ABSTRACT
Eukaryotic nuclei are essential organelles, storing the majority of the cellular DNA, comprising the site of most DNA and RNA synthesis, controlling gene expression and therefore regulating cellular function. The majority of mammalian cells retain their nucleus throughout their lifetime, however, in three mammalian tissues the nucleus is entirely removed and its removal is essential for cell function. Lens fibre cells, erythroblasts and epidermal keratinocytes all lose their nucleus in the terminal differentiation pathways of these cell types. However, relatively little is known about the pathways that lead to complete nuclear removal and about how these pathways are regulated. In this review, we aim to discuss the current understanding of nuclear removal mechanisms in these three cell types and expand upon how recent studies into nuclear degradation in keratinocytes, an easily accessible experimental model, could contribute to a wider understanding of these molecular mechanisms in both health and pathology.
Nuclei are the major membrane-bound organelles of eukaryotic cells and are essential for cellular function, storing the cellular DNA, acting as the main sites of DNA and RNA synthesis, regulating gene expression and therefore cellular function [Citation1]. However, in some mammalian cell types, programmed removal of the entire nuclear structure is essential for cellular function: lens fibre cells remove the nucleus and other organelles to produce the transparent lens structure, erythroblasts extrude the nucleus to form erythrocytes which can fit through capillary trees and in the skin keratinocytes terminally differentiate into enucleate cells devoid of all intracellular organelles to form the tough cornified layer, an essential component of the epidermal water barrier [Citation2–Citation5].
Yeast cells and some mammalian cells are known to undergo partial removal of nuclear material, by targeted autophagy of the nucleus or ‘nucleophagy’; micronuclei detach from the nucleus and fuse with LC3-positive autophagosomes, or autophagosomes can form directly at the nuclear envelope [Citation6,Citation7]. Lens fibre cells, erythroblasts and keratinocytes in mammals undergo programmed removal of their entire nucleus in the eye, bone marrow and epidermis respectively (). The mammalian nucleophagic mechanisms have until recently been relatively unclear and whether these processes are involved, perhaps with several other mechanisms, for complete nuclear loss remains to be characterised [Citation8]. These three cell types are the only cells in mammalian tissues known to entirely remove their nucleus under normal physiological conditions, yet, little is known about nuclear removal in these cell types, the regulation of these pathways and whether they share common features. In this review, we aim to discuss what is known about nuclear removal in the eye, bone marrow and skin and consider areas which await definition.
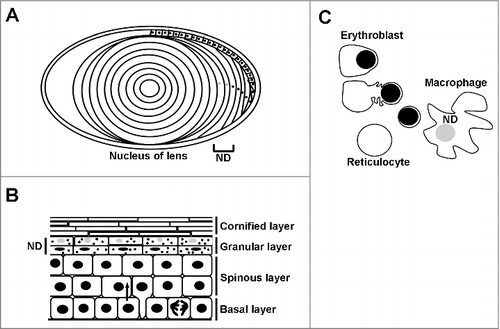
Figure 1. Nuclear degradation occurs during normal homeostasis. Degradation of the nucleus is a part of normal cellular homeostasis in three tissues. Grey nuclei and ND denotes where nuclear degradation occurs in each tissue. A) During the development of the lens, the lens epithelial cells migrate along the lens periphery before flattening out and synthesising crystallins. The middle portion, or nucleus of the lens is devoid of both organelles and the nucleus. B) Keratinocytes proliferate in the basal layer of the epidermis prior to terminal differentiation, where cells come off of the basal lamina and express different structural keratins forming the spinous layer. The nucleus is degraded in the upper layers of the epidermis called the granular layer, prior to the synthesis of the enucleate cornified layer which confers the majority of epidermal barrier function. C) Erythroblasts (red blood cell precursors) are formed by a process of nuclear condensation and extrusion, forming a body called a pyrenocyte, which is engulfed and degraded by adjacent macrophages.
Lens fibre cell nuclear removal
In the eye, lens formation requires the differentiation of lens fibre cells from epithelial cells on the outside of the lens with a complete complement of intracellular organelles into cells in the middle of the lens that are transparent, devoid of intracellular organelles and mainly filled with proteins known as ‘crystallins’ [Citation3]. The process seems to vary between different eukaryotes but involves rounding of the nucleus, formation of a smaller pyknotic nucleus before DNA degradation and nuclear breakdown, ‘karyolysis’ with release of DNA into the cytoplasm [Citation3,Citation9,Citation10].
Together with these architectural changes of the nucleus, indentations in the nuclear shape and irregularities in DNA staining have also been observed [Citation11]. The distribution of sub-nuclear structures including nucleoli and Cajal bodies alters, the nuclear lamina is degraded and karyolysis can be observed due to the presence of DNA in the cytoplasm [Citation3,Citation12,Citation13]. Costello et al. observed that close to indentations in nuclear structures in the chick embryo were complex macromolecular aggregates including membranous structures [Citation11]. They termed these structures ‘excisosomes’ which appear to be important for degradation of the nuclear envelope, and have reported preliminary results that they are also present in developing primate lenses [Citation11,Citation14].
