Abstract
The pear psylla, Cacopsylla pyricola (Hemiptera: Psyllidae), is a serious insect pest of commercial pear crops. The species, which resides on pear trees throughout its life cycle, is rapidly spreading in some regions of the world. The population genetic structure of the species collected from several pear orchards in Korea was studied to understand the nature of dispersal and field ecology of the species. The 658-bp region of mitochondrial COI gene and the 716-bp long complete internal transcribed spacer 2 (ITS2) of the nuclear ribosomal DNA were sequenced. Unlike other previously studied insect pests, the COI-based genetic diversity of the pear psylla was extremely low (maximum sequence divergence of 0.15%). This finding allowed us to conclude that the species may have been introduced in Korea relatively recently. ITS2 sequence-based analyses of phylogeny, population differentiation, gene flow, and hierarchical population structure all concordantly suggested that the pear psylla populations in Korea are neither genetically isolated nor hampered for gene flow. These genetic data are concordant with the dispersal of an overwintering winterform morph outside the non-pear habitat in the fall.
Introduction
The pear psylla, Cacopsylla pyricola (Hemiptera: Psyllidae), which is distributed in Korea, Japan, Europe, and North America, is a serious insect pest of commercially grown pears. Both nymphs and adults damage leaves and fruits by injecting a toxin that causes blackening and burning of the foliage (psylla shock), by secretion of honeydew that causes sooty mould and marks the fruit, and by transmitting mycoplasma-like organisms that are responsible for pear decline disease (Burts et al. Citation1989).
One of the most notable characteristics of the species is seasonal morphological dimorphism, which involves the conversion from a light-colored form (summerform) to a dark overwintering form (winterform) in late summer and autumn in response to the shortening photoperiod (Horton et al. Citation1994). In Korea, the approximate calendar date for the advent of the winterform has been calculated to be early September on the basis of local data (An et al. Citation1996).
In a study to determine the precise locations of overwintering in a typical pear orchard, Kim et al. (Citation2000b) reported that pear psylla overwinters mainly under the bark scale of pear trees, with some minor occurrence under circumjacent weeds and fallen leaves. This observation signifies the importance of pear tree as an overwintering habitat and has led us to assume that the winterform strictly overwinters only in pear trees. However, this finding does not conclusively establish pear trees as being the sole overwintering habitat for the winterform. Furthermore, it is unknown whether individuals dispersed from a pear orchard have other habitats.
The winterform can exit pear orchards in fall to overwinter in surrounding non-pear habitats including apple orchards (Horton et al. Citation1992), although the pear is the sole source of development and reproduction (Follett et al. Citation1985). Horton and Lewis (Citation1996) reported that the fall movement of the winterform to adjacent non-pear habitats is due to short tethered flight, implying non-true migratory flight. On the other hand, overwintering adults were reported to disperse widely, which may have driven the rapid spread of the species in the eastern United States and Canada (Westigard and Zwick Citation1972).
Table 1. List of trapping locality, collection date, sample number, COI haplotypes, ITS2 sequence types, GenBank accession numbers, ITS2 size, and G + C contents of Cacopsylla pyricola.
Provided that active dispersal of winterform is combined with the pesticide resistance that has been developed locally, nationwide resistance would be an urgent issue, as the number of registered and effective pesticides for pear psylla control diminishes (Croft et al. Citation1989). Thus, knowledge of the population genetic structure, particularly for the gene flow of the species, is important for the eventual control of the pear pest.
To address the issue, the present study analyzed the population genetic structure of the species on the basis of sequence-based information. This information enabled the analyses of genetic diversity, geographic variation, and genetic relationships with the inference of gene flow. We partially sequenced the mitochondrial cytochrome c oxidase subunit I (COI) gene and completely sequenced the nuclear internal transcriber spacer 2 (ITS2) for the purpose of study.
Materials and methods
Insects
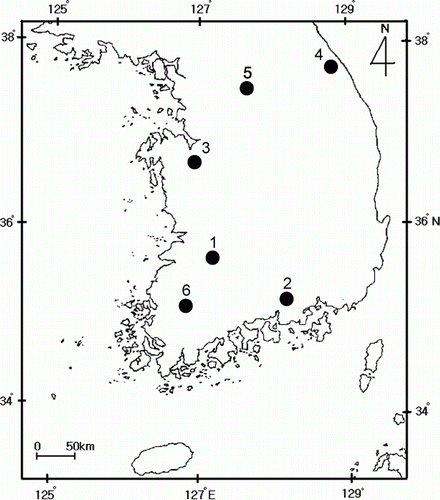
A total of 74 adults of pear psylla, Cacopsylla pyricola (Hemiptera: Psyllidae), were collected from six localities in Korea (; ). Each adult specimen was collected from different pear trees to avoid possible overrepresentation of siblings from each locality. The samples were frozen at –70°C until being used in molecular analysis.
