Abstract
Pacific abalone Haliotis discus discus is an important fisheries resource in Jeju, Korea. For basic information about its current genetic status in relation to stock enhancement, the level and distribution of genetic variation between wild and released stocks of Pacific abalone in Jeju were examined at nine cross-species microsatellite markers including the use of two novel primers. High levels of polymorphism were observed between the two populations. A total of 146 different alleles were found at all loci, with some alleles being unique. The allelic variability ranged from five to 27 in the wild population and from four to 16 in the released sample. The average observed and expected heterozygosities were estimated to be 0.74 and 0.84 in the wild sample and 0.70 and 0.78 in the released sample, respectively. Although a considerable loss of rare alleles was observed in the released sample, no statistically significant reductions were found in heterozygosity or allelic diversity in the released sample compared to the wild population. Low but significant genetic differentiation was found between the wild and released populations. These results suggest that the intensive breeding practices for stock enhancement may have resulted in a further decrease in genetic diversity, and that the cross-species microsatellite markers used in this study represent a potentially efficient means for further genetic studies, providing beneficial information for the protection and management of H. discus discus.
Introduction
Pacific abalone Haliotis discus discus is distributed mainly along the coastal waters of Jeju Island, Korea, and the warm south-western waters of Japan. On Jeju Island, this species has long been considered an important species in both fisheries and aquaculture. Recently, stock enhancement programs have been introduced to increase harvest yields, and many Pacific abalone seeds were released into coastal areas of Pyungdae in the island in 2007 by the Jeju province fisheries institute (JPFI). H. discus discus (Camac-jeonbok) is considered a sister subspecies of H. discus hannai (Cham-jeonbok), from which it is difficult to clearly discriminate based on morphologic characteristics (Ino Citation1952; An et al. Citation2005; Hara and Sekino Citation2005). Nevertheless, some fisheries scientists and fishermen from both the government and private hatcheries sectors in Jeju claim that Camac-jeonbok H. discus discus is quite different from the Cham-jeonbok H. discus hannai; thus, H. discus discus was used in this study.
The Food and Agriculture Organization of the United Nations (FAO) recommends the genetic characterization of both the stock population used in population enhancement programs and the target population, so that any genetic changes can be monitored for the sustainable management of exploited fish populations (FAO Citation1993). Thus, for the successful management and sustainable use of such species, the genetic variation between hatchery and wild populations and how this variation is maintained during hatchery rearing should be monitored from the initiation of stocking (Ståhl Citation1987; Primmer et al. Citation1999). Detailed information on the genetic status of exploited species can assist in the development of comprehensive long-term management and protection plans. Therefore, many studies have investigated the genetic variability of wild and farmed populations of marine species using several types of molecular markers (Hindar et al. Citation1991; Sekino et al. Citation2002; Hara and Sekino Citation2003; Alarcón et al. Citation2004; Skaala et al. Citation2004; Chen et al Citation2008; Gum et al. Citation2009). Currently, however, no information is available on the genetic diversity of wild and released populations of H. discus discus in relation to stock enhancement in Jeju, Korea.
Microsatellites, short tandem repeats of nuclear DNA, are highly polymorphic co-dominant markers. They are expected to be particularly useful for monitoring changes in genetic variation of farmed stocks, parentage assignment, and fine-scale studies of population structure in marine species (Jeong et al. Citation2003; Li et al. Citation2004; An et al. Citation2008). Novel microsatellite markers have been developed from Pacific abalone (Sekino and Hara Citation2001; Li et al. Citation2002; An and Han Citation2005; Sekino et al. Citation2005). Typically, the development of microsatellites for each new species demands considerable time, effort, and cost. Cross-species comparisons can identify polymorphic microsatellite loci, so that fewer species-specific markers are needed for studies. In an effort to increase the potential use of cross-species microsatellite markers for H. discus discus populations and pedigree analyses, we used previously developed primers (An et al. Citation2010) and two unpublished microsatellite loci from H. discus hannai. In the present study, we conducted a baseline survey of the genetic diversity of wild and released populations of H. discus discus in Jeju, Korea, using cross-species microsatellite markers.
