Abstract
Ketosis is a condition where ketone bodies are used as the energy source instead of glucose. The state of ketosis becomes dangerous if the fat metabolism in the body is high. When the glucose availability gets limited, liver activates fat metabolism to make glucose for energy. As the results of these, ketone bodies will form resulting the state of ketosis. In the present study, we analyzed the in vitro ketosis condition and the effect of glucose level in ketosis induction. The natural organic sulfur containing compound methylsulfonylmethane (MSM) was analyzed for anti-ketosis activity in normal mouse hepatocyte, FL83B cell model system. Ketosis condition was induced by modulating glucose concentration. Growth hormone receptor (GHR) has a great role in the ketosis condition. We hypothesize that MSM has the ability to regulate GHR signaling through signal transducer and activator of transcription 5b (STAT5b). The whole cell lysate analysis and DNA binding activity analysis also suggested the critical role of STAT5b in ketosis condition. Angiopoietin-like 3 (ANGPTL3) protein is a molecular target to treat ketosis. STAT5b is a transcription factor for ANGPTL3. We elucidated the treatment of ketosis through the control of molecular targets such as ANGPTL3, STAT5b, and GHR by MSM, and we also evaluated the relation between ANGPTL3 and STAT5b signaling molecules as critical regulators.
Keywords:
Introduction
Most of the energy in the body is provided by blood glucose by means of glycolysis. However, when liver glucogen stores are depleted, the system tends to switch on the fat metabolism. As a result, ketone bodies are formed by ketogenesis. Thereafter, most of the body’s energy comes from ketone bodies in a metabolic state known as ketosis. If the body is said to be in ketosis condition, then the concentration of ketone bodies in serum is over 0.5 mM with low and consistent blood glucose level. In hyperketonemia condition, there is an increased level of ketone bodies. The ketone bodies that take part in the energy formation are acetate, acetoacetate, and β-hydroxybutyrate (BHB; Laffel Citation1999) where insulin and glucagon regulate their level (Johnston et al. Citation1982). The state of ketosis is more related with carbohydrate and fat metabolism. When the body is in fasting or running with low glucose, the metabolism shifts to ketosis until the body gets a carbohydrate-rich meal to change the metabolism to glycolysis. Ketosis is considered to be in a fat-burning mode because fat reserves are readily released and consumed in ketosis.
High level of ketosis is very dangerous, as it makes the blood more acidic and affects the normal functioning of kidney and liver. The regulation of growth hormone signaling occurs via growth hormone receptor (GHR) which is found to have a critical role in subclinical ketosis (Loor et al. Citation2007). As found by Loor et al., the expression of GHR should downregulate with ketosis. Signal transducer and activator of transcription 5b (STAT5b) is an essential factor for the cell growth (Nosaka et al. Citation1999) which is a transcription factor for GHR, and the expression patterns of these molecules is found to be the same. Henceforth, the ketosis state also affects the level of STAT5b. Angiopoietin-like 3 (ANGPTL3), which is considered as a pharmacological target for treatment ketosis, can take part in the metabolic disorder of lipid metabolism and carbohydrate metabolism by suppressing lipoprotein lipase (Xiao et al. Citation2012). STAT5b is a transcription factor for ANGPTL3, and the relationship between ANGPTL3 and STAT5b in ketosis can be elucidated by their expression level during the state of ketosis.
Ketosis mainly happens due to the feed restriction (Drackley et al. Citation1991), as lack of availability of glucose leads to the formation of ketone bodies. Thereafter, the body tends to change to fat metabolism from carbohydrate metabolism. This condition remains the same until the body utilizes glucose-rich feed. Construction of ketosis model can be done by restricting the feed in vivo. For the analysis of ketosis in vitro, the induction of ketosis or construction of ketosis model is required. Ketosis model in cells can be created by restricting the level of glucose in the media (Koeslag et al. Citation1982). The usage of low glucose or no glucose media, according to the type of cell line, can induce the ketosis condition in vitro. Due to this unavailability of glucose, the cell system undergoes fat metabolism as its energy source. Hence, the ketosis condition can be formed due to glucose starvation.