An important stage in the process of nuclear removal is DNA degradation. This step occurs in the nucleus of developing lens fibre cells, as illustrated by the presence of TUNEL staining, which recognises free 3’-OH ends of DNA [Citation12,Citation13]. Expression of the DNA degrading enzyme, DNaseIIβ is upregulated in mouse lens fibre cell differentiation and mice deficient for DNaseIIβ develop cataracts and have DNA present in the mature lens, indicative of incomplete nuclear removal [Citation13,Citation15,Citation16]. Another DNA degrading enzyme may also be required for this process as in DNaseIIβ deficient mouse lenses, fragmentation and clumping of DNA is still observed, suggesting some DNA reorganisation and degradation may be occurring [Citation15].
DNaseIIβ has been localised to lysosomes closely associated with the nucleus and suggested to be delivered to nuclear material by fusion of lysosomes with the nucleus [Citation13,Citation15]. However, it has been suggested that the autophagy and apoptosis pathways of eukaryotic cells are not co-opted to perform nuclear removal. Nuclear removal was not affected by knockout of the apoptotic caspase-3, caspase-6 or caspase-7 enzymes, or a double knockout of caspase-3 and caspase-6 [Citation17]. No autophagosomes were observed close to the degrading nucleus in chick lenses and ATG5 has also been shown to be dispensable for nuclear removal [Citation11,Citation18]. However, ATG5 independent autophagy pathways have been reported and although lysosomes were also not observed close to the nucleus in the chick lenses cells, this has been reported in mouse lenses [Citation11,Citation19,Citation20]. The ubiquitin proteasome pathway has also been identified in the nucleus of developing lens cells where it may account for degradation of the nucleoplasm [Citation21].
The variety of proteins identified as important for nuclear removal in the lens may indicate the variety of pathways required to regulate a process that should only be activated in this specific differentiation process. Regulation of lens nuclear removal has been shown to require both the suppression of mTORC1 signalling to induce the expression of autophagy related proteins such as ULK1 and LC3 and the activation of CDK1; without CDK1 signalling phosphorylation of nuclear lamina proteins lamin A/C was decreased and nuclear degradation was affected [Citation20,Citation22]. Additionally, there are some clues from defects in transcriptional regulators, such as GATA-3, HSF4 and BRG1, with defects in these regulators leading to defective lens nuclear removal and defects such as cataracts [Citation23–Citation25]. However, other components of this regulatory pathway and how this process is initiated is currently unclear and may also involve calcium signalling, as the cytoplasmic calcium ion concentration increases in lens fibre cell differentiation [Citation20,Citation22,Citation26].
Erythroblast nuclear removal
In the bone marrow, erythropoiesis involves the differentiation of hematopoietic stem cells through several erythroid progenitor cells to mature erythrocytes [Citation27]. Prior to the formation of mature erythrocytes, erythroblasts extrude their nuclei through a protrusion of plasma membrane which is pinched off, forming an enucleate reticulocyte and a ‘pyrenocyte’, containing the condensed nucleus surrounded by a thin layer of cytoplasm [Citation2]. The reticulocyte forms the mature erythrocyte and the pyrenocyte is engulfed by macrophages of the bone marrow and degraded by fusion with lysosomes [Citation28,Citation29].
Erythrocyte enucleation occurs throughout mammalian life span at a rate of approximately 2.5 million times per second, however relatively little is known about how this process occurs and is regulated [Citation30]. In the space of ten minutes, chromosomes inside the nucleus condense, with loss of discernible nucleoli structures and the nucleus decreases in size and becomes rounder [Citation31–Citation33]. DNA condensation through histone deacetylation by HDAC2 has been implicated in enucleation, and nuclear condensation has been suggested to occur through the leakage of DNA into the cytoplasm, through caspase-3 dependent nuclear openings, and through E2F-dependent transcriptional regulation of Citron Rho-interacting 60 kinase [Citation34–Citation37]. The condensed nucleus is then expelled from the erythrocyte, through the activity of an actin rich structure known as the ‘enucleosome’ behind the nucleus [Citation38]. The mechanism for the final abscission of the pyrenocyte involves intracellular vesicle fusion and potentially formation of a cleavage actomyosin ring [Citation2,Citation39].
DNA degradation is also required in erythropoiesis, however, as nuclear breakdown occurs in the macrophages, after engulfment of the pyrenocyte, DNaseIIα expression is essential in macrophages, not in the enucleating erythroblasts [Citation28].