Materials And Methods
Sample collection and DNA extraction
Two wild and released populations of H. discus discus (each n=50; shell length, over 10 cm) were sampled from the coastal waters of Pyungdae in Jeju Island, Korea in April 2009 by some fisheries scientists of the JPFI. This coastal water was the place that many hatchery-raised seeds (shell length 4 cm, 18–20 months of age) had been released in 2007 as part of a stock enhancement program by the JPFI. Hatchery-raised abalone seeds were partially produced from the brood stock captured and reared for reproduction at the JPFI and bought partially by the Jeju special self-governing province. No details regarding the founding and maintenance of the hatchery-raised seeds bought are available; however, their original parents were held at a farm on the coast of Jeju, Korea. Samples were selected by very experienced researchers, based on the shell characteristics of wild animals; for released specimens, clear green shell coloration of 4 cm length on the top of shell, which is typically observed in hatchery-raised abalone, and/or metal tags on shells (which had been applied as part of the stocking practice) were also used for identification. It can be considered that the extent to which stocking impacts the wild population is small or little in the case of abalone with shell length over 10 cm (4 years of age). Tissue samples (approximately 1 cm3) from two populations were taken and stored in 2 mL 99% ethanol at 4°C until DNA extraction. Total DNA from each sample was extracted using a MagExtractor-genomic DNA purification kit (Toyobo, Osaka, Japan). The DNA extractions were performed using an automated DNA extraction system (MagExtractor MFX-2100; Toyobo), according to the manufacturer's instructions. The extracted genomic DNA was stored at −20°C until needed.
Microsatellite genotyping
Seven highly variable microsatellite loci characterized recently (KHdh14, KHdh35, KHdh48, KHdh54, KHdh73, KHdh87 and KHdh88; An et al. Citation2010) and two unpublished loci (KHdh3 and KHdh4161) from H. discus hannai were used. A total of 100 abalones from the two populations were typed. Primer sequences, microsatellite repeat sequences, fluorescent labels, and the optimal annealing temperature for each locus are listed in . The forward primer from each primer set was 5′-fluorescent-labeled with one of three dyes: 6-FAM, HEX, or NED (PE Applied Biosystems, Foster City, CA, USA). PCR amplification of nine microsatellite loci was performed using an RTC 200 instrument (MJ Research, Waltham, MA, USA) in 10 µL solution containing 10–50 ng DNA, 1 ×ExTaq buffer, 0.2 mM dNTPs, 10 pmol each primer, and 0.25 U Taq DNA polymerase (TaKaRa Biomedical Inc., Shiga, Japan). The amplification protocol included an initial denaturation for 11 min at 95°C followed by 35 cycles of 1 min at 94°C, 1 min at the optimal annealing temperature, and 1 min at 72°C, with a final extension for 5 min at 72°C. For genotyping, 1 µL PCR product was added to each reaction containing formamide with a size standard GeneScan-400HD (ROX) (PE Applied Biosystems) and electrophoresed, using an ABI3130 DNA sequencer (PE Applied Biosystems). The fragment length of the PCR products was determined using GeneMapper software (version 4.0, PE Applied Biosystems).
Table 1. Two novel cross-species primers of used for genotyping of Haliotis discus discus.
Statistical analyses
MICRO-CHECKER 2.2.3 software (van Oosterhout et al. Citation2004) was used to detect genotyping errors due to null alleles, stuttering or allele dropout (1000 randomizations). GENEPOP 3.1b software (Rousset and Raymond Citation1995) was used to identify deviations from Hardy-Weinberg equilibrium, HWE (exact tests, 1000 iterations), allelic size range, number of unique alleles, and observed and expected heterozygosities (indicating an excess or deficiency of heterozygotes). FSTAT 2.9.3.2 (Goudet 1995) was used to calculate number of alleles, gene diversity, and F IS (Weir and Cockerham Citation1984) per locus and sample.