Methylsulfonylmethane (MSM) is a natural, organic sulfur compound mainly present in foods like fruits, vegetables, beverages, etc. It is also known as dimethyl sulfone. The consumption of MSM can be possible through feed (Morton & Siegel Citation1986; Horvath et al. Citation2002; Caron et al. Citation2010). This MSM has different abilities such as anticancer activity against various type of cancers (Ebisuzaki Citation2003; Lim et al. Citation2012; Caron et al. Citation2013a, Citation2013b; Joung et al. Citation2014), antioxidant activity, and anti-inflammatory mechanisms (Beilke et al. Citation1987; DiSilvestro et al. Citation2008). Our recent studies proved that MSM has the ability to upregulate the expression levels of GHR and STAT5b in osteoblast-like cells (Joung et al. Citation2012). That property is very significant in the role of MSM in hepatic cells.
Ketosis condition is very dangerous if the amount of ketone bodies is very high. In this present study, our aim is to construct the ketosis cell model by glucose starvation and its treatment using MSM. Also our aim is to treat ketosis condition using molecular targets such as ANGPTL3, STAT5b, and GHR by MSM and to elucidate the relation between ANGPTL3 and STAT5b signaling molecules.
Materials and methods
Antibodies and reagents
F-12K media were purchased from Life Technologies (Carlsbad, CA). D-glucose and MSM were purchased from Sigma Chemical (St. Louis, MO). Penicillin-streptomycin solution and fetal bovine serum (FBS) were purchased from Hyclone (South of Logan, UT). A total of 0.05% trypsin-EDTA was purchased from Gibco-BRL (Grand Island, NY). STAT5b, IGF-1Rβ, GHR, ANGPTL3 antibodies, and secondary antibodies (goat anti-mouse and rabbit IgG-horseradish peroxidase) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Phosphorylated IGF-1Rβ antibody was purchased from Cell Signalling Technology (Beverly, MA), and phosphorylated STAT5 was purchased from Upstate Biotechnology (Lake Placid, NY). β-actin was purchased from Sigma Chemical Co. (St. Louis, MO). The enhanced chemiluminescence (ECL plus) detection kit was purchased from Amersham Pharmacia Biotech (Piscataway, NJ). Restore™ Western Blot Stripping Buffer and NE-PER kit were purchased from Pierce (Rockford, IL). RNeasy mini kit was purchased from Qiagen (Hilden, Germany). RT-PCR Premix kit and 18s primer for reverse transcription polymerase chain reaction (RT-PCR) were synthesized by Bioneer (Daejeon, Korea). ANGPTL3 primer was purchased from Integrated DNA Technologies (Coralville, IA). Electrophoretic mobility shift assay (EMSA) kit and oligonucleotide probes (STAT5b) were obtained from Promega Corp (Madison, WI). BHB (ketone body) colorimetric assay kit was purchased from the Cayman Chemical Co. (Ann Arbor, MI).
Cell culture and treatment
Mouse hepatocyte, FL83B cell lines were maintained in F-12K (3% glucose) medium containing 10% FBS, 100 U/ml penicillin and streptomycin at 37°C in 5% CO2. The cells were placed in airtight chambers (Nu Aire, Plymouth, MN). At the beginning of each experiment, the cells were resuspended in the medium at a density of 2.5 × 105 cells/ml. Cells were treated with 9% D-glucose and MSM at 30 and 40 mM.
Cell proliferation inhibition
Cell viability was assayed by measuring blue formazan that was metabolized from 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetra-zolium bromide (MTT) by mitochondrial dehydrogenase, which is active only in live cells. The cells were resuspended in the medium one day before of drug treatment, at a density of 3 × 103 cells per well in 96-well culture plates. Liquid medium was replaced with a fresh medium for control (vehicle). Cells were incubated with various concentrations of MSM. MTT (5 mg/ml) was added to each well and incubated for 4 h at 37°C. The formazan product formed was dissolved by adding 200 μl dimethylsulfoxide to each well, and the absorbance was measured at 550 nm on Ultra Multifunctional Microplate Reader (TECAN, Durham, NC). All measurements were performed triplicate and were repeated at least three times.