Deficiency of caspase-3, an apoptotic enzyme, in mice did not lead to erythropoietic effects and pan caspase inhibitors did not affect enucleation [Citation28]. Additionally, the autophagy protein ATG5 was not required for nuclear removal [Citation18]. Suggesting that mechanisms of extrusion and nuclear breakdown are not linked to the cellular processes of apoptosis or autophagy. However, caspase-3 is required for transient nuclear openings that occur prior to nuclear extrusion and ATG5-independent autophagy pathways have been reported in mammalian cells [Citation19,Citation34]. This may indicate the complexity of the mechanisms controlling this pathway, and several mechanisms have been proposed for the scission of the pyrenocyte [Citation2,Citation39]. Indeed the regulation of these pathways and more precisely the initial mechanism that triggers nuclear removal remains unclear, although calcium signalling has been implicated; uptake of extracellular calcium causes a burst of increased intracellular calcium concentration 10 min prior to enucleation, which is required for efficient enucleation [Citation40].
Epidermal keratinocyte nuclear removal
In the epidermis, keratinocytes terminally differentiate throughout life from proliferating keratinocytes in the basal layer into spinous, granular and then cornified layer keratinocytes, or corneocytes, in the uppermost layer [Citation4]. In the process of differentiation from granular keratinocytes into corneocytes, granular cells remove all their organelles, including the nucleus, allowing them to contain a high proportion of keratin and form a rigid cell layer that is essential for formation of the epidermal water barrier [Citation4].
The nucleus is removed relatively rapidly from the uppermost granular cell layer, taking at most six hours [Citation4,Citation41]. However, although this process occurs throughout the epidermis, throughout an organism's lifetime, the mechanism by which granular keratinocytes remove their nucleus is as yet incompletely understood.
Before removal the nucleus undergoes significant morphological changes: between the basal layer and the granular layers the keratinocyte nucleus decreases in volume, becomes more elongated, rotates to become more aligned to the basement membrane and develops indentations in its structure [Citation42,Citation43]. The morphology and organisation of sub-nuclear structures also alters; decreased numbers of larger nucleoli move closer to the centre of the nucleus and the arrangement of heterochromatic structures also changes [Citation42]. However, architecture modifications beyond the granular layer have not been characterised in these studies, and indeed, transitional stages of the nuclear breakdown have yet to be characterised, perhaps due to the rapid nature of the breakdown [Citation4].
Several mechanisms have been shown to be required for keratinocyte nuclear removal, including expression of DNA-degrading enzymes, targeted degradation of nuclear lamina proteins and degradation of parts of the nucleus through nucleophagy and, accordingly different regulatory pathways have been proposed [Citation43–Citation46].
Without the expression of DNA degrading enzymes, principally DNase1L2 and the primarily lysosomal DNAse, DNase2, nuclei are retained in the cornified layer, a process known as parakeratosis [Citation44]. However, unlike in lens fibre cells, the lack of TUNEL staining suggests free 3’-OH ends of DNA are not present in this degradation, which may indicate differences in the DNA degradation mechanisms between keratinocytes and lens fibre cells [Citation12,Citation43]. In addition to DNase1L2 and DNase2 a further DNA degrading enzyme may also be required; retained nuclei of DNase1L2 and DNase2 double knockout mice were TUNEL-positive, indicating some DNA degradation is occurring [Citation44,Citation47]. This may be mediated by TREX2, an exonuclease upregulated during keratinocyte terminal differentiation, whose expression has been reported to increase in psoriatic lesion and to be essential for nuclear degradation in lingual keratinocytes [Citation47,Citation48]. In mouse cells it was recently shown how lack of DNase2 not only would lead to nuclear material intracellular accumulation but also deregulation of the autophagy degrading machinery. This further confirms that signalling pathways deriving from the nucleus can either sense DNA damage or DNA re-arrangement and trigger autophagy [Citation49].
How the DNA is accessed by these enzymes is not yet clear. The DNases would require delivery to the nucleus, and indeed filaggrin fragments have been reported in the nucleus, indicating a mechanism of protein transport into the nucleus which may not normally occur [Citation46]. Additionally, nuclear lamina degradation has been suggested to occur prior to DNase-dependent degradation; in DNase1L2 and DNase2 knockout mice lamin A/C degradation occurs without complete nuclear removal [Citation44].
Lamins are intermediate filaments, organised into the nuclear lamina beneath the nuclear envelope, important for nuclear structure and organisation of nucleus. Although loss of lamins B1 and B2, does not affect skin development, degradation of lamin A/C is required for nuclear removal [Citation45,Citation50]. AKT1 dependent phosphorylation of lamin A/C was reduced in terminally differentiating AKT1 deficient keratinocytes, with decreased lamin degradation and retention of nuclear material in the cornified layers, indicating targeted breakdown of the nuclear lamina is required for nuclear removal [Citation45].
