Abstract
The complete mitogenome of the subsocial wood-eating cockroach Cryptocercus kyebangensis was sequenced and compared with that of another Asian cockroach, Cryptocercus relictus and the primitive termite Mastotermes darwiniensis. The complete mitogenome of C. kyebangensis is a closed circular molecule of 15,720 bp, containing13 protein-coding genes, two ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA genes, and one D-loop region. Nucleotide composition of the C. kyebangensis mitogenome shows an AT bias (74.4%), in particular, 46.0% A, 28.4% T, 9.7% G, and 15.9% C. GC skews are similar among the mitogenomes of C. kyebangensis, C. relictus, and M. darwiniensis, but AT skew is stronger in Cryptocercus mitogenomes than in the M. darwiniensis mitogenome. The whole mitogenome of C. kyebangensis showed 88.8% nucleotide similarity to that of C. relictus but only 74.9% similarity to that of M. darwiniensis. Phylogenetic analyses, based on amino acid sequences of the 13 mitochondrial proteins, showed that the Cryptocercus clade has a sister relation with termites, including the primitive termite M. darwiniensis. Sister relations of Blattidae and Blattellidae, with the clade Cryptocercidae + Isoptera as a sister group, are also evident. Polyphagidae (Eupolyphaga sinensis) appears to be most basal in the phylogenetic trees, showing sister relations with the remaining cockroaches.
1. Introduction
Subsocial wood-eating cockroaches of the genus Cryptocercus show peculiar behavioral characteristics, including wood feeding, symbiotic relations with intestinal protozoa, trophallaxis by adults, and obligatory associations among adults and their offspring, which are similar to those among lower termites (Nalepa Citation1984; Park et al. Citation2002; Park & Choe Citation2008; Maekawa & Nalepa Citation2011). In addition, recent phylogenetic studies involving various mitochondrial or/and nuclear molecular markers showed that Cryptocercus is a sister group of a primitive termite species, Mastotermes darwiniensis (Lo et al. Citation2000; Legendre et al. Citation2008; Cameron et al. Citation2012; Djernæs et al. Citation2012). Because of these behavioral characteristics and their close relation with termites, wood-eating cockroaches of Cryptocercus have often been used as model species to elucidate the early stages of social evolution in termites (Klass et al. Citation2008; Djernæs et al. Citation2015).
Insect mitogenomes, which are a compact circular molecule (typically 15–18 kb in size), tend to have evolved faster than nuclear genomes did (Cameron Citation2014). Thus, they are widely used as a source of sequence data for molecular phylogenetic studies on evolutionary trends of social behavior among related lineages and for molecular evolutionary studies on patterns and trends in the genomic evolution of insects (Chandra et al. Citation2006; Cameron Citation2014).
In the present study, we sequenced and characterized the complete mitogenome of Cryptocercus kyebangensis from South Korea and compared it with previously published mitogenomes of Cryptocercus relictus and M. darwiniensis. To date, complete mitogenome sequences of seven species from four families (Cryptocercidae, Blattidae, Blattellidae, and Polyphagidae) of the order Blattodea have been published (Yamauchi et al. Citation2004; Zhang et al. Citation2010; Cameron et al. Citation2012; Xiao et al. Citation2012; Chen Citation2013). We show phylogenetic relations among cockroach families based on the amino acid sequences derived from 13 mitochondrial protein-coding genes (PCGs) in the whole mitogenome. We also confirm the sister relation between Cryptocercus species and the primitive termite M. darwiniensis.
