Abstract
The objective of this study is to elucidate the evolutionary rate of Cryptocercus SSU rDNAs based on the relative rate test (RRT) and comparative analysis of the primary sequences. In the RRTs, the evolutionary rate differences (dKs) of the Cryptocercus SSU rDNAs were not statistically significant (0.107 <P<0.996) in all cases, although an Asian Cryptocercus appeared to evolve more slowly than a North American Cryptocercus as indicated by the negative signs of dKs. Contrary to the RRT, the comparative analysis of SSU rDNA sequences revealed some interesting aspects in the evolutionary rates of Cryptocercus SSU rDNAs. In comparison with those of closely (Mastotermes darwiniensis and Tenodera aridifolia) or more distantly related taxa (Locusta migratoria), the SSU rDNAs of the genus Cryptocercus showed that insertions are progressing in a variable region (HV2). The SSU rDNA of Calymmaderus punctulatus species complex (Mountain Lake Biological Station, VA), a North American species, was unusually more elongated in the HV2 than those of the reference taxa or the other congeneric species.
Introduction
Wood-feeding Cryptocercus cockroaches, which live in high mountainous forests, have been well known because of the peculiar characteristics of their life history and geographic distribution. Cryptocercus adults live with their offspring over an extended period of 3 years and provide their young with proctodeal feeding (Seelinger & Seelinger Citation1983; Nalepa Citation1984; Park & Choe Citation2003a, Citation2003b).
Cryptocercus species have been found only in a few forests of the Palearctic and Nearctic regions (Scudder Citation1862; Bey-Bienko Citation1950; Nalepa et al. Citation1997, Citation2001; Burnside et al. Citation1999; Grandcolas Citation1999a, Citation1999b, Citation2000; Grandcolas et al. Citation2001; Park et al. Citation2004). In Nearctic regions, they show a disjunct distribution in the USA; Cryptocercus clevelandi is distributed in the Coast and Cascade Mountains of northern California and southwestern Oregon (Nalepa et al. Citation1997), while Cryptocercus punctulatus species complex, also described as Cryptocercus darwini, Cryptocercus garciai, Cryptocercus punctulatus and Cryptocercus wrighti by Burnside et al. (Citation1999), is reported to occur along the Appalachian and Allegheny Mountains (Burnside et al. Citation1999; Nalepa et al. Citation2002). In Palearctic regions, Cryptocercus has been reported in only a few forests of Southwest China and Northeast Asia including Korea, Manchuria and Siberia (Bey-Bienko Citation1950; Grandcolas Citation2000; Grandcolas et al. Citation2001; Nalepa et al. Citation2001; Park et al. Citation2004).
Since they are wingless and live only in rotten logs, their migration appears to be restricted to local boundary zones. Thus, the causes and dynamics that shape their present-day distribution have been the subject of vigorous discussion over the last decade, associated with their taxonomic status as well as phylogeography (Burnside et al. Citation1999; Nalepa & Bandi Citation1999; Grandcolas Citation1999a, Citation1999b; Clark et al. Citation2001; Hossain & Kambhampati Citation2001; Nalepa et al. Citation2002; Park et al. Citation2004).
In addition, to assess inter- and/or intra-specific genetic variation, or to better understand the spatiotemporal history of the geographic isolates, studies using mitochondrial DNA markers have been conducted extensively for Cryptocercus species, especially for the Nearctic cockroaches (Burnside et al. Citation1999; Clark et al. Citation2001; Hossain & Kambhampati Citation2001). In karyotype analysis, four variants (male 2n=37, 39, 43, 45) were found in C. punctulatus species complex in the southern Appalachian Mountains of eastern North America, and none of the variants occurs in the same locality, displaying different distribution range (Burnside et al. Citation1999; Nalepa et al. Citation2002).
According to two evolutionary scenarios of the variants suggested by Nalepa et al. (Citation2002), the karyotype variants might have evolved independently from an ancestral population without having sequential events to one another (parallel scenario), or they might have been decreased sequentially from the ancestral karyotype (sequential scenario). As in the recent studies using karyotype and mtDNA markers (Everaerts et al. Citation2008), further molecular survey for these geographic isolates may provide clues to better understanding of the evolutionary trend of the genus Cryptocercus related to the molecular sequence evolution.
