Abstract
To examine the taxonomic status of Tscherskia triton nestor, which inhabits in Korea, we first obtained partial cytochrome oxidase I (COI) (657 bp) and cytochrome b (426 bp) sequences of T. t. nestor from the Korean Jeju Island and Tscherskia triton fuscipes from northeastern China, and these sequences were compared with the corresponding sequences of Tscherskia triton, available from GenBank. In the COI analysis, T. t. nestor from Jeju was distinct from T. t. fuscipe from northeastern China, with the average Jukes–Cantor distance of 3.45%, and in the cytochrome b analysis, T. t. nestor from Jeju was divergent from T. t. fuscipes from northeastern China and T. t. triton from the North China Plain, with the average distance of 3.88%. We conclude that these sequencing results on the distinctiveness of T. t. nestor from Jeju support the current classification of T. t. nestor from Korea, although further analyses with mitochondrial sequences of T. t. nestor from the mainland Korea and nuclear sequences of T. triton from Korea and China are necessary to confirm our findings. In addition, we considered genetically distinct T. t. triton from Jeju as an allopatric phylogroup, which needs special protection for its conservation. Additionally, we found the lack of cytochrome b sequences between T. t. fuscipes from northeastern China and T. t. triton in the North China Plain, although further systematic analyses to clarify the taxonomic status of T. t. fuscipes are necessary.
Introduction
The greater long-tailed hamster or Korean hamster (Tscherskia triton, de Winton, 1899), composed of 11 nominal subspecies, inhabits from Korea through China (Shensi to Heilongjiang) to Ussuri region, far-eastern Russia (Corbet Citation1978; Musser & Carleton Citation2005). In China, two subspecies (Tscherskia triton fuscipes Allen from northeastern region and T. triton from the North China Plain) inhabit (Zhang et al. Citation1997), and in the mainland Korean Peninsula and the Jeju Island, T. t. nestor Thomas is distributed (Jones & Johnson Citation1960).
Molecular genetic studies for taxonomic reconsideration have become widespread during the past decade, and mitochondrial DNA (mtDNA) is a highly sensitive genetic marker, which is suitable for studies of closely related taxa or populations of a variety of species (Sunnucks Citation2000). The cytochrome b gene sequences have been informative at various taxonomic levels in mammals (Arnason et al. Citation2004), and the cytochrome oxidase I (COI) gene forms the primary barcode sequence to support identification and discovery of a species (Rubinoff Citation2005).
Based on the mitochondrial cytochrome b, 12S rRNA, and nuclear vWF (von Willebrand Fctor) genes, molecular phylogeny of subfamily Cricetinae, including T. triton, was investigated (Neumann et al. Citation2006), and the population structure of T. triton from the North China Plain was examined from the analyses of random amplification of polymorphic DNA pattern (Yongqing et al. Citation2002), control region sequences (Xie & Zhang Citation2005), and microsatellites pattern (Dong et al. Citation2010). However, population study with the COI and cytochrome b genes of T. triton from Korea and northeastern China has not been carried out yet.
To examine the taxonomic status of T. t. nestor from Jeju, we obtained the COI and the cytochrome b sequences of the two subspecies of T. triton (T. t. nestor from the Korean Jeju Island and T. t. fuscipes from northeastern China), and we compared these sequences with the corresponding sequences of T. triton, available from GenBank.
Materials and methods
We used two specimens (2601 and 2602) of T. t. nestor from Moseulpo, the Jeju Island, and Korea, and five specimens of T. t. fuscipes from Heilongjiang, northeastern China (112790 and 112792 from Pingshan, 112942 from the Kelao River, and 112990 and 112992 from Forest park). Small pieces of muscle were collected and preserved in a deep freezer.
Total cellular DNA was extracted using a genomic DNA extraction kit (Intron Co., Seoul, Korea). The COI gene was polymerase chain reaction (PCR)-amplified using the RonM and NancyM primers, designed by Pfunder et al. (Citation2004), and the PCR thermal cycle for COI gene was as follows: 94°C for 5 min, 94°C for 1 min, 53°C for 1 min, 72°C for 1 min (30 cycles), and 72°C for 5 min. The cytochrome b gene was PCR-amplified using the L14724 and H15149 primers, designed by Irwin et al. (Citation1991), and the PCR thermal cycle for cytochrome b was as follows: 94°C for 5 min, 94°C for 1 min, 54°C for 1 min, 72°C for 1 min (30 cycles), and 72°C for 5 min. The amplified products were purified using a DNA PrepMate kit with a silica-based matrix (Intron Co.). The purified PCR products were analyzed with an automated DNA Sequencer (Perkin Elmer 377; Perkin Elmer, Norwalk, CT, USA) at Bioneer Co. (Seoul, Korea).