The rest of the nucleoplasm and the nuclear envelope also requires degradation and removal of these structures and parts of DNA has been hypothesized to be, at least in part, via nuclear targeted autophagy [Citation43]. However, whether canonical autophagy is important for keratinocyte nuclear removal is unclear; ATG5 and ATG7 are dispensable for epidermal nuclear removal [Citation51,Citation52]. However, ATG5/ATG7 independent autophagy pathways have been reported in mammalian cells and may be important in keratinocyte nucleophagy [Citation19]. Additionally, expression of some autophagy proteins is upregulated in keratinocyte differentiation and loss of autophagy proteins WIPI1 or ULK1 prevents nuclear removal [Citation43]. Few autophagy markers have been shown to have a nuclear localization. An elegant study has reported how nuclear LC3, which is mainly in the LC3-II form during starvation, is relocated into the cytoplasm [Citation53], and more recently nuclear LC3-II and phosphorylated Ulk1 were shown to interact with γ-H2AX, Rad51 or PARP-1, involved in maintenance of genomic stability [Citation54]. Likewise p62 has been shown to regulate chromatin ubiquitination during DNA damage response [Citation55]. In differentiating keratinocytes, LC3 co-localises close to the nucleus with a histone binding protein, HP1α, suggesting autophagosomal breakdown of nuclear contents [Citation43]. Interestingly, in differentiating keratinocytes LC3 can also interact with lamin B1, which accumulates in proximity of the perinuclear region where LC3/p62 double-positive aggregates where identified, suggesting nuclear targeted autophagy may also be important for nuclear lamina breakdown [Citation43].
However, this process has only been documenting early stages of nuclear removal and there may be other mechanisms essential for complete degradation of the nucleus [Citation8].
In nuclear envelopathies, diseases with defects in lamin genes, and mice with mutations in the gene encoding lamin A/C partial degradation of the nucleus occurs [Citation56]. Vesicular structures were observed perinuclearly, and in mice with a lamin A/C mutation these structures were identified as perinuclear autophagosomes and lysosomes and contained nuclear material, indicating alterations to the structure of the nuclear lamina is required for nuclear degradation [Citation56].
Similarly, to erythroblast and lens fibre cell nuclear removal, the apoptotic machinery is not implicated in keratinocyte nuclear removal [Citation17,Citation28]. Caspase-3 is not activated upon differentiation and the protein iASPP prevents activation of apoptotic pathways in differentiating keratinocytes [Citation43,Citation57].
Again, how this programmed removal of the nucleus is activated and regulated is incomplete. AKT1 and mTORC1 are required for regulation of nuclear removal and are both involved in growth, survival and differentiation signalling pathways, however, not much is known about how these proteins are activated and controlled in the specific case of nuclear removal [Citation43,Citation45,Citation58]. Calcium has again been postulated as a possible regulating factor, although its role in nuclear removal has not been studied [Citation4].
Mammalian nuclear removal – Commonalities and differences
Erythroblasts, lens fibre cells and keratinocytes all undergo rapid nuclear removal as part of their highly regulated terminal differentiation programs. All three processes involve condensation of nuclear DNA, reductions in nuclear volume, changes to nuclear morphology and requirement of DNA degrading enzymes (). However, current knowledge suggests they have evolved distinct processes for complete removal of the nucleus, the key processes understood to be important in the nuclear removal of these three tissues are summarised in . Erythroblasts expel a condensed nucleus from the cell, whereas, in lens fibre cells and keratinocytes the nucleus is broken down whilst still contained within the differentiating cell [Citation2–Citation4].
Table 1. Commonalities and differences in the key processes of mammalian nuclear removal; Comparison of known nuclear degradation processes and signalling pathways activated in keratinocytes, lens fibre cells and erythroblasts. A tick denotes that process or phenomenon is active in that cell type, a cross denotes that it is not, and – not determined in that cell type.
In both lens fibre cells and keratinocytes, the appearance of nuclear indentations increases with differentiation and macromolecular and membrane bound aggregates closely associated with the nuclear membrane are reported in these indentations [Citation11,Citation14,Citation20,Citation43]. Although autophagy is not activated in a ‘classical’ manner in these cells, targeted autophagy of the nucleus, ‘nucleophagy’, may occur [Citation43]. In keratinocytes lysosomal and autophagosomal proteins localised close to the nuclear membrane, co-staining with DNA binding proteins and vesicles of lysosomal appearance were visualised close to the nucleus in murine lens fibre cells. However, macromolecular aggregates termed the excisosome have also been observed at this location without vesicles of lysosomal appearance in chick and preliminary experiments in primate lenses [Citation11,Citation14,Citation20,Citation43]. Whether the excisosome and the autophagic bodies seen in proximity to the nucleus in terminally differentiating keratinocytes are analogous or even identical structures is open to debate, however the removal of portions of nuclear materials concomitant with lamin degradation appear to be common between these two tissues.