2. Materials and methods
2.1. Samples
A complete mitogenome (KP872847) of the Korean wood-feeding cockroach C. kyebangensis (Blattodea: Cryptocercidae) was sequenced and characterized. Mitogenome sequences of C. relictus (Blattodea: Cryptocercidae) and M. darwiniensis (Isoptera: Mastotermitidae) were downloaded from GenBank for comparison with the C. kyebangensis mitogenome. Mitogenomes of Periplaneta fuliginosa and Periplaneta americana (Blattidae), Blattella germanica and Blattella bisignata (Blaberoidea: Blattellidae or Ectobiidae), and Eupolyphaga sinensis (Corydioidea: Polyphagidae or Corydiidae) were also downloaded from GenBank for analyses of interfamilial relations within Blattodea. A few termite mitogenomes – from Zootermopsis angusticollis, Z. nevadensis, Porotermes adamsoni, and Neotermes insularis – were included in the phylogenetic analyses to confirm the sister relation between the wood-eating cockroach Cryptocercus species and the primitive termite M. darwiniensis. Tamolanica tamolana (Mantidae) and Locusta migratoria (Acrididae) were used as outgroups.
2.2. Genomic-DNA extraction, primer design, PCR, and DNA sequencing
Total genomic DNA was extracted from leg tissues using the DNeasy® Blood & Tissue Kit (Qiagen, Valencia, CA, USA). The primers were designed based on the complete mitogenome of C. relictus (JX144941) available from GenBank. PCR was performed in a final reaction volume of 25 μL, which contained 10 mM Tris-HCl (pH 8.4), 50 mM KCl, 4 mM MgCl2, 200 mM each dNTP, 50 pmol of each primer, 2 U of TaKaRa Ex Taq® DNA Polymerase, and 1 μL of a DNA sample, under the following cycling conditions: 94°C for 5 min (initial denaturation); then 94°C for 1 min (denaturation), 48–62°C for 30 s (annealing), and 72°C for 1.3 min (extension) for 30 cycles; and a final extension at 72°C for 10 min. The PCR products were resolved by electrophoresis in a 1.0% agarose gel, and then extracted by means of the DNA Gel Extraction Kit (Qiagen, Valencia, CA, USA), and sent to Biomedic Co., Ltd. (Bucheon, South Korea), for sequencing by primer walking in both directions.
2.3. Genome annotation
The complete mitogenome of C. kyebangensis was annotated and characterized based on the mitogenome of C. relictus in the Geneious Pro 5.5.9 software (Auckland, New Zealand). The mitochondrial PCG sequences were translated in the invertebrate mitogenome genetic code and identified by sequence similarity to C. relictus. Both the tRNAscan-SE search server and MITOS web server (with default parameters) were used for the identification of tRNA genes and possible stem-loop secondary structures within these tRNA sequences (Lowe & Eddy Citation1997; Bernt et al. Citation2013).
2.4. Phylogenetic analysis
The phylogenetic trees were constructed on the basis of the 13 mitochondrial protein sequences by the Bayesian inference (BI) procedures in MrBayes 3.2.2 (Ronquist et al. Citation2012) and the maximum likelihood (ML) criterion in MEGA v 6.06 (Tamura et al. Citation2011). For the BI analysis, ‘MtREV' (Adachi & Hasegawa Citation1996) was selected as the best substitution model, and two independent runs of four heated Markov chain Monte Carlo (MCMC) chains (one cold chain and three hot chains) were simultaneously conducted for one million generations, with sampling once every 100 generations. Stationarity was analyzed by plotting log likelihood values against generation numbers, and the first 25% from each run were discarded as burn-in. In the ML analysis, the mtREV with Freqs (+F) and gamma distributed invariant sites (G + I) was selected as the best model of evolution by the MEGA v6.06. The number of discrete categories (five) was specified by default. The ML heuristic method for tree search was the nearest-neighbor interchange, starting with the initial tree selected automatically under the default neighbor joining/BioNJ. The reliability of the phylogenetic tree was supported by 1000 bootstrap replications.
3. Results and discussion
3.1. Genome organization and nucleotide composition
The complete mitogenome of C. kyebangensiscontains 13 PCGs, two ribosomal RNA genes (12S rRNA and 16S rRNA), 22 tRNA genes, and one D-loop region (), as is the case in other insects. The total length of the C. kyebangensis mitogenome is 15,720 bp: bigger than that of C. relictus (15,373 bp). Such a difference in mitogenome size is mainly due to variation in the control region size ().