We first obtained partial COI sequences (657 bp) and cytochrome b sequences (426 bp) of the seven specimens from T. t. nestor and T. t. fuscipes, and the COI sequences were compared with the corresponding four sequences of T. t. triton from the North China Plain (AJ973388, EU031048, EU584001, and EU584098), obtained from GenBank.
Sequence alignment, detection of parsimony informative sites, model selection, calculation of nucleotide distances, and tree constructions with 1000 bootstrapped replications were carried out using MEGA5 (Tamura et al. Citation2011). The Jukes–Cantor (JC) model, which showed the lowest Bayesian information criterion score, was chosen and neighbor joining and maximum likelihood trees were constructed. However, the two trees from the two different methods were congruent, and maximum likelihood trees with the two markers are shown in this paper. For COI analysis, Microtus fortis (JN311714) and M. pennsylvanicus (JN311714) were used as outgroup, and for cytochrome b analysis, M. thomasi (JN019776) and M. agrestis (AF119271) were used as outgroup.
Results
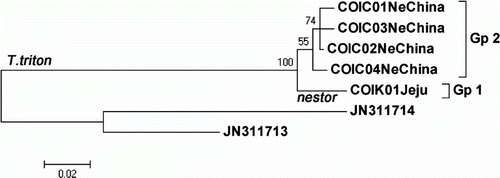
From the seven partial COI sequences (657 bp) of the two T. triton subspecies, one haplotype (COIK01) was identified from the two specimens of T. t. nestor in the Jeju Island, and four haplotypes (COIC01 from specimen 112790, COIC02 from 112792, COIC03 from 112942 and 112990, and COIC04 from 112992) were revealed from the five specimens of T. t. fuscipes in northeastern China (five COI haplotypes of two T. triton subspecies are deposited in GenBank under accession nos. KC152882, JF444259, JF444261, JF444269, and JF444273). A maximum likelihood tree with the five haplotypes of two T. triton subspecies is shown in . The one haplotype of T. t. nestor from Jeju (Gp 1) was different from the other four haplotypes of T. t. fuscipes in northeastern China (Gp 2). The JC distance between the two subgroups (Gps 1 and 2) was 3.45%. In addition, within the five COI haplotypes of T. triton, 29 sites (4.41%) were variable, and eight sites (1.22%) were parsimoniously informative.
Figure 1. Maximum likelihood tree with five partial cytochrome oxidase I (COI) haplotypes (657 bp) of two T. triton subspecies. One haplotype (COIK01) of T. t. nestor from the Korean Jeju Island and four haplotypes (COIC01–COIC04) of T. t. fuscipes from northeastern China were obtained from this study. Location name follows haplotype name in each haplotype, as given in the Materials and methods section, and the bootstrap values>50% are reported at the internodes. Microtuss fortis (JN311714) and M. pennsylvanicus (JN311714) were used as outgroup.
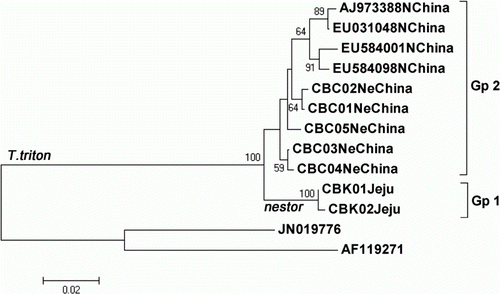
From the seven partial cytochrome b sequences (426 bp), two haplotypes (CBK01 from specimen 2601 and CBK02 from 2602) were identified from T. t. nestor in Jeju, and five haplotypes (CBC01 to CBC05 from specimens 112790 to 112992, respectively) were recognized from T. t. fuscipes in northeastern China (seven cytochrome b haplotypes of two T. triton subspecies are deposited in GenBank under accession nos. KC152875–KC152881). A maximum likelihood tree with the 11 haplotypes of three T. triton subspecies (seven haplotypes of two subspecies from this study and four haplotypes of one subspecies from GenBank) is shown in . The two haplotypes of T. t. nestor from Jeju (Gp 1) were different from the five haplotypes (CBC01 –CBC05) of T. t. fuscipes in northeastern China (Gp 2, in part) and the four haplotypes (AJ973388, EU031048, EU584001, and EU584098) of T. t. triton from the North China Plain (Gp 2, the rest) as well. The JC distance between the two subgroups (Gps 1 and 2) was 3.88%. In addition, within the 11 cytochrome b haplotypes of T. triton, 25 sites (5.87%) were variable, and 18 sites (4.22%) were parsimoniously informative.