The extent of population subdivision was examined by calculating fixation indices. ARLEQUIN 3.0 software (Excoffier et al. Citation2005) was used to assess linkage disequilibrium for all pairs of loci (Slatkin and Excoffier Citation1996) and calculate single-locus F ST and global multilocus F ST values (1000 permutations) (Weir and Cockerham Citation1984). R ST (Slatkin Citation1995; Michalakis and Excoffier Citation1996) estimates were calculated using the computer program GENEPOP version 3.4, available on the internet. R ST incorporates the correlation of the weighted mean allele size expressed as the number of tandem repeats. Significance levels were adjusted for multiple tests using the sequential Bonferroni correction technique (Rice Citation1989).
Results
Cross-species amplification
The majority of cross-species amplifications from the closely related abalone species resulted in PCR fragments with the expected microsatellite morphology and correct allele sizes, indicating homologous loci. Homology was not tested through sequence comparison. More than 50% of all individuals were reanalysed to ensure the repeatability of allele scoring. Samples that failed to show amplification products were reamplified. There was no indication that genotyping errors affected allele scoring due to allele dropouts or stuttering at any of the markers in any of the samples. Samples that failed to amplify were not included and this makes it unlikely that poor DNA quality affected our results.
Genetic variability comparisons
All primers were polymorphic in all samples, and the level of polymorphism varied depending on the locus. The genetic variability in each population is presented in . In total, 146 different alleles were observed over nine loci, and some alleles were unique to each population. The number of alleles per locus varied from four at locus KHdh54 to 27 at locus KHdh14; fewer alleles were found in the released population than in the wild one, but the difference was not statistically significant (P>0.05).
Table 2. Summary statistics for nine microsatellite loci between abalone collections.
The MICRO-CHECKER analysis showed that some loci may have been influenced by one or more null alleles in both populations; loci KHdh3, KHdh54, and KHdh73 in the released samples and KHdh3 and KHdh73 in the wild population were affected. KHdh3 and KHdh73 appeared to have been influenced in both populations, which indicate that using these two loci for population genetic analyses that assume HWE may be problematic. Thus, a global multilocus F ST value was estimated with and without this locus. However, locus KHdh54 was affected by null alleles in only one sample; thus, this was included in further analyses.
The levels of observed (Ho) and expected (He) heterozygosity varied between the two sample populations, ranging from 0.30 at KHdh3 to 1.00 at KHdh14, KHdh48, and KHdh87, and from 0.50 at KHdh3 to 0.97 at KHdh4161, respectively. No significant difference between Ho and He was found between samples at most loci (Kruskal-Wallis test, P>0.05).
Wild samples had higher genetic diversity (G d) and a wider range of allele sizes (S) at most loci (). In total, 89 alleles were unique to the two populations: 63 in wild samples and 26 in released ones. For most loci, wild abalone had more unique alleles than the released population. The allele frequencies at all nine selected loci in each sample are shown in and . These data reveal differences between the populations. The inbreeding coefficients (F IS) varied among markers, from −0.318 (KHdh88) to 0.454 (KHdh54) in the released samples, and from −0.253 (KHdh87) to 0.524 (KHdh3) in the wild samples. The average F IS, including all markers, was 0.17 in the released population and 0.15 in the wild one.
Figure 1. Allele size frequency distributions of the nine microsatellite loci in two Haliotis discus discus populations. HddR, released abalone; HddN, wild abalone.