Table 1. Characterization of the C. kyebangensis mitogenome and its sequence similarity to C. relictus and M. darwiniensis mitogenomes.
Table 2. Size, nucleotide composition (%) and skewness of C. kyebangensis, C. relictus and M. darwiniensis mitogenomes.
Nucleotide composition of the C. kyebangensis mitogenome shows an AT bias (74.4%), in particular, 46.0% A, 28.4% T, 9.7% G, and 15.9% C. AT content of the C. kyebangensis mitogenome (73.5%) is similar to that of the C. relictus mitogenome, whereas AT content of the C. kyebangensis mitogenome (68%) is higher than that of the M. darwiniensis mitogenome (). Compared to the M. darwiniensis mitogenome, there is a stronger AT skew in the two Cryptocercus mitogenomes, but GC skews are similar between M. darwiniensis and Cryptocercus mitogenomes (). Among the gene regions, AT and GC skews of the other three gene regions (PCGs, rRNA genes, and tRNA genes), with the exception of the control region, are similar between C. kyebangensis and C. relictus. In the control region, however, the AT skew value of C. kyebangensis is much higher than that of C. relictus, whereas the GC skew of C. kyebangensis is much weaker than that of C. relictus.
3.2. PCGs
The total length of the 13 mitochondrial PCGs of C. kyebangensis is 11,184 bp, which are translated into 3718 amino acid residues, with the exception of stop codons (30 bp).
The C. kyebangensis mitogenome includes five kinds of start codon (ATG, TTG, ATA, GTG, and ATT) for translation initiation, a TAA stop codon, and two incomplete stop codons for translation termination (). ATG is the most common start codon and occurs in the other eight PCGs except for COX1 (TTG), ATP8 and ND3 (ATA), ND5 (GTG), and ND6 (ATT). The stop codon TAA is used for termination of eight PCGs. Incomplete stop codons are found in ND2 (TA*) and in COX2, ND1, ND5, and CYTB (T**) and may be completed by polyadenylation of the 3′ end of an mRNA after transcription (Boore Citation1999).
Some compact and overlapping regions were found among PCGs of animal mitogenomes (Fernandez-Silva et al. Citation2003). We also observed a 7-bp overlap between ATP8 and ATP6, a single-base pair overlap between ATP6 and COX3, and a 7-bp overlap between ND and ND4L (). Together with the incomplete stop codons, the gene overlaps may be a product of the selective pressure to reduce genome size, noted in mitochondria (Rand Citation1993).
The total number of mitochondrial amino acid residues is the same for C. kyebangensis and C. relictus whereas the M. darwiniensis mitogenome encodes six amino acid residues fewer than Cryptocercus mitogenomes do (Supplementary ). The differences in the number of mitochondrial amino acid residues were found in COX1, ND1, and ND6. Cryptocercus mitogenomes encode seven amino acid residues more in ND1 and one amino acid residue more in ND6 in comparison with the M. darwiniensis mitogenome, whereas Cryptocercus mitogenomes encode two amino acid residues fewer in COX1 than the M. darwiniensis mitogenome does.
3.3. The tRNA genes and rRNA genes
The total length of 22 tRNA genes was 1451 bp, with AT content 74.6% (). The standard cloverleaf secondary structure was observed in all tRNA sequences, except for tRNACys(AGY), which lacks the DHU-arm. Most tRNA sequences could fold into the canonical cloverleaf secondary structure. The total lengths of 22 tRNA genes were only single-base difference between the two Cryptocercus species but slightly smaller than that of M. darwiniensis tRNA genes (1469 bp). The total length of C. kyebangensis rRNA genes (12S rRNA and 16S rRNA) was 2067 bp, with AT content 76.5%; this length was similar to that of the C. relictus rRNA genes (2063 bp). In contrast, the total length of the Cryptocercus rRNA genes was smaller than that of the M. darwiniensis rRNA genes (2117 bp).
