. In addition, RNF20 could selectively suppress pro-oncogenic gene expression by inhibiting transcription elongation factor TFIIS-mediated transcriptional elongation.Citation40,41 RNF20/RNF40 cooperates with the (FACT) component suppressor of Ty homolog-16 (SUPt16H) to induce chromatin structure changing, which contributes to DSB repair.Citation42 Whether these partners/substrates are involved in meiosis are still unknown, and the investigation on if there are meiotic specific RNF20 partners/substrates is still ongoing. Finally, some unique features have been evolved during mammalian cell meiosis, such as meiotic sex chromosome inactivation during spermatogenesis, germinal vesicle breakdown and meiosis resumption during oogenesis, whether Rnf20-mediated H2Bub or any other partners/substrates are involved in these special processes are open questions in the field. The elucidation of the above questions definitely will highlight the epigenetic regulation of meiosis and might provide new therapeutic target to treat some related infertility patients.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Nature Science of China (Grant No. 31471277 and 91649202) and National key R&D program of China (Grant No. 2016YFA0500901).
References
- Ohkura H. Meiosis: An Overview of Key Differences from Mitosis. Cold Spring Harbor Perspectives Biol 2015; 7:a015859; PMID:25605710; https://doi.org/10.1101/cshperspect.a015859
- Bomblies K, Higgins JD, Yant L. Meiosis evolves: adaptation to external and internal environments. New Phytologist 2015; 208:306-23; PMID:26075313; https://doi.org/10.1111/nph.13499
- Kato Y, Alavattam KG, Sin HS, Meetei AR, Pang QS, Andreassen PR, Namekawa SH. FANCB is essential in the male germline and regulates H3K9 methylation on the sex chromosomes during meiosis. Hum Mol Genetics 2015; 24:5234-49; PMID:26123487; https://doi.org/10.1093/hmg/ddv244
- Kagami A, Sakuno T, Yamagishi Y, Ishiguro T, Tsukahara T, Shirahige K, Tanaka K, Watanabe Y. Acetylation regulates monopolar attachment at multiple levels during meiosis I in fission yeast. EMBO Reports 2011; 12:1189-95; PMID:21979813; https://doi.org/10.1038/embor.2011.188
- Wang Q, Wei HJ, Du J, Cao Y, Zhang NN, Liu XY, Chen D, Ma W. H3 Thr3 phosphorylation is crucial for meiotic resumption and anaphase onset in oocyte meiosis. Cell Cycle 2016; 15:213-24; PMID:26636626; https://doi.org/10.1080/15384101.2015.1121330
- Vigodner M. Sumoylation precedes accumulation of phosphorylated H2AX on sex chromosomes during their meiotic inactivation. Chromosome Res 2009; 17:37-45; PMID:19156530; https://doi.org/10.1007/s10577-008-9006-x
- Ooga M, Suzuki MG, Aoki F. Involvement of histone H2B monoubiquitination in the regulation of mouse preimplantation development. J Reprod Develop 2015; 61:179-84; https://doi.org/10.1262/jrd.2014-137
- Luo MC, Zhou J, Leu NA, Abreu CM, Wang JL, Anguera MC, de Rooij DG, Jasin M, Wang PJ. Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during male Meiosis. PLoS Genetics 2015; 11:e1004954; PMID:25634095; https://doi.org/10.1371/journal.pgen.1004954
- Crichton JH, Playfoot CJ, Adams IR. The Role of chromatin modifications in progression through mouse Meiotic Prophase. J Genetics Genomics 2014; 41:97-106; PMID:24656230; https://doi.org/10.1016/j.jgg.2014.01.003
- Xu Z, Song Z, Li G, Tu H, Liu W, Liu Y, Wang P, Wang Y, Cui X, Liu C, et al. H2B ubiquitination regulates meiotic recombination by promoting chromatin relaxation. Nucleic Acids Res 2016; 44:9681-97; PMID:27431324; https://doi.org/10.1093/nar/gkw652