The DNA degrading enzymes required for DNA breakdown do also differ. DNaseIIβ is necessary for lens fibre cell nuclear degradation, DNase1L2 and DNase2 are required for keratinocyte nuclear breakdown and DNaseIIα is required for pyrenocyte degradation by macrophages [Citation13,Citation15,Citation44]. How DNases access the nucleus from lysosomes in lens fibre cells and keratinocytes is not yet clear, nucleus-lysosome fusion has been suggested, although this process has not be observed and DNA staining is not clearly visible in the lysosomal structures [Citation13,Citation16,Citation43].
Implications and outlook – Piecing together the nuclear degradation process
How entire nuclei are removed from mammalian cells has been a long-standing question, and we are beginning to characterise the processes that regulate controlled nuclear removal. There appear to be several varied mechanisms that regulate these events, intracellularly in lens fibre cells and keratinocytes and by extrusion in erythroblasts () [Citation2–Citation4]. There may also be additional mechanisms for the removal of other cellular organelles in the differentiation of these cell types. In keratinocytes, increased numbers of lysosomes concomitant with the removal of organelles such as mitochondria and the Golgi and the requirement for autophagy in keratinocyte differentiation suggests autophagy-dependent removal [Citation41,Citation59,Citation4,Citation43]. Nucleophagy in keratinocytes could be linked to this ‘macro-autophagy’ of other organelles, but this remains to be established [Citation43]. However, in lens fibre cells degradation of the nucleus can be inhibited without affecting other organelles and in erythrocytes autophagic pathways have been shown to clear mitochondria in a separate pathway to the expulsion of the nucleus suggesting that nuclear removal is likely to be a distinct pathway to organelle degradation [Citation60,Citation61,Citation15].
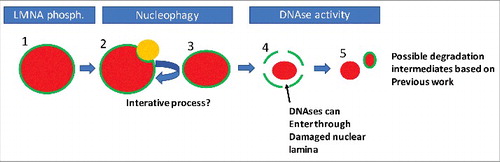
Whether the initial pathways of nuclear remodelling, and subsequent breakdown of the nuclear envelope and degradation of nuclear DNA in lens fibre cells and keratinocytes, are common to these cell types has yet to be determined, however based on our experimental findings and the work of other groups we could propose the following order of known processes of nuclear degradation in keratinocytes. Firstly, AKT1 dependent phosphorylation of LMNA occurs (, Step 1). We hypothesise that this marks a region that is targeted for nucleophagy. Also, DNase2 may act during this part of the process if it is present in the autophagolysosome (, Steps 2 and 3). This process is iterative, but a point is reached where integrity of the nuclear lamina cannot be maintained (, Step 4). At this point various DNases can enter the damaged nucleus to degrade the DNA. What is not clear is whether the remainder of the nuclear lamina is degraded prior, during or after this process.
Figure 2. A possible order of events in nuclear degradation in keratinocytes. Possible stages of nuclear degradation based on our and other's data. To begin, the nucleus is intact but is marked by phosphorylation of Lamin A/C (1). This targets an autophagolysosome (LC3-positive/LAMP2-positive body, orange) to that region of the nuclear lamina (2). The autophagolysosome removes some of the nuclear content, reducing nuclear size (3). Steps 1–3 are repeated iteratively until the nuclear lamina is sufficiently damaged to allow ingress of DNases. Then large scale degradation of the nuclear material occurs, potentially concomitant with further degradation of the nuclear lamina (5). Red colour denotes nuclear material, while green denotes the nuclear lamina.
The later stages of nuclear removal in lens fibre cells and keratinocytes, beyond remodelling of nuclear structure and initial association with lysosomes or other macromolecular aggregates, remain to be characterised in both cell types. Erythroblast nuclear removal has been characterised with a variety of methods including microarray analysis of gene expression, flow cytometry analysis of morphology with pharmacological treatments and fluorescently labelled nuclear components [Citation39,Citation62–Citation64]. Lens fibre cell differentiation in vitro is complex and does not fully recapitulate the formation of a lens, however, well established assays have been determined for keratinocyte differentiation in culture, and nuclear removal could perhaps be followed in these cells using the aforementioned tools [Citation20].
Future directions
Nuclear removal, particularly in lens fibre cell and keratinocyte differentiation, is a complex process which is as yet incompletely understood. However, some key questions that arise from studies of nuclear removal in these cell types and erythroblasts include:
Do lens fibre cells and keratinocytes undergo cycles of nuclear opening, is this controlled by Lamin degradation?
How do lysosomal DNases get delivered to the nucleus? And how do filaggrin fragments access keratinocyte nuclei?
The organisation of the nuclear lamina can affect heterochromatin organisation – does nuclear remodelling alter DNA structure in a targeted way to alter gene expression and how long during the process can transcription occur?