Primer design
To sequence the COI gene corresponding to the ‘DNA barcode region’ (Hebert et al. Citation2003), we eventually used the novel primer set CPF4 and CPR4 (Supplementary ). The schematic illustration of primer locations and detailed strategy to design primers are presented in Supplementary . For the amplification of ITS2 of nuclear rDNA, primers NG02955 and AB052895 located on the 5.8S and 28S rDNAs, respectively, were successfully used (Ji et al. Citation2003; Supplementary ).
DNA extraction, PCR, cloning, and sequencing
Total DNA was extracted with a Wizard Genomic DNA Purification Kit using the manufacturer's instructions (Promega, USA). For the amplification of the 658-bp region of COI gene, polymerase chain reaction (PCR) was conducted under the following conditions: an initial denaturation step at 94°C for 7 min, a 35-cycle amplification (94°C for 1 min, 50–60°C for 1 min, and 72°C for 1 min), and the final extension step for 7 min at 72°C. For PCR of ITS2, an initial denaturation step at 94°C for 4 min, a 35-cycle amplification (94°C for 40 sec, 50–60°C for 20 sec, and 72°C for 40 sec), and the final extension step for 2 min at 72°C were conducted. To confirm successful DNA amplification, electrophoresis was conducted using 1.0× TAE buffer on a 1.0% agarose gel. The PCR product was purified with a PCR purification Kit (Bioneer, Korea). The COI gene amplicons were directly sequenced, whereas those from ITS2 were cloned into a pGEM-T Easy vector (Promega). For the cloning process, XL1-Blue competent cells (Stratagene, USA) were transformed with the ligated DNA, and the resultant plasmid DNA was isolated using a Wizard Plus SV Minipreps DNA Purification System (Promega). DNA sequencing was conducted using the ABI PRISM® BigDye® Terminator ver. 3.1 Cycle Sequencing Kit with an ABI 377 Genetic Analyzer (PE Applied Biosystems, USA). All products were sequenced from both strands.
Sequence analysis and phylogenetic inference
Sequence delimitation and alignment for both COI gene and ITS2 were conducted using MAFFT ver. 6 (Katoh et al. Citation2002). The boundary of the ITS2 sequence was delimited using the Hidden Markov Model-based ITS2 annotation software, with the model selection set to either Metazoa or Diptera and the maximum E-value set as the default value, E < 0.001 (Keller et al. Citation2009). When the homologous sequences from two individuals differed by one or more nucleotide base (for both COI and ITS2) or one insertion/deletion (indel) position (for ITS2), the sequences were considered different haplotypes (for COI) or sequence types (for ITS2). Haplotype or sequence type designations were applied to new sequences as they were discovered (i.e. BARCP01 and BARCP02 for COI, and ITSCP01, ITSCP02, ITSCP03 and so on for ITS2). Ultimately, two COI gene haplotypes and 26 sequence types from the ITS2 region were obtained and individual sequence information was deposited in the GenBank database ().
Phylogenetic analysis among the ITS2 sequence types was conducted via the maximum-parsimony (MP) method (Fitch Citation1971) using Phylogenetic Analysis Using Parsimony and Other Method (PAUP) ver. 4.0b10 (Swofford Citation2002). The analysis was conducted by heuristic search using a tree-bisection-reconnection for branch-swapping algorithm, steepest descent option not in effect, stepwise addition option for starting tree, number of trees held at each step during stepwise addition for one, and initial ‘MaxTrees’ set for 100. Branches were collapsed if the maximum branch length was zero. Characters were given an equal weighting with the gaps treated as a fifth character. The reliability of the trees was assessed by 1000 iterations of bootstrapping (Felsenstein Citation1985). To root trees, the homologous region of one within-superfamilial species, Bactericera cockerelli (GenBank accession number GQ249868.1) and one within-familial species, Anomoneura mori, sequenced for this study were used as outgroups.