- Weake VM, Workman JL. Histone Ubiquitination: Triggering gene activity. Mol Cell 2008; 29:653-63; PMID:18374642; https://doi.org/10.1016/j.molcel.2008.02.014
- Sadowski M, Suryadinata R, Tan AR, Roesley SNA, Sarcevic B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life 2012; 64:136-42; PMID:22131221; https://doi.org/10.1002/iub.589
- Suresh B, Lee J, Hong S-H, Kim K-S, Ramakrishna S. The role of deubiquitinating enzymes in spermatogenesis. Cell Mol Life Sci 2015; 72:4711-20; PMID:26350476; https://doi.org/10.1007/s00018-015-2030-z
- Fuchs G, Oren M. Writing and reading H2B monoubiquitylation. Biochim Et Biophys Acta (BBA) - Gene Regulatory Mechan 2014; 1839:694-701; https://doi.org/10.1016/j.bbagrm.2014.01.002
- Song YH, Ahn SH. A Bre1-associated Protein, Large 1 (Lge1), promotes H2B ubiquitylation during the early stages of transcription elongation. J Biol Chem 2009; 285:2361-7; PMID:19923226; https://doi.org/10.1074/jbc.M109.039255
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 2003; 11:261-6; PMID:12535538; https://doi.org/10.1016/S1097-2765(02)00826-2
- Foglizzo M, Middleton AJ, Day CL. Structure and Function of the RING Domains of RNF20 and RNF40, Dimeric E3 Ligases that Monoubiquitylate Histone H2B. J Mol Biol 2016; 428:4073-86; PMID:27569044; https://doi.org/10.1016/j.jmb.2016.07.025
- Zhang F, Yu X. WAC, a functional partner of RNF20/40, regulates Histone H2B Ubiquitination and gene transcription. Mol Cell 2011; 41:384-97; PMID:21329877; https://doi.org/10.1016/j.molcel.2011.01.024
- Gu X, Jiang D, Wang Y, Bachmair A, He Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J 2009; 57:522-33; PMID:18980658; https://doi.org/10.1111/j.1365-313X.2008.03709.x
- Liu Y, Koornneef M, Soppe WJJ. The Absence of Histone H2B Monoubiquitination in the Arabidopsis hub1 (rdo4) Mutant Reveals a Role for Chromatin Remodeling in Seed Dormancy. Plant Cell 2007; 19:433-44; PMID:17329563; https://doi.org/10.1105/tpc.106.049221
- Jordan PW, Klein F, Leach DRF. Novel roles for selected genes in meiotic DNA processing. PLoS Genetics 2007; 3:2368-80; https://doi.org/10.1371/journal.pgen.0030222
- Peterson CL. Chromatin: A ubiquitin crowbar opens chromatin. Nat Chem Biol 2011; 7:68-9; PMID:21245855; https://doi.org/10.1038/nchembio.514
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol 2011; 7:113-9; PMID:21196936; https://doi.org/10.1038/nchembio.501
- Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev 2011; 25:2254-65; https://doi.org/10.1101/gad.177238.111
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B Monoubiquitination Functions Cooperatively with FACT to Regulate Elongation by RNA Polymerase II. Cell 2006; 125:703-17; PMID:16713563; https://doi.org/10.1016/j.cell.2006.04.029
- Jung I, Kim SK, Kim M, Han YM, Kim YS, Kim D, Lee D. H2B monoubiquitylation is a 5′-enriched active transcription mark and correlates with exon–intron structure in human cells. Genome Res 2012; 22:1026-35; PMID:22421545; https://doi.org/10.1101/gr.120634.111
- Roest HP, vanKlaveren J, deWit J, vanGurp CG, Koken MHM, Vermey M, van Roijen JH, Hoogerbrugge JW, Vreeburg JT, Baarends WM, et al. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 1996; 86:799-810; PMID:8797826; https://doi.org/10.1016/S0092-8674(00)80154-3
- Yamashita K, Shinohara M, Shinohara A. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc Natl Acad Sci 2004; 101:11380-5; https://doi.org/10.1073/pnas.0400078101