Figure 2. Maximum likelihood tree with 11 partial cytochrome b haplotypes (426 bp) of three T. triton subspecies. Two haplotypes (CBK01 and CBK02) of T. t. nestor from the Korean Jeju Island and five haplotypes (CBC01–CBC05) of T. t. fuscipes from northeastern China were obtained from this study, whereas four haplotypes (AJ973388, EU031048, EU584001, and EU584098) of T. t. triton from the North China Plain were obtained from GenBank. Location name follows haplotype name or accession number in each haplotype, as given in the Materials and methods section, and the bootstrap values >50% are reported at the internodes. Microtus thomasi (JN019776) and M. agrestis (AF119271) were used as outgroup.
Discussion
Kim et al. (Citation2010) collected 13 greater long-tailed hamsters (0.47%) among 2742 rodents captured during 2003–2004 from northern Gyeonggi-do in Korea, and Kim et al. (Citation2011) noted that the number of T. s. triton specimen captured during 2001–2005 from Gyeonggi-do was one (0.01%) among 7245 rodents. Thus, T. t. nestor from Korea appears to be very rare, and we have not caught T. t. nestor from the mainland Korea for more than 10 years. However, we have succeeded to catch two T. t. nestor specimens from the Jeju Island and used them for this study because this subspecies is rare in Korea, although the number of samples is two.
In addition, Johnson et al. (Citation2000) noted that the island populations should diverge over time (genetically and morphologically) from the respective mainland species populations, indicating that T. t. nestor from the Korean Jeju Island may or may not be genetically divergent from T. t. nestor in the mainland Korea. We, however, could not catch the greater long-tailed hamster specimens from the mainland Korea, and thus, we compared the sequences of Jeju T. t. nestor to the sequences of other subspecies of T. triton, although in future it is necessary to collect and examine T. t. nestor from the mainland Korea to confirm our findings on T. t. nestor in this paper.
A subspecies is an aggregate of phenetically similar populations of a species differing taxonomically from other populations of that species (Mayr & Ashlock Citation1991), and it was advocated that a classification should reflect all available characters distributed as widely and evenly as possible over the organisms studied (Huelsenbeck et al. Citation1996). Thomas (Citation1907) noted that the size of Cricetulus nestor (=T. t. nestor) from Korea, including the Korean Jeju Island, is larger in morphological characteristics than that of other C. triton from China. In this sequencing study with the COI gene (), T. t. nestor from Jeju (Gp 1) was different from T. t. fuscipes in northeastern China (Gp 2), with an average JC distance of 3.45%, and in this cytochrome b sequence analysis (), T. t. nestor from Jeju (Gp 1) was also different from the other two T. triton subspecies in China (T. t. fuscipes in northeastern China [Gp 2, in part] and T. t. triton from the North China Plain [Gp 2, the rest]), with an average distance of 3.88%. Thus, we conclude that these genetic results on the distinctiveness of T. t. nestor from Jeju support the current classification of T. t. nestor. Additionally, the importance of examining both the nuclear and mitochondrial loci when attempting to elucidate the patterns of genetic structure has been previously mentioned (Rubinoff & Holland Citation2005), and further analyses with nuclear DNA sequences of T. triton from Korea and China are necessary to confirm our findings on T. t. nestor.
Natural barriers to dispersal, which limit species distribution, include mountain range and rivers (Goldberg & Lande Citation2007). The northern boundary of the Korean Peninsula is formed naturally by the Yalu River, the Baitou Mountain (the main peak of the Changbai Mountains in northeastern China, 2744 m above sea level), and the Tumen River, and three other endemic species or subspecies of mammals from Korea have been reexamined by nuclear and mtDNA analyses: Myodes regulus (Koh et al. Citation2011), Lepus coreanus (Koh & Jang Citation2010), Hydropotes inermis argyropus (Koh et al. Citation2009b), and Tamias sibiricus barberi (Koh et al. Citation2009a; Citation2010). From this study ( and ), the endemic T. t. nestor from Jeju (Gp 1) was found to be concordantly divergent from the other two subspecies of T. triton from China (Gp 2).
At the end of the last glacial, large areas of continental shelf were dry land, facilitating an exchange of plant and animal species by land-bridge connections to what are now isolated islands (Lomolino et al. Citation2010), and T. t. nestor is found in the Korean Peninsula, including the Jeju Island (Jones & Johnson Citation1960), indicating that T. t. nestor from the mainland Korea had naturally dispersed into the Jeju Island during or before the last glacial period. Thus, we considered that the divergence of Jeju T. t. nestor was mainly due to the isolation caused by geographical barriers near the northern boundary of the Korean Peninsula, once they dispersed from China through the mainland Korea to the Korean Jeju Island, although it is necessary to examine DNA sequences of the mainland Korean T. t. nestor in order to confirm our results.