Estimation of genetic diversity, distance, and gene flow
Nucleotide diversity for ITS2, which is reflective of genetic diversity within each locality, was acquired using Arlequin ver. 3.0 (Excoffier et al. Citation2005). Maximum sequence divergence within populations was estimated via the extraction of the within-locality estimates of unrooted pairwise distance from PAUP (Swofford Citation2002). Genetic distance and migration rate were estimated with Arlequin ver. 3.0 (Excoffier et al. Citation2005). Population pairwise genetic distance (F ST) and a permutation test of the significant differentiation of the pairs of localities (1000 bootstraps) were determined in accordance with the approach described by Excoffier et al. (Citation1992), wherein the distance between DNA sequences was calculated via the Kimura 2-parameters method (Kimura Citation1980). Pairwise F ST values were used to estimate the per generation migration rate, Nm (the product of the effective population size N e and migration rate, m), based on the equilibrium relationship: F ST=1/(4Nm+1).
Hierarchical genetic structure
Genetic relationships among populations and sets of populations were assessed by the Holsinger and Mason-Gamer (H-MG) method (1996). Unlike other variance analyses, this approach generated the hierarchical relationships of the groups without specifying the hierarchical structure of the populations before the analysis.
Results
Sequence analysis
Only two haplotypes (BARCP01 and BARCP02) were obtained by sequencing 658 bp of COI gene from 40 individuals of C. pyricola (). Sequence alignment revealed only one variable nucleotide between the two haplotypes, revealing a transition (T↔C) without amino acid replacement. Thirty-nine specimens accounted for BARCP01, but a single individual collected from Yangyang-gun (locality 4) only possessed BARCP02. In the case of ITS2, a total of 26 sequence types (ITSCP01–ITSCP26) was acquired by sequencing of ITS2 rDNA from 34 C. pyricola individuals (). Uncorrected pairwise distance among the 26 sequence types revealed the sequence divergence ranging from 0.14% to 2.10% (1–15 bp; Supplementary ). The highest sequence divergence was obtained when each ITSCP04 and ITSCP26 was respectively compared to ITSCP18, ITSCP20, and ITSSCP24 (Supplementary ). One of the reasons for the higher variability in ITS2 might possibly be attributable to the higher evolutionary ratio and relative freedom from structural and functional constrains of ITS2 (Tang et al. Citation1996; Navajas et al. Citation1998), but this interpretation may require more generalization.
Table 2. Within-locality diversity estimates of Cacopsylla pyricola.
Regional distribution of sequence type
Most of the sequence types were found in a single locality as a single individual, but the sequence type ITSCP01 was found in four localities (localities 2, 4, 5, and 6) as six individuals, ITSCP02 in two localities (localities 5 and 6) as two individuals, and ITSCP02 in a single locality (locality 2; Jinju-city) as two individuals (). Thus, the distribution of C. pyricola sequence types can be summarized as a restricted local distribution in most sequence types, with a wide distribution in only a limited number of sequence types (i.e. ITSCP01).
Phylogenetic analysis
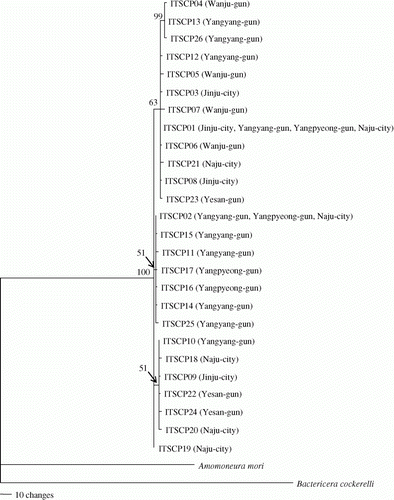
Phylogenetic analysis was conducted to determine the relationships among the 26 ITS2 sequence types to detect any discernible group in connection with geographic distribution (). The majority of the sequence types (n=23) were weakly associated or unresolved, due mainly to moderate to low genetic divergence among them, but one phylogenetic group was highly supported (99% of bootstrap value). This group was composed of three sequence types that were derived from two localities (locality 1, Wanju-gun; and locality 4, Yangyang-gun). Considering that the geographic distance between localities 1 and 4 is more than 350 km (), the clustering of the sequence types derived from these localities may reflect a non-locality-based clustering, which is indicative of a random association caused by a strong gene flow. Furthermore, phylogenetic analyses with each single outgroup has not proved the clustering of the sequence types, lacking geographic association (data not shown).