Abbreviations
| DNA | = | deoxyribonucleic acid |
| ND | = | nuclear degradation |
| RNA | = | ribonucleic acid |
| TUNEL | = | terminal deoxynucleotidyl transferase dUTP (deoxyuridine triphosphate) nick end labelling. |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. Garland Science: 2002.
- Keerthivasan G, Wickrema A, Crispino JD. Erythroblast Enucleation. SAGE-Hindawi Access to Res Stem Cells Int. 2011;2011:139851:
- Bassnett S.. On the mechanism of organelle degradation in the vertebrate lens. Exp Eye Res. 2009;88:133–9. doi:10.1016/j.exer.2008.08.017.
- Eckhart L, Lippens S, Tschachler E, et al. Cell death by cornification. Biochim Biophys Acta – Mol Cell Res. 2013;1833:3471–80. doi:10.1016/j.bbamcr.2013.06.010.
- Ji P. New Insights into the Mechanisms of Mammalian Erythroid Chromatin Condensation and Enucleation. Int Rev Cell Mol Biol. 2015;316:159–82. doi:10.1016/bs.ircmb.2015.01.006.
- Mijaljica D, Devenish RJ. Nucleophagy at a glance. J Cell Sci. 2013;126:4325–30. doi:10.1242/jcs.133090.
- Chen K, Huang C, Yuan J, et al. Long-term artificial selection reveals a role of TCTP in autophagy in mammalian cells. Mol Biol Evol. 2014;31:2194–211. doi:10.1093/molbev/msu181.
- Peng H, Lavker RM. Nucleophagy: A New Look at Past Observations. J Invest Dermatol. 2016;136:1316–8. doi:10.1016/j.jid.2016.04.019.
- Modak SP, Perdue SW. Terminal lens cell differentiation I. Histological and microspectrophotometric analysis of nuclear Degeneration. Exp Cell Res. 1970;59:43–56. doi:10.1016/0014-4827(70)90622-1.
- Vrensen GF, Graw J, De Wolf A. Nuclear breakdown during terminal differentiation of primary lens fibres in mice: a transmission electron microscopic study. Exp Eye Res. 1991;52:647–59. doi:10.1016/0014-4835(91)90017-9.
- Costello MJ, Brennan LA, Mohamed A, et al. Identification and ultrastructural characterization of a novel nuclear degradation complex in differentiating Lens Fiber cells. PLoS One. 2016;11:e0160785. doi:10.1371/journal.pone.0160785.
- Dahm R, Gribbon C, Quinlan RA, et al. Changes in the nucleolar and coiled body compartments precede lamina and chromatin reorganization during fibre cell denucleation in the bovine lens. Eur J Cell Biol. 1998;75:237–46. doi:10.1016/S0171-9335(98)80118-0.
- De Maria A, Bassnett S. DNase IIβ distribution and activity in the mouse lens. Investig Opthalmology Vis Sci. 2007;48:5638. doi:10.1167/iovs.07-0782.
- Costello MJ, Mohamed A, Gilliland K, et al. High resolution confocal microscopy of potential newly described nuclear excisosomes in primate lenses. Invest Ophthalmol Vis Sci. 2017;58:1213.
- Nishimoto S, Kawane K, Watanabe-Fukunaga R, et al. et al. Nuclear cataract caused by a lack of DNA degradation in the mouse eye lens. Nature. 2003;424:1071–4. doi:10.1038/nature01895.
- Nakahara M, Nagasaka A, Koike M, et al. Degradation of nuclear DNA by DNase II-like acid DNase in cortical fiber cells of mouse eye lens. FEBS J. 2007;274:3055–64. doi:10.1111/j.1742-4658.2007.05836.x.
- Zandy AJ, Lakhani S, Zheng T, et al. Role of the executioner caspases during lens development. J Biol Chem. 2005;280:30263–72. doi:10.1074/jbc.M504007200.
- Matsui M, Yamamoto A, Kuma A, et al. Organelle degradation during the lens and erythroid differentiation is independent of autophagy. Biochem Biophys Res Commun. 2006;339:485–9. doi:10.1016/j.bbrc.2005.11.044.
- Nishida Y, Arakawa S, Fujitani K, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. doi:10.1038/nature08455.
- Rowan S, Chang ML, Reznikov N, et al. Disassembly of the lens fiber cell nucleus to create a clear lens: The p27 descent. Exp Eye Res. 2017;156:72–8. doi:10.1016/j.exer.2016.02.011.
- Girão H, Pereira P, Taylor A, et al. Subcellular redistribution of components of the ubiquitin-proteasome pathway during lens differentiation and maturation. Investig Ophthalmol Vis Sci. 2005;46:1386–92. doi:10.1167/iovs.04-0563.
- Basu S, Rajakaruna S, Reyes B, et al. Suppression of MAPK/JNK-MTORC1 signaling leads to premature loss of organelles and nuclei by autophagy during terminal differentiation of lens fiber cells. Autophagy. 2014;10:1193–211. doi:10.4161/auto.28768.