In this paper, we used nearly a complete sequence of small subunit nuclear ribosomal DNA (SSU rDNA or 18S rDNA) to infer the evolutionary trend of Cryptocercus. The 18S rDNA sequence has been generally known to include both very highly conserved and variable regions in their sequences. In the conserved regions high level of sequence identity is observed even among distantly related taxa (Turbeville et al. Citation1991; Whiting et al. Citation1997), while nucleotide insertions and deletions (indels) and/or nucleotide substitution are often found in the relatively fast-evolving variable regions even among closely related taxa (Hancock Citation1995).
These features make the utility of SSU rDNA as one of the most widely used molecular markers in multidisciplinary studies of the evolutionary trend in the nucleotide sequence and phylogenetic relationships among both closely and more distantly related taxa (Vogler et al. Citation1997; Whiting et al. Citation1997). In this study, we analysed primary structures of SSU rDNA sequences from Northeast Asian and North American Cryptocercus. Based on the comparative analyses of the sequences, we discussed the evolutionary trends between Cryptocercus of Northeast Asia and eastern North America.
Materials and methods
Sample collection
This study includes C. punctulatus species complex from the USA and two species (Cryptocercus kyebangensis and Cryptocercus relictus) from Northeast Asia. We collected C. kyebangensis from Mt. Yongmun in South Korea, C. relictus from Vladivostok in eastern Russia and C. punctulatus species complex from the Mountain Lake Biological Station (MLBS), Virginia in eastern North America. Although SSU rDNA has been known to be highly conserved at within-species level, the target gene fragments for two individuals from each locality were sequenced in order to detect potential sequence variation within the species.
DNA extraction, amplification, purification and sequencing
Genomic DNA was extracted from legs of the samples preserved in 100% ethanol using AccuPrep Genomic DNA Extraction Kit (Bioneer). Nearly complete sequences of 18S gene were amplified using polymerase chain reaction (PCR). Using a Gene®Amp PCR system 9700 (Perkin-Elmer), the polymerase chain reaction was carried out in 20 µl volumes, using 2 µl of genomic DNA as template. The PCR reactions included 10 pmol of each primer, dNTPs at 250 µM, 2.5 mM MgCl2 and 1 unit of Taq Polymerase (Promega). Primers used for the amplification of SSU rDNA sequences are as follows: (18e) 5′-CTGGTTGATCCTGCCAGT-3′ (Hillis & Dixon Citation1991) and (18P) 5′-TAATGATCCTTCCGCAGGTTCACCT-3′ (Halanych et al. Citation1998). The temperature profile for amplifying target gene sequence was as follows: a denaturation at 94°C for 3 min and 30 cycles of 94°C for 1 min, 58°C for 90s and 72°C for 2 min followed by a final extension of 72°C for 10 min. PCR products were electrophoresed on a 1% agarose gel and purified using Cleanmix DNA Purification Kit (TA 200 CLN). Purified products were used as templates for sequencing. Sequencing reactions were performed using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer), according to the manufacturer's instructions, and a Gene®Amp PCR system 9700 (Perkin-Elmer). Sequences were determined by automatic sequencing on a 3730 DNA Sequencer (ABI) through 8% polyacrylamide gel. Both complementary strands were sequenced for all individuals. All of the sequences obtained in the current study are deposited at the Genbank under the accession numbers of JX091733 for C. relictus, JX091734 for C. kyebnagnesis and JX091735 for C. punctulatus species complex (MLBS).
Data analysis
For comparative sequence analysis, sequences of SSU rDNAs were aligned using Clustal X program package (Thompson et al. Citation1997) (Supplementary 1). As references for comparative analysis of the aligned Cryptocercus sequences, we used previously published sequences of Mastotermes darwiniensis (AF220568; Genbank accession numbers) (Isoptera) (Lo et al. Citation2000) and Tenodera aridifolia (AF423805) (Mantodea) (Whiting Citation2002) closely related to the genus Cryptocercus and that of Locusta migratoria (AF370793) (Orthoptera) (Giribet et al. Citation2001) distantly related to the genus. We also used a previously published sequence of C. punctulatus species complex (AF220571) (Lo et al. Citation2000) to compare with C. punctulatus species complex (MLBS).