In addition, genetically distinct, geographic subdivisions were given the term ‘phylogroups’ (Avise & Walker Citation1999). In this study, we found the genetic distinctness of T. t. nestor from the Jeju Island ( and ) and that T. t. triton from Jeju is a discrete subpopulation and an allopatric phylogroup as well. Pennock and Dimmick (Citation1997) noted that the US Endangered Species Act grants protection to species, subspecies, and ‘distinct population segments.’ Thus, we considered that genetically distinct, endemic T. t. nestor from Jeju needs special protection for its conservation, although the International Union for Conservation of Nature (IUCN) conservation status for all populations of T.triton is in the least concern category (Musser & Carleton Citation2005).
Additionally, from the cytochrome b sequence analysis (), we found that T. t. fuscipes from northeastern China (Gp 2, in part) is not divergent from T. t. triton from the North China Plain (Gp 2, the rest), and we propose further systematic analyses to clarify the taxonomic status of T. t. fuscipes.
References
- Arnason , U , Gullberg , A and Janke , A. 2004 . Mitogenomic analyses provide new insights into cetacean origin and evolution . Gene , 333 : 27 – 34 . doi: 10.1016/j.gene.2004.02.010
- Avise , JC and Walker , D. 1999 . Species realities and numbers in sexual vertebrates: perspectives from an asexual transmitted genome . Proc Nat Acad Sci USA , 96 : 992 – 995 . doi: 10.1073/pnas.96.3.992
- Corbet , GB. 1978 . The mammals of the Palaearctic region: a taxonomic review , 170 London : British Museum (Natural History), Cornell University Press .
- Dong , J , Li , C and Zhang , Z. 2010 . Density-dependent genetic variation in dynamic populations of the greater long-tailed hamster (Tscherskia triton) . J Mamm , 91 : 200 – 217 . doi: 10.1644/09-MAMM-A-098R1.1
- Goldberg , EE and Land , L. 2007 . Species and dispersal barriers . Amer Naturalist , 170 : 297 – 304 . doi: 10.1086/518946
- Huelsenbeck , JP , Bull , JJ and Cunningham , CW. 1996 . Combining data in phylogenetic analysis . Tree , 11 : 152 – 158 .
- Irwin , DM , Kocher , TD and Wilson , AC. 1991 . Evolution of the cytochrome b gene of mammals . J Mol Evol , 32 : 128 – 144 . doi: 10.1007/BF02515385
- Johnson , KP , Adler , FR and Cherry , JL. 2000 . Genetic and phylogenetic consequences of island biogeography . Evolution , 54 : 387 – 396 .
- Jones , JK and Johnson , DH. 1960 . Review of the insectivores of Korea. University of Kansas Publication . Museum Natural History , 9 : 549 – 578 .
- Kim , HC , Yang , YC , Chong , ST , Ko , SJ , Lee , SE , Klein , TA and Chae , JS. 2010 . Detection of Rickettsia typhi and seasonal prevalence of fleas collected from small mammals in the Republic of Korea . J Wildlife Disease , 46 : 165 – 172 .
- Kim , HC , Terry , AK , Kang , HJ , Gu , SH , Moon , SS , Baek , LJ , Chong , ST , O'Guinn , ML , Lee , JS , Turell , MJ and Song , JW. 2011 . Ecological surveillance of small mammals at Dagmar north training area, Gyeonggi Province, Republic of Korea, 2001–2005 . J Vector Ecol , 36 : 42 – 54 . doi: 10.1111/j.1948-7134.2011.00139.x
- Koh , HS and Jang , KH. 2010 . Genetic distinctness of the Korean hare, Lepus coreanus (Mammalia, Lagomorpha), revealed by nuclear thyroglobulin gene and mtDNA control region sequences . Biochem Genet , 48 : 706 – 710 . doi: 10.1007/s10528-010-9353-0
- Koh , HS , Wang , J , Lee , BK , Yang , BG , Heo , SW , Jang , KH and Chun , TY. 2009a . A phylogroup of the Siberian chipmunk from Korea (Tamias sibiricus barberi) revealed from the mitochondrial DNA cytochrome b gene . Biochem Genet , 47 : 1 – 7 . doi: 10.1007/s10528-008-9200-8
- Koh , HS , Lee , BG , Wang , J , Heo , SW and Jang , KH. 2009b . Two sympatric phylogroups of the Chinese water deer (Hydropotes inermis), identified by mitochondrial DNA control region and cytochrome b gene analyses . Biochem Genet , 47 : 860 – 867 . doi: 10.1007/s10528-009-9285-8
- Koh , HS , Zhang , M , Bayarlkhava , D , Ham , EJ , Kim , JS , Jang , KH and Park , NJ. 2010 . Concordant genetic distinctness of the phylogroup of the Siberian chipmunk from the Korean Peninsula (Tamias sibiricus barberi), reexamined with nuclear DNA c-myc gene exon 2 and mtDNA control region sequences . Biochem Genet , 48 : 696 – 705 . doi: 10.1007/s10528-010-9352-1
- Koh , HS , Yang , BK , Heo , SW , Jang , KH and In , ST. 2011 . Genetic distinctiveness of the Korean red-backed vole (Myodes regulus) from Korea, revealed by mitochondrial cytochrome b gene sequences . Biochem Genet , 49 : 153 – 160 . doi: 10.1007/s10528-010-9395-3
- Lomolino , MV , Riddle , BR , Whittaker , RJ and Brown , JH. 2010 . Biogeography , 4th ed , 1 – 878 . Sunderland , MA : Sinauer Associates Inc .