BHB colorimetric assay
Measurements of BHB were done by using the BHB (ketone body) colorimetric assay kit following the manufacturer’s protocol. Briefly, FL83B cells (~18 × 106) were collected by centrifugation and sonicated in cold diluted assay buffer. Removed the supernatant and resuspended again in cold diluted assay buffer. BHB standard wells and sample wells were prepared as per the protocol and initiated the reaction by adding developer solution. Incubation was done under 25°C in the dark for 30 min and absorbance was measured at 450 nm.
Western blotting
The FL83B cell lines were treated with high glucose (9%) and MSM of 30 and 40 mM for determined times. Whole cells were lysed on ice with radioimmunoprecipitation lysis buffer, containing phosphatase and protease inhibitors. Cells were disrupted by aspiration through a 23-gauge needle and centrifuged at 15,000 rpm for 10 min at 4°C to remove cellular debris. Protein concentrations were measured using the Bradford method. Equal amounts of proteins were resolved on SDS-PAGE and transferred onto nitrocellulose membrane. The blots were blocked 1 h with 5% skim milk. The membrane was probed over night at 4°C with primary antibody followed by HRP conjugated secondary antibodies. Detection was performed using the ECL plus detection kit and LAS-4000 imaging device (Fujifilm, Japan).
Reverse transcription polymerase chain reaction (RT-PCR)
The FL83B cell lines were treated with high glucose (9%) and MSM of 30 and 40 mM for 24 h. Total RNA was extracted using RNeasy Mini Kit (Qiagen) and quantified spectrometrically at 260 nm. RT-PCR analysis for ANGPTL3 and 18s RNA was performed. cDNA was synthesized from total RNA by reverse transcription at 42°C for 1 h and 80°C for 15 min using first strand cDNA synthesis kit (Bioneer, Korea). PCR was conducted by using cDNA. The PCR condition for ANGPTL3 consisted of 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C plus 40 s at 60°C. PCR products were analyzed by 1% agarose gel stained with ethidium bromide.
Electrophoretic mobility shift assay (EMSA)
The DNA binding activity of STAT5b was detected using EMSA, in which a labeled double-stranded DNA used as a DNA probe to bind active STAT5b protein in nuclear extracts. Nuclear protein extracts were prepared with the Nuclear Extract Kit (Panomics, AY2002). EMSA experiment is performed by incubating a biotin-labeled transcription factor (TF-STAT5b probe) with treated and untreated nuclear extracts. Proteins were resolved on a nondenaturing 6% PAGE gel (Bio-Rad, Korea). The proteins in the gel transferred to a nylon membrane and detected using streptavidin-HRP and a chemiluminescent substrate.
Small interference RNA (siRNA) analysis
FL83B cells (1 × 105) were cultured on to 6 well plate and maintained in complete media for 12 hours. The cells were then transfected with ON-TARGETplus SMARTpool siRNA targeting STAT5b or ON-TARGETplus Non-targeting siRNA (Dharmacon, Chicago) using Fugene-6 (Roche, IN) according to the manufacturer’s instruction. Following transfection with this mixture for 24 h, cells were treated with 40 mM MSM for an additional 24 h. Following this, total proteins were prepared and the expression levels of ANGPTL3 and STAT5b were analyzed using western blotting.
Statistical analysis
All the experiments were repeated three times, and the results were expressed as mean ± SEM. Statistical analysis was done with Student’s t-test or analysis of variance (ANOVA) test of the SAS program.
Results
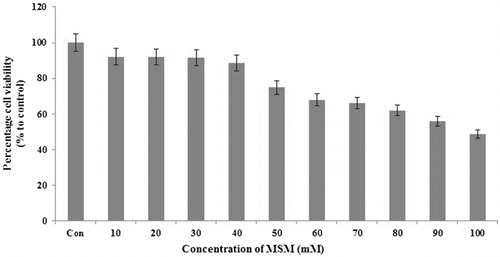
MSM inhibited the viability of mouse hepatocyte, FL83B cells in a dose dependent manner
Anti-ketosis therapeutics should be designed in such a way that it should have a little or no effect on the viability of cells. The effect of MSM on the proliferation of cultured mouse hepatocyte, FL83B cells was studied using the MTT assay. MSM as an organic sulfur containing compound believed to have anti-ketosis effect on cultured mouse hepatocytes, FL83B. In order to decipher the cellular effects of MSM on hepatocytes, we require higher viability cell counts. Hence, the concentration of MSM given 89% (40 mM) and 91% (30 mM) viable cell counts were opted for further analysis ().