The relative rate test (RRT) was conducted to estimate relative rates of nucleotide substitution of the SSU rDNAs sequences using the RRTree software (Relative Rate Test with Tree; Version 1.1) (Robinson & Huchon Citation2000). For the RRT, the SSU rDNA target sequences of five taxa were included as follows: C. relictus, C. kyebangensis, C. punctulatus species complex (AF220571), C. punctulatus species complex (MLBS) and Tenodera aridifolia (outgroup). The RRT has often been employed to determine whether the observed substitution rates are constant by comparing two sequences relative to the third as an outgroup (Sarich & Wilson Citation1973; Wu & Li Citation1985).
According to Dictyopteran phylogeny, cockroaches (Blattaria) and termites (Isoptera) are a sister and mantids (Mantodea) are positioned basal to the two former groups (). Thus, for the RRT of the genus Cryptocercus, we used T. aridifolia (AF423805) (Mantodea) (Whiting Citation2002) as an outgroup.
Figure 1. Phylogeny of the taxa used in this study. The phylogenetic tree was a modification of the tree that Lo et al. (Citation2000) inferred from 18S r DNA and COII. Each species of Isoptera, Mantodea and Orthoptera was used as references for comparative analysis of nucleotide sites in the aligned sequences of the genus Cryptocercus. T. aridifolia was used as an outgroup in the relative rate test.

In the RRT, substitution rates were corrected by Kimura 2-parameter model (Kimura Citation1980) implemented in the RRTree software. SPSS (ver. 17) package was used for general statistical analyses of base composition frequencies.
Results
Sequence alignment
We obtained nearly complete SSU rDNA sequences for each of the two Cryptocercus individuals from South Korea, eastern Russia and eastern North America, respectively. There was no individual variation in SSU rDNA sequences for Crypotocercus collected from these localities. Thus, one SSU rDNA genotype representing each collection locality was used for further analyses. The aligned SSU rDNA sequences with reference taxa were 1772 nucleotides in length (Supplementary 1).
There were three relatively well-conserved regions (at the sites 1–140, 157–642 and 771–1772) across the aligned sequences, whereas two variable regions with many indels were found at the sites 141–156 (HV1) and 643–770 (HV2) covering 144 sites in their total length (Figure 2 and Supplementary 1).
Nucleotide composition
Base composition was homogenous across the taxa (Chi-square test; 0.77<P<1) (). In reference sequences, the chi-square values of SSU rDNAs were higher in the conserved regions (2.03) than in the variable regions (0.74), while in Cryptocercus, the values were higher in the variable regions (5.67) than in the conserved regions (0.07).
Table 1. Chi-square test of homogeneity of base frequencies across taxa.
In the entire regions of SSU rDNAs, the GC contents were a little higher than that of AT in both reference and Cryptocercus sequences (Mann–Whitney U; P<0.05) (). The GC contents were also a little higher than that of AT in both conserved regions and variable regions (Mann–Whitney U; P<0.05 in both cases). Average GC/AT ratio was significantly higher in the variable regions (2.79±0.34) than in the conserved region (1.11±0.03) (Mann–Whitney U; P<0.05).
Table 2. Base composition of the whole in the regions of SSU rRNA sequences.
Variable regions
In the HV1, the SSU rDNAs of Cryptocercus showed a high degree of length variability by insertions and deletions (). In comparison with those of reference taxa, SSU rDNAs of Cryptocercus were inserted by an average of 7.2 nucleotides (±3.0, n=12) in the HV1 and an average of 31.8 nucleotides (±20.2, n=12) in the HV2, respectively. One of the most interesting results is that the HV2 region of C. punctulatus species complex (MBLS) was exceptionally more elongated than that of any other congeneric members or reference taxa. It was 35.0 (±2.6, n=3, range=33–38) nucleotides longer than that of the other Cryptocercus and 58.0 (±14.5, n=3, range=44–73) longer than that of the references.
Figure 2. HV1 (at the sites 141–156) and HV2 (at the sites 643–770) in the SSN rRNA sequences of the genus Cryptocercus aligned with those of references (L. migratoria, T. aridifolia and M. darwinensis). An unusual expansion by insertion occurs in the HV2 of the SSU rRNA of C. punctulatus species complex (MLBS). Numbers above the alignment indicate the positions of the nucleotide sites in the full sequence alignment (see Supplementary 1).