- Mayr , E and Ashlock , PD. 1991 . Principles of systematic zoology , 1 – 475 . New York , NY : McGraw-Hill Inc .
- Musser , GG and Carleton , MD. 2005 . “ Order Rodentia ” . In Mammal species of the world: a taxonomic and geographic reference , 3rd ed , Edited by: Wilson , DE and Reeder , DM . 1046 Baltimore : Jones Hopkins University Press .
- Neumann , K , Michaux , J , Lebedev , V , Yight , N , Colak , E , Ivanova , N , Poltoraus , A , Surov , A , Markov , G Maak , S . 2006 . Molecular phylogeny of the Cricetinae subfamily based on the mitochondrial cytochrome b and 12S rRNA genes and the nuclear vWF gene . Mol Phylogenet Evol , 39 : 135 – 148 . doi: 10.1016/j.ympev.2006.01.010
- Pennock , DS and Dimmick , WW. 1997 . Critique of the evolutionary significant unit as a definition for ‘distinct population segments’ under the U.S. Endangered Species Act . Conservation Biol , 11 : 611 – 619 . doi: 10.1046/j.1523-1739.1997.96109.x
- Pfunder , M , Holzgang , O and Frey , JE. 2004 . Development of microarray-based diagnostic of voles and shrews for use in biodiversity monitoring studies, and evaluation of mitochondrial cytochrome oxidase I vs. cytochrome b as genetic markers . Mol Ecol , 13 : 1277 – 1286 . doi: 10.1111/j.1365-294X.2004.02126.x
- Rubinoff , D. 2005 . Utility of mitochondrial DNA barcodes in species conservation . Conservation Biol , 20 : 1026 – 1033 . doi: 10.1111/j.1523-1739.2006.00372.x
- Rubinoff , D and Holland , BS. 2005 . Between two extremes: mitochondrial DNA is neither the panacea nor the nemesis of phylogenetic and taxonomic inference . Syst Biol , 54 : 952 – 961 . doi: 10.1080/10635150500234674
- Sunnucks , P. 2000 . Efficient genetic markers for population biology . TREE , 15 : 199 – 203 .
- Tamura , K , Peterson , D , Peterson , N , Stecher , G , Nei , M and Kumar , S. 2011 . MEGA5: molecular evolutionary genetic analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods . Mol Biol Evol , 28 : 2731 – 2739 . doi: 10.1093/molbev/msr121
- Thomas , O. 1907 . The Duke of Bedford's zoological explorations in eastern Asia. V. Second list of mammals from Korea . Proc Zool Soc London , 77 : 464
- Xie , J and Zhang , Z. 2005 . Mitochondrial and phylogeography of populations of Cricetulus triton in the North China Plain . J Mamm , 86 : 833 – 840 . doi: 10.1644/1545-1542(2005)086[0833:MDPOPO]2.0.CO;2
- Yongqing , W , Zhang , Z and Laixiang , X. 2002 . The genetic diversity of central and peripheral populations of ratlike hamster (Cricetulus triton) . Chinese Sci Bulletin , 47 : 201 – 206 . doi: 10.1360/02tb9048
- Zhang , Y , Sangke , J , Guoqiang , J , Shibau , L , Zhongyao , Y , Genggui , W and Manli , Z. 1997 . Distribution of mammalian species in China , 227 – 233 . Beijing : China Forestry Publishing House .