Ketosis cell model construction and inhibition of glucose starvation induced ketosis by MSM in FL83B cells
We constructed a ketosis cell model using glucose starvation in mouse hepatocytes. The level of ketosis was analyzed using the formation of ketone bodies. BHB colorimetric assay was used for the analysis of ketosis. We measured the BHB level by keeping high glucose level media (9%) as the negative control. Low glucose (3%) nontreated media kept as the ketosis group. MSM of 30 and 40 mM were used in low glucose media to treat against ketosis. The assay was done as per the protocol and the standard graph was plotted (). The levels of BHB calculated as per the equation given.
Note: Analysis of ketone body formation using BHB colorimetric assay: (a) BHB standard graph. (b) Estimation of BHB concentration (mM) after glucose starvation (3%), treatment with high glucose (9%), low glucose plus 30 or 40 mM MSM for 24 h. (c) Relative expression (%) of BHB with respect to the low glucose ketosis group. Statistical analysis was done by using ANOVA test. L Glc, low glucose (3%); H Glc, high glucose (9%); *P < 0.05 and **P < 0.001.
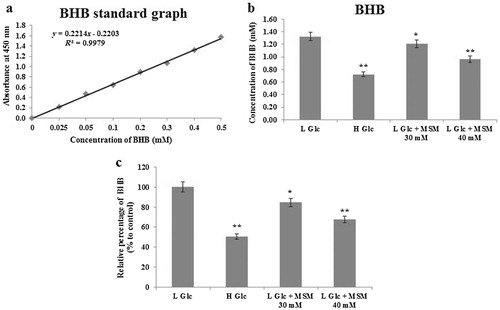
showed the measurement of BHB levels, and it is found that MSM has the ability to inhibit the formation of ketone bodies in ketosis condition. It is found that the level of BHB decreased almost 50% in high glucose positive control group. A total of 30 mM MSM produced 15% inhibition of BHB, whereas 40 mM MSM produced 32% inhibition in the level of BHB when compared to the low glucose ketosis group (). These results indicated that MSM has the capacity to inhibit ketosis in vitro.
MSM upregulated the expression of GHR in ketosis condition
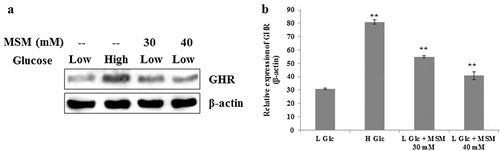
The expression levels of GHR showed to have significant role in subclinical ketosis conditions (Loor et al. Citation2007). We also evaluated the role of GHR in experimental ketosis induced cells treated with MSM. The FL83B cells were categorized into four groups as low glucose ketosis positive control group (experimental ketosis), high glucose negative control group, and two groups of experimental ketosis induced cells treated with MSM of 30 and 40 mM. The total proteins were isolated after 24 h and analyzed using western blotting. Our obtained result showed a significant enhancement on the expression of GHR in MSM treated cells comparing to the nontreated experimental ketosis induced positive control group (). The graphical representation showed a significant result for anti-ketosis by MSM treatment ().
Note: (a) Western blotting analysis of GHR expression in whole cell lysate after treatment with high glucose, low glucose plus 30 or 40 mM MSM for 24 h. (b) Relative expression of GHR with respect to β-actin. Statistical analysis was done by using ANOVA test. L Glc, low glucose (3%); H Glc, high glucose (9%); **P < 0.001.