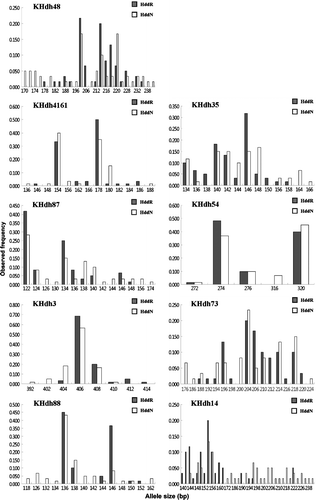
Table 3. Frequency of each microsatellite allele in wild and hatchery Haliotis discus discus samples in Jeju, Korea.
No significant linkage disequilibrium was found for all pairs of loci after sequential Bonferroni for each of 36 simultaneous tests in the wild and hatchery populations.
Significant departures from HWE (P<0.0055) were found after Bonferroni correction at six loci (KHdh3, KHdh14, Khdh54, Khdh73, Khdh87, and KHdh88) in the released samples and at two loci (Khdh3 and KHdh73) in the wild ones, indicating that deviations from HWE were due to heterozygosity deficit or excess.
Genetic differentiation between the two populations was estimated using F ST and R ST estimates. The global multilocus F ST, including all loci, was 0.021 (P <0.001), and the R ST value was 0.018 (P <0.001). When loci KHdh3 and Khdh73 were excluded, the global multilocus F ST was 0.022 (P <0.001). Low but significant F ST and R ST estimates indicate the presence of genetic differentiation between the released and wild populations.
Discussion
Despite the importance of H. discus discus as a highly valuable fishery resource in Jeju, Korea, there is little information regarding the genetic background of this species. The present study is the first report on the genetic status of this species using microsatellite markers.
We used previously developed microsatellite markers from H. discus hannai to evaluate the genetic variability of H. discus discus; these cross-species primers could serve as a valuable tool, sufficient to detect genetic differences between the wild and released populations. Although many more samples are needed to confirm these results, the cross-species primers might be valuable during the initial phase of genetic analyses, while taking into account the relatedness of species. Previously, Hara and Sekino (2005) reported that the microsatellite markers developed from H. discus hannai provided high resolution for studying the genetic differences between H. discus hannai and H. discus discus populations.
We found that both the number of alleles and expected heterozygosity are reduced in the released hatchery-raised abalone population, showing only 76% of the average number of alleles and 90% of the expected heterozygosity expressed in the wild abalone population, with considerable loss of rare alleles, although no statistically significant reduction was found. A reduction in genetic diversity relative to wild populations seems to be a characteristic of hatchery-raised organisms. It may be caused by founder effects, as well as artificial and natural selection in the culture environment (Tessier et al. Citation1997; Coughlan et al. Citation1998; Was and Wenne Citation2002; Li et al. Citation2004, 2007; Porta et al. Citation2006; Marchant et al. Citation2008). Allelic frequency in wild populations is not always reflected in hatchery populations due to random changes in the production process of a new generation. In fact, allele loss has a greater impact on genetic diversity than changes in allele frequency, because the latter may change again by random genetic drift, but there is no way to recover a lost allele. High levels of polymorphism were detected in the released population (mean He=0.76) over all nine microsatellite loci examined. Similar results have been reported for H. discus hannai and other cultivated fish species (Elliott and Reilly Citation2003; Li et al. Citation2007; An et al. Citation2008; Marchant et al. Citation2008).
Significant deviations from Hardy-Weinberg (H-W) expectations were observed in both populations. In general, the causes of such H-W disequilibrium, found frequently in microsatellite loci, probably include the substructuring of the population sample, inbreeding, homoplasy, or the presence of null alleles (Estoup et al. Citation1995; Banks et al. Citation1999; Li et al. Citation2003). In addition, a higher occurrence of H-W disequilibrium events occurred in the hatchery-raised stock than in the wild one (six versus two, respectively; ). This may have resulted from admixture of more than two independent populations, non-random mating, or artificial and natural selection during seed production and cultivation in the hatchery stock. The occurrence of higher F IS values at all loci in the released sample suggests that non-random mating might have occurred.