Figure 2. Phylogenetic analysis among 26 ITS2 sequence types of of C. pyricola. The single-most parsimonious tree (tree length = 651, consistent index = 0.972, retention index = 0.895, homoplasy index = 0.028) was obtained from an unweighted parsimony analysis. The numbers on the branches represent bootstrap values of 1000 replications. The sequences of one within-superfamilial species, Bactericera cockerelli (GenBank accession number GQ249868.1) and one within-familial species, Anomoneura mori, sequenced for this study were used as outgroups. Parentheses indicate the locality name from which the particular sequence type was obtained.
Genetic diversity indices
In a range of 0–1 in haplotype diversity (H), most localities displayed substantially higher estimates of H, ranging from 0.8667 (locality 2; Jinju-city) to 1.0000 (localities 1, 3, 5, and 6). In terms of π, the estimates were highest in the order locality 3 (Yesan-gun) as 0.011173, locality 1 (Wanju-gun) as 0.009777, locality 6 (Naju-city) as 0.009218, locality 4 (Yangyang-gun) as 0.009142, locality 5 (Yangyang-gun) as 0.005587, and locality 2 (Jinju-city) as 0.004935 (). Considering the maximum difference between the highest and lowest estimates was only about 2.3-fold, no single particular locality could be attributable as a center for genetic diversity. Nevertheless, it is noteworthy that the three localities with higher π (localities 3, 1, and 6; ) are all located in the western region of the Korean peninsula, where pear is more extensively cultivated geographically, possibly reflecting a larger population size of the pear psylla in those areas.
Gene flow
A test of statistical significance of pairwise F ST estimates resulted in a single case between localities 4 and 5 (Yangyang-gun and Yangpyeong-gun) (). Except for this pair, the ITS2 sequence data did not show any significant differentiation among populations of C. pyricola on the Korean peninsula. Consistent with the population differentiation data, the estimate of gene flow (Nm) showed the least gene flow estimate between localities 4 and 5 (Nm = 0.85372), whereas the remaining pairs of populations showed values ranging from 1.2222 to infinity. This was indicative of an exchange of approximately one ~ infinite numbers of individual migrants per generation.
Table 3. Fixation indices (F ST) and migration rate (Nm) between pairs of populations of Cacopsylla pyricola.
Hierarchical population genetic structure
As might be expected from the pairwise F ST, no statistically significant population subdivision was observed among populations or among groups of populations (Supplementary ). Populations could generally be subdivided into two groups: one composed of localities 5, 1, and 2; and another composed of localities 4, 3, and 6, although the clustering was not statistically supported. Considering the geographic distance of the within-group localities, the clustering of the localities reflected a non-distance-based clustering. For example, locality 4 (Yangyang-gun) is approximately 340 km and 490 km distant from locality 3 (Yesan-gun) and 6 (Naju-city), but locality 4 clustered together with these localities, excluding locality 5 (Yangpyeong-gun), which is only approximately 180 km distant. Considering the F ST data, Nm, and the phylogenetic analysis together with the result of the hierarchical relationships, the geographical populations of C. pyricola displayed high potential/actual dispersal.
Discussion
Genetic homogeneity
Genetic analysis of the COI gene of the pear psylla showed a maximum sequence divergence of only 0.15%, with only a single nucleotide difference between the two haplotypes. Considering other similar studies that utilized a homologous region of the mitochondrial COI gene sequence, the maximum sequence divergence of C. pyricola was very low. Estimates were 0.2% for the domestic silkworm (Kim et al. Citation2000a), 0.2% and 1.2% for two species of mushroom fly (Bae et al. Citation2001), 0.45% for the tiny dragonfly (Kim et al. Citation2007), 0.76% for the mason bee (Kim et al. Citation2008), 0.9% for the diamondback moth (Li et al. Citation2006), 0.91% for swallowtail butterfly (Jeong et al. Citation2009), and 1.67% for cabbage butterfly (Jeong et al. Citation2009). Thus, it was approximately ≤ 1.0% in the insect mitochondrial COI gene, showing that the magnitude of sequence divergence of C. pyricola is very low compared with other insect species.