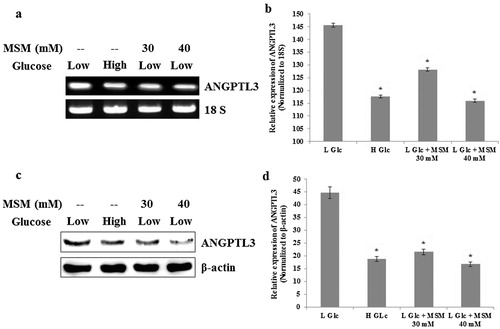
MSM downregulated the expression of ANGPTL3 in ketosis condition
ANGPTL3 is an important molecular target to treat ketosis (Hong-Bo et al.). We analyzed the level of ANGPTL3 by RT-PCR and western blotting. The samples were made from low glucose ketosis group, high glucose negative control group, and two groups of induced ketosis treated with MSM of 30 and 40 mM. The total RNA and total protein were isolated after 24 h of treatment with MSM. The expression level of ANGPTL3 should upregulate in ketosis condition. Obtained results showed the downregulated expression in high glucose negative control group when compared with the low glucose ketosis group ( and ). Decreased expression of ANGPTL3 in MSM 30 and 40 mM treated groups also indicated the downregulated condition of ketosis. and represented the graphical representation of ANGPTL3 expression level by RT-PCR and western blotting, respectively. ANGPTL3 inhibition ability by MSM denotes its inhibition ability of ketosis.
Note: (a) RT-PCR analysis of ANGPTL3 after the treatment with high glucose, low glucose plus 30 or 40 mM MSM for 24 h. (b) Relative decreases of ANGPTL3 mRNA level by high glucose, low glucose plus 30 or 40 mM MSM with respect to 18s RNA. (c) Western blotting analysis showing protein level inhibition of ANGPTL3 by high glucose, low glucose plus 30 or 40 mM MSM. (d) Relative decreases of ANGPTL3 protein level by high glucose, low glucose plus 30 or 40 mM MSM in whole cell lysates. Statistical analysis was done by using ANOVA test. L Glc, low glucose (3%); H Glc, high glucose (9%); *P < 0.001.
MSM enhanced STAT5b/IGF-1R signaling similar to GHR signaling
STAT5b is a transcription factor of ANGPTL3. We elucidated the levels of activated STAT5b and its downstream target molecule IGF-1Rβ. The total proteins were isolated from the low glucose ketosis group, high glucose group, and experimental ketosis induced cells treated with MSM of 30 and 40 mM. Western blotting analysis showed a characteristic increase in the expression levels of activated STAT5b and IGF-1Rβ in negative control and in MSM treated groups (). These results are similar to the expression of GHR that indicates the confirmed upregulation pattern for GHR. So from these results, it is showing that STAT5b is a negative regulator for ANGPTL3.
Note: (a) Western blotting analysis of whole cell lysate after treatment with high glucose, low glucose plus 30 or 40 mM MSM for 24 h. (b) Nuclear protein analysis after the treatment with high glucose, low glucose plus 30 or 40 mM MSM for 24h by western blotting. (c) DNA binding activities of STAT5b to the GAS element was upregulated by the MSM, analyzed by gel shift assay. Data shown are one representative of three independent experiments.
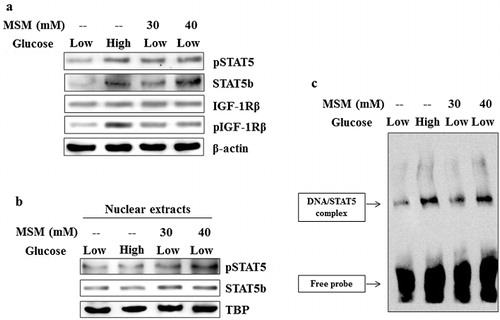
MSM upregulated STAT5b/DNA binding activity in vitro
The expression of STAT5b was upregulated by the MSM treatment in whole cell lysates. So we checked STAT5b/DNA (GAS element) binding activity by EMSA. The nuclear extracts were prepared using nuclear extraction kit and the assay was carried out. The obtained results showed significant increases in the DNA binding activities in high glucose group and 40 mM MSM group when compared with the low glucose ketosis groups (). This activity was confirmed in western blotting with enhanced STAT5b expression levels (). It also showed an increase in phospho-STAT5 levels with respect to TATA binding protein expression (). The elevated expression, phosphorylation, and DNA binding of STAT5b confirmed the ability of MSM to inhibit ketosis condition in vitro.