Based on F ST and R ST values, there is significant genetic differentiation between the populations. This difference is probably a result of reduced genetic variation due to a lower effective number of founding individuals in the hatchery, and the effects of artificial selection on the hatchery progeny ().
It has been suggested that reduced genetic variation can result in reduced performance in aquaculture, because this is the source of variation for important traits such as growth rate and disease resistance (Allendorf and Phelps Citation1980; Vuorinen Citation1984). There are several reports of reduced fitness in hatchery-bred samples when exposed to natural environments (Hansen Citation2002; Li et al. Citation2004). Release of genetically maladapted fishery reduces the effectiveness of enhancement, and is most detrimental overall if the fitness of hatchery raised fishery is only moderately lower than in the wild population. Thus, for an adequate stocking strategy for H. discus discus in Jeju, Korea, periodic genetic evaluations of broodstock, progeny, and target populations are necessary, on behalf of the health of the wild fishery resource.
For the proper management of stock enhancement programs, genetic monitoring for structure and diversity must be considered in addition to biological, ecological, and fishery factors. Before carrying out a large-scale release for stock enhancement, genetic differences between cultured and wild populations must be identified and quantified if effective monitoring of gene flow is to occur. It is therefore important that adequate amounts of this kind of genetic information be available both for hatchery-reared populations and for wild populations that may be affected by migration from them. This information will be useful for evaluating the feasibility of the enhancement program to maintain the genetic diversity of the wild population, as well as to improve the management of the hatchery population for the following season.
In summary, we investigated the genetic diversity of wild and released populations of H. discus discus in Jeju, Korea, for the first time, using cross-species microsatellite markers developed from H. discus hannai. Our results show that the markers used can detect changes in genetic composition between hatchery-raised and wild populations, and that hatchery-raised abalone has genetically differentiated from the wild population. This in turn indicates that genetic monitoring is a useful tool for the preservation and further genetic management of H. discus discus in Korea.
Acknowledgements
This work was funded by a grant from the National Fisheries Research and Development Institute (NFRDI), contribution number RP-2010-BT-34. The views expressed herein are those of the authors and do not necessarily reflect the views of NFRDI.
References
- Alarcón , JA , Magoulas , A , Georgakopoulos , T , Zouros , E and Alvarez , MC. 2004 . Genetic comparison of wild and cultivated European populations of the gilthead sea bream (Sparus aurata) . Aquaculture. , 230 : 65 – 80 .
- Allendorf , FW and Phelps , SR. 1980 . Loss of genetic variation in a hatchery stock of cutthroat trout. Trans . Am Fish Soc. , 109 : 537 – 543 .
- An , HS and Han , SJ. 2005 . Isolation and characterization of microsatellite DNA markers in the Pacific abalone, Haliotis discus hannai . Mol Ecol Notes. , 6 : 11 – 13 .
- An , HS , Jee , YJ , Min , K-S , Kim , B-L and Han , SJ. 2005 . Phylogenetic analysis of six species of Pacific abalone (Haliotidae) based on DNA sequences of 16s rRNA and cytochrome c oxidase subunit I mitochondrial genes . Mar Biotec. , 7 : 373 – 380 .
- An , HS , Kim , MJ and Hong , SW. 2008 . Genetic diversity of rock bream Oplegnathus fasciatus in southern Korea . Genes Genomics. , 30 : 451 – 459 .
- An HS Lee J-H Noh JK Kim HC Park CJ Min BH Myeong J-I. 2010 New polymorphic microsatellite markers in pacific abalone Haliotis discus hannai and their application to genetic characterization of wild and aquaculture populations. Genes Genomics 32 413 418
- Banks , MA , Blouin , MS , Baldwin , BA , Rashbrook , VK , Fitzgerald , HA , Blankenship , SM and Hedgecock , D. 1999 . Isolation and inheritance of novel microsatellites in Chinook salmon (Oncorhynchus tschawytscha) . J Hered. , 90 : 281 – 288 .