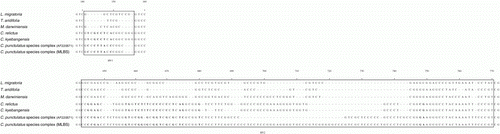
In all HV1 and HV2 (), with the exception of the difference in gap number, there was no nucleotide substitution between Asian C. relictus and C. kyebangensis and only one substitution at the site 759 (transition; A↔T) between North American C. punctulatus species complex (AF220571) and C. punctulatus species complex (MLBS), while a total of 26 substitutions (6 sites in the HV1 and 20 sites in the HV2) were detected between the Asian and North American Cryptocercus (shown in boldface nucleotides in ).
Conserved regions
In three relatively well-conserved regions (1–140, 157–642 and 771–1772 sites), excluding the variable regions, four substitutions were found (three transitions and one transversion) between two Asian Cryptocercus species, but no substitution between two individuals of North American C. punctulatus species complex. Substitutions occurred in all 13 sites (transition at five sites and transversion at eight sites) between Asian and North American Cryptocercus.
Nucleotide sequences at 17 variable sites detected in the conserved regions can distinguish the genus Cryptocercus from the reference taxa. Nucleotide sequences at these variable sites were highly conserved in all reference taxa (no substitutions detected), but substitutions occurred only in the genus Cryptocercus (). Seven of 17 sites were substitutions specific to all species of the genus Cryptocercus (as indicated in the boxes in ). Substitutions between Asian Cryptocercus (C. relictus and C. kyebangensis) occurred in only two sites (at sites 3 and 786), while there was no substitution between the two individuals of North American C. punctulatus species complex. Substitutions between Asian and North American Cryptocercus species occurred in an average of 14 sites (±1.4, n=2).
Figure 3. The selected 17 sites whose nucleotides were well conserved in the SSU rDNAs of references, but substituted in those of the genus Cryptocercus. Bold nucleotides in the sites of the genus Cryptocercus SSN rRNAs indicate that they were different from those in the corresponding sites of the references SSU rRNAs. The boxed sites in the genus Cryptocercus indicate that at the sites the nucleotides were different from those of references, but same within the genus.

C. relictus was substituted in 10 of the 17 sites. In case of C. kyebangnesis substitution occurred in 12 sites. Thus, the substitution between two Asian Cryptocercus and the references occurred on average in 11 sites (±1.4, n=2), while the substitution between North American C. punctulatus species complex and the references occurred in total 13 sites.
Relative rate test
For the RRT, the data-set is composed of the entire sequences (1772 nucleotide sites) including both conserved and variable regions (HV1 and HV2). However, total nucleotide sites used in the RRT were 1675 of the 1772 sites, probably since all sites with gaps were excluded in the RRT using the RRTree software (Robinson & Huchon Citation2000).
Results from the RRT showed that the differences (dKs=K ar −K ap , K ar −K am , K ak −K ap and K ak −K am ) of the substitution rate (K1 and K2; substitution rates between Asian Cryptocercus and T. aridifolia and between Northeast American Cryptocercus and T. aridifolia, respectively) were all negative signs, indicating a slower substitution rate in Asian than in North American Cryptocercus (). The substitution rate of C. relictus was higher than that of C. kyebangensis since the difference of substitution rates between each of two Asian Cryptocercus and T. aridifolia (dK=K ar −K ak ) has a positive value. The difference of substitution rates (dK=K ap −K am ) between each of two individuals of C. punctulatus species complex and T. aridifolia was a negative sign, implying a slower substitution rate in C. punctulatus species complex (AF220571) than C. punctulatus species complex (MLBS).