STAT5b is a negative regulator for ANGPTL3 in ketosis condition
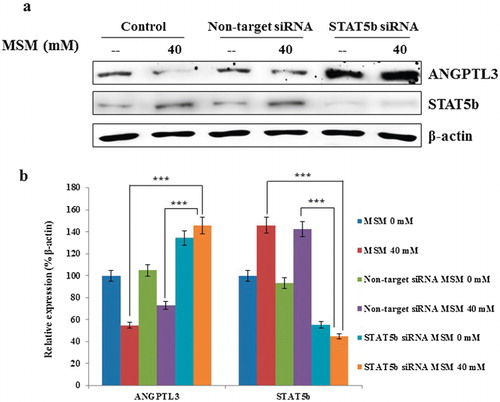
We have confirmed the downregulation of ANGPTL3 expression and upregulation of STAT5b expression, phosphorylation, and its DNA binding activity. So next we checked the relation between STAT5b and ANGPTL3. As we had a hypothesis that STAT5b is a negative regulator for ANGPTL3, in order to confirm this, we silenced the STAT5b using specific siSTAT5b. Following the silencing using specific siSTAT5b RNA, we analyzed the expression of ANGPTL3 using western blotting. Obtained results confirmed the role of STAT5b in the transcription of ANGPTL3 (). In this siSTAT5b assay, it showed statistical significant increases than the expression for both controls and 40 mM MSM sample (). The nontargeted siRNA gave the similar result for the non-siRNA treated control and 40 mM MSM samples. The expression levels of STAT5b silenced in siSTAT5b ketosis condition and 40 mM MSM sample. So these results proved the critical role of STAT5b in ketosis condition.
Note: (a) On-target inhibition of STAT5b reversed the expression pattern of ANGPTL3 after silenced STAT5b. (b) Graphical representation of the relative expression of ANGPTL3 with respect to β-actin. Statistical analysis was done by using Student’s t-test; ***p < 0.001.
Discussion
Ketogenesis is a condition where ketone bodies are formed for the energy production instead of glucose (Manninen Citation2004). Induction of ketosis condition in vitro is a challenging process. We assumed the ketosis induction could be done by means of glucose starvation in mouse hepatic cells, FL83B. For its analysis of ketosis induction, we quantified the amount of a ketone body, BHB, because BHB can be detected in cultured cells by utilizing β-hydroxybutyrate colorimetric assay kit (Persson Citation1970).
We estimated the amount of BHB formed during the glucose starvation and glucose starved MSM treated condition. As in , the high glucose (9%) showed a significant inhibition in BHB level when compared to the low glucose (3%) ketosis induced group. The amount of BHB level in low glucose clearly indicated that it is a nutritional ketosis condition. Our desired drug, MSM of 40 mM, gave 32% inhibition in the amount of BHB, which defined the ability of MSM to inhibit the ketosis condition in vitro.
The development of an anti-ketosis drug is mainly based by comparing their action with high glucose treated cultured cells. This group can be considered as a negative control for ketosis. MSM inhibited the BHB levels as same as negative control. So it can be used for further analysis. The process of ketosis depends on many molecular targets. One of these, GHR, is a protein programmed by GHR gene (Meinhardt et al. Citation2003). This GHR was found to have a regulation in subclinical ketosis condition from the normal condition (Loor et al. Citation2007). So we analyzed the expression level of GHR in experimental ketosis condition to check the role of GHR in ketosis condition. As per Loor et al., the expression level of GHR should be less compared with the level of GHR in nonketosis. Our results also gave a clear indication of upregulated expression of GHR in high glucose treated negative control group and MSM treated ketosis group. This shows the role of GHR in ketosis in vitro, so we suggest that our FL83B mouse model can be an optimal ketosis cell model.