- Chen , L , Li , Q and Yang , J. 2008 . Microsatellite genetic variation in wild and hatchery populations of the sea cucumber (Apostichopus japonicas Seleka) from northern China . Aqua Res. , 39 : 1541 – 1549 .
- Coughlan JP Imsland AK Galvin PT Fitzgerald RD Naevdal G Cross 1998 Microsatellite DNA variation in wild populations and farmed strains of turbot from Ireland and Norway: a preliminary study J Fish Biol 52 916 922
- Elliott , NG and Reilly , A. 2003 . Likelihood of a bottleneck even in the history of the Australian population of Atlantic salmon (Salmo salar L.) . Aquaculture. , 215 : 31 – 44 .
- Estoup , A , Tailliez , C , Cornuet , J and Solignac , M. 1995 . Size homoplasy and mutational processes of interrupted microsatellites in two bee species, Apis mellifera and Bombus terrestris (Apidae) . Mol Biol Evol. , 12 : 1074 – 1084 .
- Excoffier , L , Laval , G and Schneider , S. 2005 . ARLEQUIN version 3.0: an integrated software package for population genetics data analysis . Evol Bioinform Online. , 1 : 47 – 50 .
- FAO 1993 Report of the expert consultation on utilization and conservation of aquatic genetic resources FAO Fish Rep 491 : 1 58
- Goudet , J. 2002 . FSTAT: a computer program to calculate F-statistics . J Hered. , 86 : 485 – 486 .
- Gum , B , Gross , R and Geist , J. 2009 . Conservation genetics and management implications for European grayling, Thymallus thymallus: synthesis of phylogeography and population genetics . Fish Manag Ecol. , 16 : 37 – 51 .
- Hansen , MM. 2002 . Estimating the long-term effects of stocking domesticated trout into wild brown trout (Salmo trutta) populations: an approach using microsatellite DNA analysis of historical and contemporary samples . Mol Ecol. , 11 : 1003 – 1015 .
- Hara , M and Sekino , M. 2003 . Efficient detection of parentage in a cultured Japanese flounder Paralichthys olivaceus using microsatellite DNA marker . Aquaculture. , 217 : 107 – 114 .
- Hara , M and Sekino , M. 2005 . Genetic difference between Ezo-awabi Haliotis discus hannai and Kuro-awabi Haliotis discus discus populations: microsatellite-based population analysis in Japanese abalone . Fish Sci. , 71 : 754 – 766 .
- Hindar , K , Ryman , N and Utter , FM. 1991 . Genetic effects of cultured fish on natural fish populations . Can J Fish Aqua Sci. , 48 : 945 – 957 .
- Ino T. 1952 Biological study on the propagation of Japanese abalone (genus Haliotis) Bull Tokai Reg Fish Res Lab 5 1 102
- Jeong , DS , Umino , T , Kuroda , K , Hayashi , M , Nakagawa , H , Kang , JC , Morishima , K and Arai , K. 2003 . Genetic divergence and population structure of black sea bream Acanthopagrus schlegeli inferred from microsatellite analysis . Fish Sci. , 69 : 896 – 902 .
- Li , Q , Park , C and Kijima , A. 2002 . Isolation and characterization of microsatellite loci in the Pacific abalone, Haliotis discus hannai . J Shell Res. , 212 : 811 – 815 .
- Li Q Park C Kobayashi A Kijima A 2003 Inheritance of microsatellite DNA markers in the Pacific abalone Haliotis discus hannai Mar Biotec 5 331 338
- Li , Q , Park , C , Endo , T and Kijima , A. 2004 . Loss of genetic variation at microsatellite loci in hatchery strains of the Pacific abalone (Haliotis discus hannai) . Aquaculture. , 235 : 207 – 222 .