There might be that several factors govern the genetic diversity of a given species. Along with the biogeographic history as a long-term perspective (Avise Citation1994), the maintenance of a large and stable population (Frankham Citation1996) would be an important source for the maintenance of genetic diversity. Considering the pear psylla is one of the major insect pests abundant in pear orchards (Kim et al. Citation2000b), such a substantially low sequence divergence was unexpected in the COI gene. In particular, considering that the pear tree is the sole source of feeding, development, and reproduction for the pear psylla (Follett et al. Citation1985), the evolution of regional haplotypes is likely to be expected. Thus, some other explanation might be required to account for such a low sequence divergence. Maruyama and Kimura (Citation1980) have reported that frequent extinction and colonization of populations is one factor that decreases actual genetic diversity. If this theory is applied to the mitochondrial DNA data of C. pyricola, the pear psylla populations should have undergone frequent extinction and colonization. One scenario that fulfills such a condition might be that pear psylla populations are subjected to local eradication by chemical pesticides, which is the major strategy for the control of pear psylla populations. Nevertheless, chemical control may well have limited value (Burts et al. Citation1989; Unruh Citation1990), so a frequent extinction and colonization of populations caused by control practice may not fully account for the observed genetic homogeneity in the COI gene.
Another plausible explanation may involve a relatively recent introduction. The pear psylla is generally believed to be native to southern Europe and western Asia (Burts Citation1970). In Korea, the species may have become a problematic pest around 1998, although occasional occurrence has also been reported from 1993 (Jeong et al. Citation2000). These reports coincide with the finding of low genetic diversity in C. pyricola occurring in Korea in a relatively short period of time, which may have not have allowed C. pyricola populations to evolve local haplotypes.
Dispersal
The pear psylla that was detected first in Washington in 1940 quickly spread south, reaching southern Oregon by 1950 and California by 1958, covering over 2000 km in 20 years (Westigard and Zwick Citation1972). These findings are consistent with the view that the species is capable of active dispersal, possibly with strong flight ability. Similarly, only during the past approximately 10 years has pear psylla in Korea become ranked as the most notorious and abundant pest in pear orchards (Jeong et al. Citation2000). Indeed, these field observations are concordant with our data: wide distribution of two ITS2 sequences types (ITSCP02 and ITSCP01; ), clustering of individuals collected from distant localities in the phylogenetic analysis () and overall statistically insignificant F ST estimates (high gene flow) between the majority of population pairs (). These data collectively suggest that the pear psylla populations in Korean peninsula are very well connected to each other.
It has been thought that the rapid geographic spread of pear psylla in certain regions of the world is due to movement by the winterform. These inferences are largely based on the finding of the winterform outside pear orchards, in contrast to the summerform (Horton et al. Citation1994). In fact, Horton et al. (Citation1992) found that large numbers of winterform pear psylla leave pear orchards to overwinter in surrounding non-pear habitats (i.e. apple orchards) and these recolonize pears the following spring as the temperature warms. The main reason for the exodus is to find another moisture source, which disappears in the pear orchard as the leaves fall from the trees. However, the fall movement of the winterform to adjacent non-pear habitats was ascribed to tethered flight (Horton and Lewis Citation1996). Thus, the majority of pear psylla is expected to return to pear orchards. Nevertheless, for genetically homogeneous populations, it has been theorized that a small number of migrants may be sufficient to prevent drift (Hartl and Clark Citation1989). Thus, even a small portion of populations that have sought out non-pear habitats for overwintering might be a sufficient source of long-distance dispersal, and these may eventually be the source of the successful colonization of new pear orchards. Therefore, the current observation of close genetic relationships among geographic populations of pear psylla appears to be the result of dispersal by the winterform, even though we do not know the exact magnitude and extensiveness of the dispersal. For further scrutinized examination of the dispersal behavior of the species, co-dominant markers such as microsatellite DNA (e.g. An et al. Citation2010) would be a good candidate.
tacs_a_607511_sup_20659081.zip
Download Zip (107.5 KB)Acknowledgements
This study was supported by the Korean Pear Export Research Organization, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea. We authors also express thanks to H.U. Jeong for helping with sample collection.
Additional information
Notes on contributors
Jee Yeon Baek
These authors contributed equally to this paperReferences
- An , JH , Yiem , MS and Kim , DS. 1996 . Effects of photoperiod and temperature on formation and fecundity of two seasonal forms of Psylla pyricola (Homoptera: Psyllidae) . Korean J Appl Entomol. , 35 : 205 – 208 .
- An , HS , Hong , SW , Kim , EM , Lee , J-H , Noh , JK , Kim , HC , Park , CJ , Min , BH and Myeong , J-I. 2010 . Comparative genetic diversity of wild and released populations of Pacific abalone Haliotis discus discus in Jeju, Korea, based on cross-species microsatellite markers including two novel loci . Anim Cells Syst. , 14 : 305 – 313 .