3.4. The control region
The control region, which is located between the 12S rRNA gene and tRNAIle, is 1009 bp long, with AT content 80.2% (). In comparison with the control regions of C. relictus and M. darwiniensis, there is a long insertion (∼300 bp) in the control region of C. kyebangensis. In all three species, the control region's AT content is higher than that of the tree gene regions (13 PCGs, 22 tRNA genes, and two rRNA genes). AT content of control regions in the two Cryptocercus species was much higher than that of the M. darwiniensis control region (72.9%).
3.5. Sequence similarity
The whole mitogenome of C. kyebangensis shows 88.8% nucleotide similarity to that of C. relictus (). Nucleotide similarity between corresponding genes, with the exception of the control region (57.5%), ranges from 90.1% (13 PCGs) to 94.5% (22 tRNA genes). Compared to other genes, there is relatively high similarity between corresponding tRNA genes, especially tRNAArg, tRNALys, and tRNACys.
The amino acid sequence of C. kyebangensis shows 93.7% similarity to that of C. relictus but only 75.8% similarity to that of M. darwiniensis. Relatively high variation was observed in amino acid sequences of ATP8 and ND6, whereas the amino acid sequence of COX1 is well conserved ().
3.6. Interfamilial relations
Mitochondrial phylogenomics produced highly resolved phylogenetic trees with all nodes supported by high bootstrap values and revealed interfamilial relations within Blattodea as well as a relation between Blattodea and Isoptera (). Korean C. kyebangensisis is a sister to C. relictus, which occurs in Siberia. The Cryptocercus clade has a sister relation with the primitive termite M. darwiniensis and with the remainder of the termite. Thus, these results strongly support the paraphyly of Blattodea in relation to termites, as suggested in other studies (e.g. Lo et al. Citation2000; Pellens et al. Citation2007; Legendre et al. Citation2008; Cameron et al. Citation2012; Djernæs et al. Citation2012, Citation2015).The current phylogenetic tree supports the sister relation of Blattidae and Blattellidae, with the clade Cryptocercidae + Isoptera as a sister group. Polyphagidae (E. sinensis) is placed as the most basal family in the phylogenetic trees, forming a sister group toward the remaining cockroaches including Cryptocercidae, with termites as a sister group.
Figure 1. The phylogenetic relation between the wood-eating cockroach Cryptocercus species and the primitive termite M. darwiniensis as well as the interfamilial relations within Blattodea inferred from BI and ML analyses based on amino acid sequences derived from the 13 mitochondrial PCGs. Only the BI tree is shown. The numbers at the nodes indicate bootstrap values of BI (above diagonal) and ML (below diagonal).
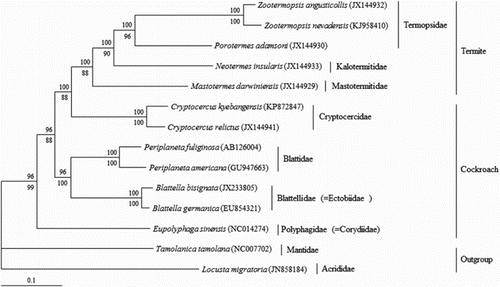
Although the present study is based on several representatives from only four families, it may provide rationale for further studies of cockroach phylogeny based on mitogenomes. In addition, the present study offers preliminary data on the interfamilial relations in Blattodea, especially phylogenetic relations among Polyphagidae, Blattidae, Blattellidae, and Cryptocercidae. In future studies, to obtain more complete cockroach phylogeny, additional mitogenomes need to be included from key genera of Blattodea.