Discussion
The Cryptocercus SSU rNDAs were not different from the general features that have ever been known from many metazoan animals, in that they have both slowly evolving conserved regions and fast evolving variable regions with relatively higher GC content (Tautz et al. Citation1988; Hancock Citation1995; Crease & Colbourne Citation1998; Choe et al. Citation1999). The comparative analysis of SSU rDNA sequences provided insights into evolutionary trends of the genus Cryptocercus. Length variations in aligned SSU rDNA sequences became much more prominent in the hypervariable regions, especially in the HV2. The unexpected expansion in the region occurred most remarkably in C. punctulatus species complex (MLBS) but not in the other Cryptocercus species. More insertions were found in the HV2 (an average of 35 nucleotides) of C. punctulatus species complex (MLBS) than that of Cryptocercus punctulatus species complex (AF220571). The high GC content in this variable region indicates that length expansion was caused mainly by insertions of G and C. With the exclusion of the insertion part, only a single transitional change was detected between HV2 of C. punctulatus species complex (MLBS) and C. punctulatus species complex (AF220571) (: A↔T in the site 759 in HV2).
Comparative sequence analysis of the conserved regions showed a peculiar substitution patterns in a total of the 17 sites () where nucleotide sequences are the same all across reference taxa, but some substitutions were found between the Cryptocercus species. Among the 17 sites, seven were transitional substitutions specific to all Cryptocercus species, leading us to conjecture that these sites may be derived characters to distinguish the genus Cryptocercus from the reference taxa. In the conserved 17 sites, nucleotides were less substituted in Asian than in North American Cryptocercus, relative to reference sequences.
In the RRT, the substitution rates did not represent statistically significant differences in all cases, although the Asian Cryptocercus tend to show slower substitution rate than the North American Cryptocercus as indicated by the negative signs of dKs (). The homogeneity test of base frequencies also showed that the sequences of SSU rDNAs have evolved with the same rate of nucleotide substitution in their evolutionary history (homogeneity assumption, 0.77 <P <1 in ). According to the molecular clock hypothesis (Zuckerkandl & Pauling Citation1965), substitutions occur at the same rate for all across the sites in closely related species. As in the RRT as well as the homogeneity test, thus, the substitution rate of Cryptocercus SSU rDNAs might not show statistically significant difference, probably because the sequences compared were too closely related to each other.
Table 3. RRT of the aligned 1675 sites excluding gaps in the entire sites of SSU rDNA sequences.
In conclusion, nucleotides of the conserved sites were less substituted in Asian than in North American Cryptocercus, relative to reference sequences, perhaps suggesting that the Asian Cryptocercus might be more slowly evolved than the North-east American Cryptocercus. This interpretation is well in congruence with earlier arguments on the Dictyopteran phylogeny that Asian C. relictus is positioned basal to the North American Cryptocercus species (Clark et al. Citation2001; Lo et al. Citation2003). However, further examination based on different genetic markers and more extensive samplings is required to elucidate the evolutionary rate of the genus Cryptocercus.
Acknowledgements
We would like to thank Dr Christine A. Nalepa (Department of Entomology, North Carolina State University) very much for providing the specimen of North American Cryptocercus. This work was carried out with the support of ‘Forest Science & Technology Projects (Project No. S211012L030110)’ provided by Korea Forest Service.
References
- Bey-Bienko G. 1950 . Blattodea. In: Faune de I New Series 40 . St. Petersburg : Akad. Nauk. SSSR ; 332 – 336 . (In Russian) .
- Burnside , CA , Smith , PT and Kambhampati , S. 1999 . Three new species of the woodroach, Cryptocercus (Blattodea: Cryptocercidae), from the eastern United States . J Kansas Entomol. Soc , 72 : 361 – 378 .
- Choe , CP , Hancock , JM , Hwang , UW and Kim , W. 1999 . Analysis of the primary sequence and secondary structure of the unusually long SSN rRNA of the soil bug, Armadillidium vulgare . J Mol Evol , 49 : 798 – 805 . doi: 10.1007/PL00006602
- Clark , JW , Hossain , S , Burnside , C and Kambhampati , S. 2001 . Coevolution between a cockroach and its bacterial endosymbiont: a biogeographical perspecitve . Proc R Soc Lond. B , 268 : 393 – 398 . doi: 10.1098/rspb.2000.1390
- Crease , TJ and Colbourne , JK. 1998 . The unusually long small-subunit ribosomal RNA of the Crustacean, Daphnia pulex: sequence and predicted secondary structure . J Mol Evol , 46 : 307 – 313 . doi: 10.1007/PL00006307
- Everaerts , C , Maekawa , K , Farine , JP , Shimada , K , Luykx , P , Brossut , R and Nalepa , CA. 2008 . The Cryptocercus punctulatus species complex (Dictyoptera: Cryptocercidae) in the eastern United States: comparsion of cuticular hydrocarbons, chromosome number, and DNA sequences . Mol Phylogenet Evol , 47 : 950 – 959 . doi: 10.1016/j.ympev.2008.03.011
- Giribet , G , Edgecombe , GD and Wheeler , WC. 2001 . Arthropod phylogeny based on eight molecular loci and morphology . Nature , 413 : 157 – 161 . doi: 10.1038/35093097
- Grandcolas , P. 1999a . Systematics, endosymbiosis and biogeography of Cryptocercus clevelandi and C. punctulatus (Blattaria: Polyphagidae) from North America: a phylogenetic perspective . Ann. Entomol Soc Am , 92 : 285 – 291 .