- Robzyk K, Recht L, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science 2000; 287:501-4; PMID:10642555; https://doi.org/10.1126/science.287.5452.501
- Cao Y, Dai Y, Cui S, Ma L. Histone H2B Monoubiquitination in the Chromatin of flowering locus C regulates flowering time in Arabidopsis. Plant Cell 2008; 20:2586-602; PMID:18849490; https://doi.org/10.1105/tpc.108.062760
- Schmitz RJ, Tamada Y, Doyle MR, Zhang X, Amasino RM. Histone H2B Deubiquitination is required for transcriptional activation of flowering locus C and for proper control of flowering in Arabidopsis. Plant Physiol 2008; 149:1196-204; PMID:19091875; https://doi.org/10.1104/pp.108.131508
- Xuan T, Xin T, He J, Tan J, Gao Y, Feng S, He L, Zhao G, Li M. dBre1/dSet1-dependent pathway for histone H3K4 trimethylation has essential roles in controlling germline stem cell maintenance and germ cell differentiation in the Drosophila ovary. Dev Biol 2013; 379:167-81; PMID:23624310; https://doi.org/10.1016/j.ydbio.2013.04.015
- Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 2001; 11:82-8; PMID:11166216; https://doi.org/10.1016/S0962-8924(00)01898-5
- Nie WC, He F, Yuan SM, Jia ZW, Wang RR, Gao XD. Roles of an N-terminal coiled-coil-containing domain in the localization and function of Bem3, a Rho GTPase-activating protein in budding yeast. Fungal Genetics Biol 2017; 99:40-51; PMID:28064039; https://doi.org/10.1016/j.fgb.2016.12.010
- Ren P, Sheng Z, Wang Y, Yi X, Zhou Q, Zhou J, Xiang S, Hu X, Zhang J. RNF20 promotes the polyubiquitination and proteasome-dependent degradation of AP-2 protein. Acta Biochim Et Biophys Sinica 2013; 46:136-40; PMID:24374663; https://doi.org/10.1093/abbs/gmt136
- Liu Z, Oh SM, Okada M, Liu X, Cheng D, Peng J, Brat DJ, Sun SY, Zhou W, Gu W, et al. Human BRE1 Is an E3 Ubiquitin Ligase for Ebp1 Tumor suppressor. Mol Biol Cell 2008; 20:757-68; PMID:19037095; https://doi.org/10.1091/mbc.E08-09-0983
- Duan Y, Huo DW, Gao J, Wu H, Ye Z, Liu Z, Zhang K, Shan L, Zhou X, Wang Y, et al. Ubiquitin ligase RNF20/40 facilitates spindle assembly and promotes breast carcinogenesis through stabilizing motor protein Eg5. Nat Commun 2016; 7:12648; PMID:27557628; https://doi.org/10.1038/ncomms12648 10.1038/ncomms13462
- Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of Histone H2B by Rad6 is required for efficient Dot1-mediated Methylation of Histone H3 Lysine 79. J Biol Chem 2002; 277:34655-7; PMID:12167634; https://doi.org/10.1074/jbc.C200433200
- Xie W, Nagarajan S, Baumgart SJ, Kosinsky RL, Najafova Z, Kari V, Hennion M, Indenbirken D, Bonn S, Grundhoff A, et al. RNF40 regulates gene expression in an epigenetic context-dependent manner. Genome Biol 2017; 18:32; PMID:28209164; https://doi.org/10.1186/s13059-017-1159-5
- Jääskeläinen T, Makkonen H, Visakorpi T, Kim J, Roeder RG, Palvimo JJ. Histone H2B ubiquitin ligases RNF20 and RNF40 in androgen signaling and prostate cancer cell growth. Mol Cell Endocrinol 2012; 350:87-98; PMID:22155569; https://doi.org/10.1016/j.mce.2011.11.025
- Shema E, Kim J, Roeder Robert G, Oren M. RNF20 inhibits TFIIS-Facilitated transcriptional elongation to suppress Pro-oncogenic gene expression. Mol Cell 2011; 42:477-88; PMID:21596312; https://doi.org/10.1016/j.molcel.2011.03.011
- Kari V, Shchebet A, Neumann H, Johnsen SA. The H2B ubiquitin ligase RNF40 cooperates with SUPT16H to induce dynamic changes in chromatin structure during DNA double-strand break repair. Cell Cycle 2014; 10:3495-504; https://doi.org/10.4161/cc.10.20.17769