- Maeda A, Moriguchi T, Hamada M, et al. Transcription factor GATA-3 is essential for lens development. Dev Dyn. 2009;238:2280–91. doi:10.1002/dvdy.22035.
- Cui X, Wang L, Zhang J, et al. et al. HSF4 regulates DLAD expression and promotes lens de-nucleation. Biochim Biophys Acta – Mol Basis Dis. 2013;1832:1167–72. doi:10.1016/j.bbadis.2013.03.007.
- He S, Pirity MK, Wang W-L, et al. et al. Chromatin remodeling enzyme Brg1 is required for mouse lens fiber cell terminal differentiation and its denucleation. Epigenetics Chromatin. 2010;3:21. doi:10.1186/1756-8935-3-21.
- Gao J, Sun X, Martinez-Wittinghan FJ, et al. Connections between connexins, calcium, and cataracts in the lens. J Gen Physiol. 2004;124:289–300. doi:10.1085/jgp.200409121.
- Nandakumar SK, Ulirsch JC, Sankaran VG. Advances in understanding erythropoiesis: Evolving perspectives. Br J Haematol. 2016;173:206–18. doi:10.1111/bjh.13938.
- Yoshida H, Kawane K, Koike M, et al. Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature. 2005;437:754–8. doi:10.1038/nature03964.
- Toda S, Nishi C, Yanagihashi Y, et al. Clearance of apoptotic cells and pyrenocytes. Curr Top Dev Biol. 2015;114:267–95. doi:10.1016/bs.ctdb.2015.07.017.
- Palis J.. Primitive and definitive erythropoiesis in mammals. Front Physiol. 2014;5:3. doi:10.3389/fphys.2014.00003.
- Muir AR, Kerr DNS. Erythropoiesis: an electron microscopical study. Q J Exp Physiol Cogn Med Sci. 1958;43:106–14.
- McGrath KE, Catherman SC, Palis J. Delineating stages of erythropoiesis using imaging flow cytometry. Methods. 2017;112:68–74. doi:10.1016/j.ymeth.2016.08.012.
- Krauss SW, Lo AJ, Short SA, et al. Nuclear substructure reorganization during late-stage erythropoiesis is selective and does not involve caspase cleavage of major nuclear substructural proteins. Blood. 2005;106:2200–5. doi:10.1182/blood-2005-04-1357.
- Zhao B, Mei Y, Schipma MJ, et al. Nuclear condensation during mouse erythropoiesis requires caspase-3-mediated nuclear opening. Dev Cell. 2016;36:498–510. doi:10.1016/j.devcel.2016.02.001.
- Simpson CF, Kling JM. The mechanism of denucleation in circulating erythroblasts. J Cell Biol. 1967;35:237–45. doi:10.1083/jcb.35.1.237.
- Ji P, Yeh V, Ramirez T, et al. Histone deacetylase 2 is required for chromatin condensation and subsequent enucleation of cultured mouse fetal erythroblasts. Haematologica. 2010;95:2013–21. doi:10.3324/haematol.2010.029827.
- Swartz KL, Wood SN, Murthy T, et al. E2F-2 promotes nuclear condensation and enucleation of terminally differentiated erythroblasts. Mol Cell Biol. 2017;37:e00274–16. doi:10.1128/MCB.00274-16.
- Nowak RB, Papoin J, Gokhin DS, et al. Tropomodulin 1 controls erythroblast enucleation via regulation of F-actin in the enucleosome. Blood. 2017;130:1144–55. doi:10.1182/blood-2017-05-787051.
- Konstantinidis DG, Pushkaran S, Johnson JF, et al. Signaling and cytoskeletal requirements in erythroblast enucleation. Blood. 2012;119:6118–27. doi:10.1182/blood-2011-09-379263.
- Wölwer CB, Pase LB, Russell SM, et al. Calcium signaling is required for erythroid enucleation. PLoS One. 2016;11:1–12. doi:10.1371/journal.pone.0146201.
- Lavker RM, Matoltsy AG. Formation of horny cells: the fate of cell organelles and differentiation products in ruminal epithelium. J Cell Biol. 1970;44:501–12. doi:10.1083/jcb.44.3.501.
- Gdula MR, Poterlowicz K, Mardaryev AN, et al. Remodeling of three-dimensional organization of the nucleus during terminal keratinocyte differentiation in the epidermis. J Invest Dermatol. 2013;133:2191–201. doi:10.1038/jid.2013.66.
- Akinduro O, Sully K, Patel A, et al. et al. Constitutive autophagy and nucleophagy during epidermal differentiation. J Invest Dermatol. 2016;136:1460–70. doi:10.1016/j.jid.2016.03.016.