- Grandcolas , P. 1999b . Reconstructing the past of Cryptocercus (Blattaria: Polyphagidae): phylogenetic histories and stories . Ann. Entomol Soc Am , 92 : 303 – 307 .
- Grandcolas , P. 2000 . Cryptocercus matilei n.sp., du Sichuan de Chine [Sichuan of China] (Dictyoptera, Blattaria, Polyphaginae) . Revue Fr Ent , 22 : 223 – 226 .
- Grandcolas , P , Park , YC , Choe , JC , Piulachs , MD , Belles , X , D'Haese , C , Farine , JP and Brossnt , R. 2001 . What does reveal Cryptocercus kyebangensis, n. sp. from South Korea about Cryptocercus evolution? A study in morphology, molecular phylogeny and chemistry of tergal glands (Dictyoptera, Blattaria, Polyphagidae) . Proc Acad Nat Sci Phila , 151 : 61 – 79 . doi: 10.1635/0097-3157(2001)151[0061:WDCKNS]2.0.CO;2
- Halanych , KM , Lutzr , A and Vrijenhoek , RC. 1998 . Evolutionary origins and age of vestimentiferan tubeworms . Cah Biol Mar , 39 : 355 – 358 .
- Hancock , JM. 1995 . The contribution of DNA slippage to eukaryotic nuclear 18S rRNA evolution . J Mol Evol , 40 : 629 – 639 . doi: 10.1007/BF00160511
- Hillis , DM and Dixon , MT. 1991 . Ribosomal DNA: molecular evolution and phylogenetic inference . Q Rev Biol , 66 : 411 – 453 . doi: 10.1086/417338
- Hossain , S and Kambhampati , S. 2001 . Phylogeny of Cryptocercus species (Blattodea: Cryptocercidae) inferred from nuclear ribosomal DNA . Mol Phylogent Evol , 21 : 162 – 165 . doi: 10.1006/mpev.2001.1000
- Kimura , M. 1980 . A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences . J Mol Evol , 16 : 111 – 120 . doi: 10.1007/BF01731581
- Lo , N , Bandi , C , Watanabe , H , Nalepa , C and Beninati , T. 2003 . Evidence for cocladogenesis between diverse dictyopteran lineages and their intracellular endosymbionts . Mol Biol Evol , 20 : 907 – 913 . doi: 10.1093/molbev/msg097
- Lo , N , Tokuda , G , Watanabe , H , Rose , H , Slaytor , M , Maekawa , K , Bandi , C and Noda , H. 2000 . Evidence from multiple gene sequences indicates that termites evolved from wood-feeding cockroaches . Curr Biol , 10 : 801 – 804 . doi: 10.1016/S0960-9822(00)00561-3
- Nalepa , CA. 1984 . Colony composition, protozoan transfer and some life history characteristics of the woodroach Cryptocercus punctulatus Scudder (Dictyoptera: Cryptocercidae) . Behav Ecol Sociobiol , 14 : 273 – 279 . doi: 10.1007/BF00299498
- Nalepa , CA , Byers , GW , Bandi , C and Sironi , M. 1997 . Description of Cryptocercus clevelandi (Dictyoptera: Cryptocercidae) from the northwestern United States, molecular analysis of bacterial symbionts in its fat body, and notes on biology, distribution, and biogeography . Ann Entomol Soc Am , 90 : 416 – 424 .