ANGPTL3 is one of the targets to treat ketosis. It can suppress lipoprotein lipase and play a crucial role in carbohydrate metabolism and lipid metabolism (Xiao et al. Citation2012). Some studies showed that increased expression of ANGPTL3 contributes to metabolic disorder of carbohydrate and fat metabolism (Robciuc et al. Citation2010). It can also downregulate endothelial lipase and thereby inhibits high density lipoprotein (Shimamura et al. Citation2003). These properties account for ANGPTL3 as a molecular target for the treatments of ketosis. STAT5b is a transcription factor for ANGPTL3 and GHR (Xu et al. Citation1996). Here we are assuming that STAT5b plays a vital role in the regulation of ANGPTL3 and thereby the experimental ketosis condition. Evaluation of experimental ketosis with MSM showed the inhibition pattern for ANGPTL3 in both protein and RNA levels (). The ability of MSM to inhibit ketosis was confirmed with respect to the expression pattern of ANGPTL3 in high glucose group. According to our hypothesis, STAT5b should have some role in the treatment of ketosis because GHR is having a role in subclinical ketosis. So we evaluated the expression pattern of STAT5b in total protein and nuclear protein, which gave a significant increase level in MSM treated ketosis induced group as well as negative control group. We also elucidated the DNA binding activity of STAT5b to GAS element to confirm its role in ketosis condition. These results clearly point to the role of STAT5b as a molecular target to treat ketosis.
As we found out the role of STAT5b and ANGPTL3 as molecular targets in the treatment of experimental ketosis, it is very important to find out the relationship between both molecules as it is unknown. The expression patterns in different analysis showed completely opposite action in ketosis condition as well as in ketosis treatment condition. We investigated the expression level of ANGPTL3 after silenced STAT5b and the results were fascinating. Expression pattern for ANGPTL3 in STAT5b siRNA group gave a high level of expression when compared to other groups. As a result, we can conclude that STAT5b is a negative regulator for ANGPTL3. The expression levels for ANGPTL3 in the STAT5b siRNA and the MSM treated STAT5b siRNA groups remain unaltered. This clearly indicated that MSM acts well on ANGPTL3 expression through STAT5b pathway. Therefore, we suggest that STAT5b is a negative regulator for ANGPTL3 control in the treatment of ketosis by MSM.
Acknowledgment
This research was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. 010465042014)” Rural Development Administration, Republic of Korea.
References
- Beilke MA, Collins-Lech C, Sohnle PG. 1987. Effects of dimethyl sulfoxide on the oxidative function of human neutrophils. J Lab Clin Med. 110:91–96.
- Caron JM, Bannon M, Rosshirt L, Luis J, Monteagudo L, Caron JM, Sternstein GM. 2010. Methyl sulfone induces loss of metastatic properties and reemergence of normal phenotype in a metastatic cloudman S-91 (M3) murine melanoma cell line. PLoS One. 5:e11788. 10.1371/journal.pone.0011788.s002
- Caron JM, Bannon M, Rosshirt L, O’Donovan L. 2013a. Methyl sulfone manifests anticancer activity in a metastatic murine breast cancer cell line and in human breast cancer tissue–part I: murine 4T1 (66cl-4) cell line. Chemotherapy. 59:14–23.
- Caron JM, Monteagudo L, Sanders M, Bannon M, Deckers PJ. 2013b. Methyl sulfone manifests anticancer activity in a metastatic murine breast cancer cell line and in human breast cancer tissue–part 2: human breast cancer tissue. Chemotherapy. 59:24–34.
- DiSilvestro RA, DiSilvestro DJ, DiSilvestro DJ. 2008. Methylsulfonylmethane (MSM) intake in mice produces elevated liver glutathione and partially protects against carbon tetrachloride-induced liver injury. FASEB J. 22:445–448.
- Drackley JK, Veenhuizen JJ, Richard MJ, Young JW. 1991. Metabolic changes in blood and liver of dairy cows during either feed restriction or administration of 1,3-butanediol. J Dairy Sci. 74:4254–4264. 10.3168/jds.S0022-0302(91)78620-7
- Ebisuzaki K. 2003. Aspirin and methylsulfonylmethane (MSM): a search for common mechanisms, with implications for cancer prevention. Anticancer Res. 23:453–458.
- Horvath K, Noker PE, Somfai-Relle S, Glávits R, Financsek I, Schauss AG. 2002. Toxicity of methylsulfonylmethane in rats. Food Chem Toxicol. 40:1459–1462. 10.1016/S0278-6915(02)00086-8
- Johnston DG, Pernet A, McCulloch A, Blesa-Malpica G, Burrin JM, Alberti KG. 1982. Some hormonal influences on glucose and ketone body metabolism in normal human subjects. Ciba Found Symp. 87:168–191.