- Li , Q , Shu , J , Yu , R and Tian , C. 2007 . Genetic variability of cultured populations of the Pacific abalone (Haliotis discus hannai Ino) in China based on microsatellites . Aquaculture Res. , 38 : 981 – 990 .
- Marchant , S , Haye , PA , Marín , SA and Winkler , FM. 2008 . Genetic variability revealed with microsatellite markers in an introduced population of the abalone Haliotis discus hannai Ino . Aquaculture Res. , 40 : 298 – 304 .
- Michalakis , Y and Excoffier , L. 1996 . A genetic estimation of population subdivision using distances between alleles with special reference for microsatellite loci . Genetics. , 142 : 1061 – 1064 .
- Porta , J , Porta , JM , Martínez-Rodríguez , G and Alvarez , MC. 2006 . Genetic structure and genetic relatedness of a hatchery stock of Senegal sole (Solea senegalensis) inferred by microsatellites . Aquaculture. , 251 : 46 – 55 .
- Primmer , CR , Aho , T , Piironen , J , Estoup , A , Cornuet , JM and Ranta , E. 1999 . Microsatellite analysis of hatchery stocks and natural populations of Arctic charr, Salvelinus alpinus, from the Nordic region: implications for conservation . Hereditas. , 130 : 277 – 289 .
- Rice , WR. 1989 . Analyzing tables of statistical tests . Evolution. , 43 : 223 – 225 .
- Rousset , F and Raymond , M. 1995 . Testing heterozygote excess and deficiency . Genetics. , 140 : 1413 – 1419 .
- Sekino , M and Hara , M. 2001 . Application of microsatellite DNA markers to population genetics studies of Japanese flounder Paralichthys olivaceus . Mar Biotec. , 3 : 572 – 589 .
- Sekino , M , Hara , M and Taniguchi , N. 2002 . Loss of microsatellite and mitochondrial DNA variation in hatchery strains of Japanese flounder Paralichthys olivaceus . Aquaculture. , 213 : 101 – 122 .
- Sekino , M , Saido , T , Fujita , T , Kobayashi , T and Takami , H. 2005 . Microsatellite DNA markers of Ezo abalone (Haliotis discus hannai): a preliminary assessment of natural populations sampled from heavily stocked areas . Aquaculture. , 243 : 33 – 47 .
- Skaala , Ø , Høyheim , B , Glover , K and Dahle , G. 2004 . Microsatellite analysis in domesticated and wild Atlantic salmon (Salmo salar L.): allelic diversity and identification of individuals . Aquaculture. , 240 : 131 – 143 .
- Slatkin , M. 1995 . A measure of population subdivision based on microsatellite allele frequencies . Genetics. , 139 : 457 – 462 .
- Slatkin , M and Excoffier , L. 1996 . Testing for linkage disequilibrium in genotypic data using the EM algorithm . Heredity. , 76 : 377 – 383 .
- Ståhl G Genetic population structure of Atlantic salmon Population genetics and fishery management Ryman N Utter F 1987 University Washington Press Seattle 121 140
- Tessier , N , Bernatchez , L and Wright , JM. 1997 . Population structure and impact supportive breeding inferred from mitochondrial and microsatellite DNA analyses in land-locked Atlantic salmon Salmo salar L . Mol Ecol. , 6 : 735 – 750 .
- van Oosterhout , C , Hutchinson , WF , Wills , DPM and Shipley , P. 2004 . MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data . Mol Ecol Notes. , 4 : 135
- Vuorinen , J. 1984 . Reduction of genetic variability in a hatchery stock of brown trout, Salmo trutta . J Fish Biol. , 24 : 339 – 348 .
- Was , A and Wenne , R. 2002 . Genetic differentiation in hatchery and wild sea trout (Salmo trutta) in the southern Baltic at microsatellite loci . Aquaculture. , 204 : 493 – 506 .
- Weir , BS and Cockerham , CC. 1984 . Estimating F-statistics for the analysis of population structure . Evolution. , 38 : 1358 – 1370 .