- Nalepa , CA and Bandi , C. 1999 . Phylogenetic status, distribution and biogeography of Cryptocercus (Dictyoptera: Cryptocercidae) . Ann Entomol Soc Am , 92 : 292 – 302 .
- Nalepa , CA , Li , L , Li , WH and Lazell , J. 2001 . Rediscovery of the wood-eating cockroach Cryptocercus primarius (Dictyoptera: Cryptocercidae) in China, with notes on ecology and distribution . Acta Zootaxonom Sin , 26 : 184 – 190 .
- Nalepa , CA , Luykx , P , Klass , KD and Deitz , LL. 2002 . Distribution of karyotypes of the Cryptocercus punctulatus species complex (Dictyoptera: Cryptocercidae) in the southern Appalachians: relation to habitat and history . Ann Entomol Soc Am , 95 : 276 – 287 . doi: 10.1603/0013-8746(2002)095[0276:DOKOTC]2.0.CO;2
- Park , YC and Choe , JC. 2003a . Life history and population dynamics of Korean woodroach (Cryptocercus kyebangensis) populations . Korean J Biol Sci , 7 : 111 – 117 . doi: 10.1080/12265071.2003.9647691
- Park , YC and Choe , JC. 2003b . Effects of parental care on offspring growth in the Korean wood-feeding cockroach, Cryptocercus kyebangensis . J Ethol , 21 : 71 – 77 .
- Park , YC , Maekawa , K , Matsumoto , T , Santoni , R and Choe , JC. 2004 . Molecular phylogeny and biogeography of the Korean woodroaches Cryptocercus spp . Mol Phylogenet Evol , 30 : 450 – 464 . doi: 10.1016/S1055-7903(03)00220-3
- Robinson-Rechavi , M and Huchon , D. 2000 . RRTree: relative-rate tests between groups of sequences on a phylogenetic tree . Bioinformatics , 16 : 296 – 297 . doi: 10.1093/bioinformatics/16.3.296
- Sarich , VM and Wilson , AC. 1973 . Generation time and genomic evolution in primates . Science , 179 : 1144 – 1147 . doi: 10.1126/science.179.4078.1144
- Scudder , SH. 1862 . Materials for a monograph of North American Orthoptera . Boston J Nat Hist , 7 : 409 – 480 .
- Seelinger , G and Seelinger , U. 1983 . On the social organization, alarm and fighting in the primitive cockroach Cryptocercus punctulatus Scudder . Z Tierpsychol , 61 : 315 – 333 .
- Tautz , D , Hancock , JM , Webb , DA , Tautz , C and Dover , GA. 1988 . Complete sequences of the rRNA genes of Drosophila melanogaster . Mol Biol Evol , 5 : 366 – 376 .
- Thompson , JD , Gibson , TJ , Plewniak , F , Jeanmougin , F and Higgins , DG. 1997 . The Clustal X windows interface: flexible strategies of multiple sequence alignment aided by quality analysis tools . Nucleic Acids Res , 25 : 4879 – 4882 . doi: 10.1093/nar/25.24.4876
- Turbeville , JM , Pfeifer , DM , Field , KG and Raff , RA. 1991 . The phylogenetic status of arthropods, as inferred from 18S rRNA sequences . Mol Biol Evol , 8 : 669 – 686 .
- Vogler , AP , Welsh , A and Hancock , JM. 1997 . Phylogenetic analysis of slippage-like sequence variation in the V4 rRNA expansion segment in tiger beetles (Cicindelidae) . Mol Biol Evol , 14 : 6 – 19 . doi: 10.1093/oxfordjournals.molbev.a025703
- Whiting , MF. 2002 . Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera . Zool. Scr , 31 : 93 – 104 . doi: 10.1046/j.0300-3256.2001.00095.x
- Whiting , MF , Carpenter , JC , Wheeler , QD and Wheeler , WC. 1997 . The strepsiptera problem: phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology . Syst Biol , 46 : 1 – 68 .
- Wu , CI and Li , WH. 1985 . Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci . USA , 82 : 1741 – 1745 . doi: 10.1073/pnas.82.6.1741
- Zuckerkandl , E and Pauling , L. 1965 . “ Evolutionary divergence and convergence in proteins ” . In Evolving genes and proteins , Edited by: Bryson , V and Vogel , HJ . 97 – 166 . New York : Academic Press .