- Fischer H, Buchberger M, Napirei M, et al. Inactivation of DNase1L2 and DNase2 in keratinocytes suppresses DNA degradation during epidermal cornification and results in constitutive parakeratosis. Sci Rep. 2017;7:6433. doi:10.1038/s41598-017-06652-8.
- Naeem AS, Zhu Y, Di WL, et al. AKT1-mediated Lamin A/C degradation is required for nuclear degradation and normal epidermal terminal differentiation. Cell Death Differ. 2015;22:2123–32. doi:10.1038/cdd.2015.62.
- Yamamoto-Tanaka M, Makino T, Motoyama A, et al. Multiple pathways are involved in DNA degradation during keratinocyte terminal differentiation. Cell Death Dis. 2014;5:e1181. doi:10.1038/cddis.2014.145.
- Manils J, Fischer H, Climent J, et al. et al. Double deficiency of Trex2 and DNase1L2 nucleases leads to accumulation of DNA in lingual cornifying keratinocytes without activating inflammatory responses. Sci Rep. 2017;7:11902. doi:10.1038/s41598-017-12308-4.
- Manils J, Casas E, Viña-Vilaseca A, et al. et al. The Exonuclease Trex2 Shapes Psoriatic Phenotype. J Invest Dermatol. 2016;136:2345–55. doi:10.1016/j.jid.2016.05.122.
- Lan Y, Londoño D, Bouley R, et al. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep. 2014;9:180–92. doi:10.1016/j.celrep.2014.08.074.
- Yang SH, Chang SY, Yin L, et al. An absence of both lamin B1 and lamin B2 in keratinocytes has no effect on cell proliferation or the development of skin and hair. Hum Mol Genet. 2011;20:3537–44. doi:10.1093/hmg/ddr266.
- Rossiter H, König U, Barresi C, et al. et al. Epidermal keratinocytes form a functional skin barrier in the absence of Atg7 dependent autophagy. J Dermatol Sci. 2013;71:67–75. doi:10.1016/j.jdermsci.2013.04.015.
- Sukseree S, Rossiter H, Mildner M, et al. Targeted deletion of Atg5 reveals differential roles of autophagy in keratin K5-expressing epithelia. Biochem Biophys Res Commun. 2013;430:689–94. doi:10.1016/j.bbrc.2012.11.090.
- Huang R, Xu Y, Wan W, et al. et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell. 2015;57:456–67. doi:10.1016/j.molcel.2014.12.013.
- Yan S, Liu L, Ren F, et al. Sunitinib induces genomic instability of renal carcinoma cells through affecting the interaction of LC3-II and PARP-1. Cell Death Dis. 2017;8:e2988. doi:10.1038/cddis.2017.387.
- Wang Y, Zhang N, Zhang L, et al. et al. Autophagy regulates chromatin ubiquitination in DNA damage response through elimination of SQSTM1/p62. Mol Cell. 2016;63:34–48. doi:10.1016/j.molcel.2016.05.027.
- Park YE, Hayashi YK, Bonne G, et al. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009;5:795–804. doi:10.4161/auto.8901.
- Chikh A, Sanza P, Raimondi C, et al. iASPP is a novel autophagy inhibitor in keratinocytes. J Cell Sci. 2014;127:3079–93. doi:10.1242/jcs.144816.
- Naeem AS, Tommasi C, Cole C, et al. et al. A mechanistic target of rapamycin complex 1/2 (mTORC1)/V-Akt murine thymoma viral oncogene homolog 1 (AKT1)/cathepsin H axis controls filaggrin expression and processing in skin, a novel mechanism for skin barrier disruption in patients with atopic dermat. J Allergy Clin Immunol. 2017;139:1228–41. doi:10.1016/j.jaci.2016.09.052.
- Morioka K, Takano-ohmuro H, Sameshima M, et al. Extinction of organelles in differentiating epidermis. Acta Histochem Cytochem. 1999;32:465–76. doi:10.1267/ahc.32.465.
- Ney PA. Normal and disordered reticulocyte maturation. Curr Opin Hematol. 2011;18:152–7. doi:10.1097/MOH.0b013e328345213e.
- Schweers RL, Zhang J, Randall MS, et al. et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–5. doi:10.1073/pnas.0708818104.
- Rouzbeh S, Kobari L, Cambot M, et al. Molecular signature of erythroblast enucleation in human embryonic stem cells. Stem Cells. 2015;33:2431–41. doi:10.1002/stem.2027.
- Vacaru AM, Isern J, Fraser ST, et al. Analysis of primitive erythroid cell proliferation and enucleation using a cyan fluorescent reporter in transgenic mice. Genesis. 2013;51:751–62. doi:10.1002/dvg.22420.
- Wölwer CB, Pase LB, Pearson HB, et al. A chemical screening approach to identify novel key mediators of erythroid enucleation. PLoS One. 2015;10:e0142655. doi:10.1371/journal.pone.0142655.