- Joung YH, Lim EJ, Darvin P, Chung SC, Jang JW, Park KD, Lee HK, Kim HS, Park T, Yang YM. 2012. MSM enhances GH signaling via the Jak2/STAT5b pathway in osteoblast-like cells and osteoblast differentiation through the activation of STAT5b in MSCs. PLoS One. e47477.
- Joung YH, Na YM, Yoo YB, Darvin P, Sp N, Kang DY, Kim SY, Kim HS, Choi YH, Lee HK, et al. 2014. Combination of AG490, a Jak2 inhibitor, and methylsulfonylmethane synergistically suppresses bladder tumor growth via the Jak2/STAT3 pathway. Int J Oncol. 44:883–895.
- Koeslag JH, Noakes TD, Sloan AW. 1982. The effects of alanine, glucose and starch ingestion on the ketosis produced by exercise and by starvation. J Physiol. 325:363–376.
- Laffel L. 1999. Ketone bodies: a review of physiology, pathophysiology and application of monitoring to diabetes. Diabetes Metab Res Rev. 15:412–426. 10.1002/(SICI)1520-7560(199911/12)15:6<412::AID-DMRR72>3.0.CO;2-8
- Lim EJ, Hong DY, Park JH, Joung YH, Darvin P, Kim SY, Na YM, Hwang TS, Ye SK, Moon ES, et al. 2012. Methylsulfonylmethane suppresses breast cancer growth by down-regulating stat3 and stat5b pathways. PLoS One. 7:e33361.
- Loor JJ, Everts RE, Bionaz M, Dann HM, Morin DE, Oliveira R, Rodriguez-Zas SL, Drackley JK, Lewin HA. 2007. Nutrition-induced ketosis alters metabolic and signaling gene networks in liver of periparturient dairy cows. Physiol Genomics. 32:105–116. 10.1152/physiolgenomics.00188.2007
- Manninen AH. 2004. Metabolic effects of the very-low-carbohydrate diets: misunderstood “Villains” of human metabolism. J Int Soc Sports Nutr. 1:7–11. 10.1186/1550-2783-1-2-7
- Meinhardt U, Eble A, Besson A, Strasburger CJ, Sraer JD, Mullis PE. 2003. Regulation of growth-hormone-receptor gene expression by growth hormone and pegvisomant in human mesangial cells. Kidney Int. 64:421–430. 10.1046/j.1523-1755.2003.00117.x
- Morton JI, Siegel BV. 1986. Effects of oral dimethyl sulfoxide and dimethyl sulfone on murine autoimmune lymphoproliferative disease. Proc Soc Exp Biol Med. 183:227–230. 10.3181/00379727-183-42409
- Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui AL, Kitamura T. 1999. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J. 18:4754–4765. 10.1093/emboj/18.17.4754
- Persson B. 1970. Determination of plasma acetoacetate and D-β-hydroxy butyrate in new-born infants by an enzymatic fluorometric micro-method. Scand J Clin Lab Invest. 25:9–18. 10.3109/00365517009046184
- Robciuc MR, Tahvanainen E, Jauhiainen M, Ehnholm C. 2010. Quantitation of serum angiopoietin-like proteins 3 and 4 in a Finnish population sample. J Lipid Res. 51:824–831. 10.1194/jlr.M002618
- Shimamura M, Matsuda M, Kobayashi S, Ando Y, Ono M, Koishi R, Furukawa H, Makishima M, Shimomura I. 2003. Angiopoietin-like protein 3, a hepatic secretory factor, activates lipolysis in adipocytes. Biochem Biophys Res Commun. 301:604–609. 10.1016/S0006-291X(02)03058-9
- Xiao HB, Sun ZL, Zhou N. 2012. 1,3,5,8-tetrahydroxyxanthone regulates ANGPTL3-LPL pathway to lessen the ketosis in mice. Eur J Pharm Sci. 46:26–31. 10.1016/j.ejps.2012.02.001
- Xu BC, Wang X, Darus CJ, Kopchick JJ. 1996. Growth hormone promotes the association of transcription factor STAT5 with the growth hormone receptor. J Biol Chem. 271:19768–19773. 10.1074/jbc.271.33.19768