- Avise , JC. 1994 . Molecular Markers, Natural History and Evolution , New York : Chapman and Hall .
- Bae , JS , Kim , I , Kim , SR , Jin , BR and Sohn , HD. 2001 . Mitochondrial DNA sequence variation of the mushroom pest flies, Lycoelia mali (Diptera: Sciaridae), and, Coboldia fuscipes (Diptera: Scatopsidae), in Korea . App Entomol Zool Jpn. , 36 : 451 – 457 .
- Burts , EC. 1970 . The pear psylla in central Washington . Wash Agric Exp Stn Circ. , 516 : 13
- Burts , EC , van den Baan , HE and Croft , BA. 1989 . Pyrethroid resistance in pear psylla, Psylla pyricola Forster (Homoptera: Psyllidae), and synergism of pyrethroids with piperonyl butoxide . Can Entomol. , 121 : 219 – 223 .
- Croft , BA , Burts , EC , van de Baan , HE , Westigard , PH and Riedl , H. 1989 . Local and regional resistance to fenvalerate in Psylla pyricola Foerster (Homoptera: Psyllidae) in western North America . Can Entomol. , 121 : 121 – 129 .
- Excoffier , L , Smouse , PE and Quattro , JM. 1992 . Analysis of molecular variance inferred from metric distances among DNA haplotypes – Application to human mitochondrial DNA restriction data . Genetics. , 131 : 479 – 491 .
- Excoffier , L , Laval , G and Schneider , S. 2005 . Arlequin (version 3.0): an integrated software package for population genetics data analysis . Evol Bioinform Online. , 1 : 47 – 50 .
- Felsenstein , J. 1985 . Confidence limits on phylogenetics: an approach using the bootstrap . Evolution. , 29 : 783 – 791 .
- Fitch , WM. 1971 . Towards defining the course of evolution: minimum change for a specific tree topology . Syst Zool. , 20 : 406 – 416 .
- Follett , PA , Croft , BA and Westigard , PH. 1985 . Regional resistance to pesticides in Psylla pyricola from Oregon pear orchards . Can Entomol. , 117 : 565 – 573 .
- Frankham , R. 1996 . Relationship of genetic variation to population size in wildlife . Conserv Biol. , 10 : 1500 – 1508 .
- Hartl DL , Clark AG . 1989 . Principles of population genetics. , 2nd ed. . Sunderland , MA : Sinauer Associates .
- Hebert , PD , Cywinska , A , Ball , SL and de Ward , JR. 2003 . Biological identification through DNA barcodes . Proc R Soc Lond (B) , 270 : 313 – 321 .
- Holsinger , KE and Mason-Gamer , RJ. 1996 . Hierarchical analysis of nucleotide diversity in geographically structured populations . Genetics. , 142 : 629 – 639 .
- Horton , DR. 1993 . Diurnal patterns in yellow trap catch of pear psylla (Homoptera: Psylla): differences between sexes and morphotypes . Can Entomol. , 125 : 761 – 767 .
- Horton , DR and Lewis , TM . 1996 . Effects of fenoxycarb on ovarian development, spring fecundity and longevity in winterform pear psylla . Entomol Exp Appl. , 81 : 181 – 187 .
- Horton , DR , Higbee , BS , Unruh , TR and Westigard , PH. 1992 . Spatial characteristic and effects of fall density and weather on overwintering loss of pear psylla (Homoptera: Psyllidae) . Environ Entomol. , 21 : 1319 – 1332 .
- Horton , DR , Burts , EC , Unruh , TR , Krysan , JL , Coop , LB and Croft , BA. 1994 . Phenology of fall dispersal by winterform pear psylla (Homoptera: Psyllidae) in relation to leaf fall and weather . Can Entomol. , 126 : 111 – 120 .
- Jeong , H-Y , Kim , D-S , Cho , M-R and Yiem , MS. 2000 . Recent status of major fruit tree pest occurrences in Korea . J Kor Soc Hort Sci. , 41 : 607 – 612 .
- Jeong , HC , Kim , JA , Im , HH , Jeong , HU , Hong , MY , Lee , JL , Han , YS and Kim , I. 2009 . Mitochondrial DNA sequence variation of the swallowtail butterfly, Papilio xuthus, and the cabbage butterfly, Pieris rapae . Biochem Genet. , 47 : 165 – 178 .
- Ji , Y , Zhang , D and He , L. 2003 . Evolutionary conservation and versatility of a new set of primers for amplifying the ribosomal internal transcribed spacer regions in insects and other invertebrates . Mol Ecol Notes. , 3 : 581 – 585 .
- Katoh , K , Misawa , K , Kuma , K and Miyata , T. 2002 . MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform . Nucleic Acids Res. , 30 : 3059 – 3066 .
- Keller , I , Korner-Nievergelt , F and Jenni , L. 2009 . Within-winter movements: a common phenomenon in the Common Pochard Aythya ferina . J Ornithol. , 150 : 483 – 494 .
- Kim , I , Bae , JS , Sohn , HD , Kang , PD , Ryu , KS , Sohn , BH , Jeong , WB and Jin , BR. 2000a . Genetic homogeneity in the domestic silkworm, Bombyx mori, and phylogenetic relationship between B. mori and wild silkworm, B. mandarina, using mitochondrial COI gene sequences . Int J Indust Entomol. , 1 : 9 – 17 .
- Kim , DS , Cho , MR , Jeon , HY , Yiem , MS and Lee , JH. 2000b . Population trends and temperature-dependent development of pear psylla, Cacopsylla pyricola Foerster (Homoptera: Psyllidae) . Korean J Appl Entomol , 39 : 73 – 82 .
- Kim , KY , Jang , SK , Park , DW , Hong , MY , Oh , KH , Kim , KY , Hwang , JS , Han , YS and Kim , I. 2007 . Mitochondrial DNA sequence variation of the tiny dragonfly, Nannophya pygmaea (Odonata: Libellulidae) . Int J Indust Entomol. , 15 : 47 – 58 .
- Kim , HY , Lee , KY , Lee , SB , Kim , SR , Hong , MY , Kim , DY and Kim , I. 2008 . Mitochondrial DNA sequence variation of the mason bee, Osmia cornifrons (Hymenoptera: Apidae) . Int J Indust Entommol. , 16 : 75 – 86 .
- Kim , MJ , Yoon , HJ , Im , HH , Jeong , HU , Kim , MI , Kim , SR and Kim , I. 2009 . Mitochondrial DNA sequence variation of the bumblebee, Bombus ardens (Hymenoptera: Apidae) . J Asia Pacific Entomol. , 12 : 133 – 139 .
- Kimura , M. 1980 . A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences . J Mol Evol. , 16 : 111 – 120 .
- Li , J , Choi , YS , Kim , I , Sohn , HD and Jin , BR. 2006 . Genetic variation of the diamondback moth, Plutella xylostella (Lepidoptera: Yponomeutidae) in China inferred from mitochondrial COI gene sequences . Euro J Entomol. , 103 : 605 – 611 .
- Maruyama , T and Kimura , M. 1980 . Genetic variability and effective population size when local extinction and recolonization of subpopulations are frequent . Proc Natl Acad Sci USA. , 77 : 6710 – 6714 .
- Navajas , M , Lagnel , J , Gutierrez , J and Boursot , P. 1998 . Species-wide homogeneity of nuclear ribosomal ITS2 sequences in the spider mite Tetranychus urticae contrasts with extensive mitochondrial COI polymorphism . Heredity , 80 : 742 – 752 .
- Puillandre , N , Dupas , S , Dangles , O , Zeddam , J-L , Capdevielle-Dulac , C , Barbin , K , Torres-Leguizamon , M and Silvain , J-F. 2008 . Genetic bottleneck in invasive species: the potato tuber moth adds to the list . Biol Invasions. , 10 : 319 – 333 .
- Swofford , DL. 2002 . PAUP* Phylogenetic analysis using parsimony (*and other method) ver. 4.10 , Sunderland , MA : Sinauer Associates .
- Unruh , TR. 1990 . Genetic structure among 18 west coast pear psylla populations: implications for the evolution of resistance . Am Entomol. , 36 : 37 – 43 .
- Tang , J , Toé , L , Back , C and Unnasch , TR. 1996 . Intra-specific heterogeneity of the rDNA internal transcribed spacer in the Simulium damnosum (Diptera: Simuliidae) complex . Mol Biol Evol. , 13 : 244 – 252 .
- Westigard PH , Zwick RW . 1972 . The pear psylla in Oregon . Oregon Stn Agric Bull. 122 .