4. Conclusion
The complete mitogenome of the Korean wood-eating cockroach C. kyebangensis was sequenced and compared with the mitogenomes of two closely related species, C. relictus and M. darwiniensis. The complete mitogenome of C. kyebangensis is a closed circular molecule of 15,720 bp, containing a typical metazoan set of 37 mitochondrial genes and one D-loop region. General characteristics of the C. kyebangensis mitogenome are more similar to those of the C. relictus mitogenome than those of the mitogenome of the primitive termite M. darwiniensis. In the phylogenetic tree, the Korean C. kyebangensis is well grouped with C. relictus. The phylogenetic tree also strongly supports the paraphyly of Blattodea with respect to termites, with Cryptocercus as a sister taxon of termites.
Supplemental data
Supplemental data for this article can be accessed at doi:10.1080/19768354.2015.1105866.
Supplementary Table 1.docx
Download MS Word (22.7 KB)Acknowledgements
The authors would like to thank the anonymous reviewers for their valuable comments and suggestions to improve the quality of the paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by a grant from Kangwon Green Environment Center (KWGEC) in 2014.
References
- Adachi J, Hasegawa M. 1996. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J Mol Evol. 42:459–468. doi: 10.1007/BF02498640
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319. doi: 10.1016/j.ympev.2012.08.023
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780. doi: 10.1093/nar/27.8.1767
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117. doi: 10.1146/annurev-ento-011613-162007
- Cameron SL, Lo N, Bourguignon T, Svenson GJ, Evans TA. 2012. A mitochondrial genome phylogeny of termites (Blattodea: Termitoidae): robust support for interfamilial relationships and molecular synapomorphies define major clades. Mol Phylogenet Evol. 65:163–173. doi: 10.1016/j.ympev.2012.05.034
- Chandra SBC, Vlk JL, Kapatral V. 2006. Comparative insect mitochondrial genomes: differences despite conserved genome synteny. Afr J Biotechnol. 5:1308–1318.
- Chen AH. 2013. Complete mitochondrial genome of the double-striped cockroach Blattella bisignata (Insecta: Blattaria: Blaberoidea). Mitochondr DNA. 24:14–16. doi: 10.3109/19401736.2012.710228
- Djernæs M, Klass KD, Eggleton P. 2015. Identifying possible sister groups of Cryptocercidae+Isoptera: a combined molecular and morphological phylogeny of Dictyoptera. Mol Phylogenet Evol. 84:284–303. doi: 10.1016/j.ympev.2014.08.019
- Djernæs M, Klass KD, Picker MP, Damgaard J. 2012. Phylogeny of cockroaches (Insecta, Dictyoptera, Blattodea), with placement of aberrant taxa and exploration of outgroup sampling. Syst Entomol. 37:65–83. doi: 10.1111/j.1365-3113.2011.00598.x
- Fernandez-Silva P, Enriquez JA, Montoya J. 2003. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol. 88:41–56. doi: 10.1113/eph8802514
- Klass KD, Nalepa C, Lo N. 2008. Wood-feeding cockroaches as models for termite evolution (Insecta: Dictyoptera): Cryptocercus vs. Parasphaeria boleiriana. Mol Phylogenet Evol. 46:809–817. doi: 10.1016/j.ympev.2007.11.028
- Legendre F, Whiting MF, Bordereau C, Cancello EM, Evans TA, Grandcolas P. 2008. The phylogeny of termites (Dictyoptera: Isoptera) based on mitochondrial and nuclear markers: implications for the evolution of the worker and pseudergate castes, and foraging behaviors. Mol Phylgenet Evol. 48:615–627. doi: 10.1016/j.ympev.2008.04.017
- Lo N, Tokuda G, Watanabe H, Rose H, Slaytor M, Maekawa K, Bandi C, Noda H. 2000. Evidence from multiple gene sequences indicates that termites evolved from wood-feeding cockroaches. Curr Biol. 10:801–804. doi: 10.1016/S0960-9822(00)00561-3
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in gesnomic sequence. Nucleic Acids Res. 25:955–964. doi: 10.1093/nar/25.5.0955
- Maekawa K, Nalepa CA. 2011. Biogeography and phylogeny of wood-feeding cockroaches in the genus Cryptocercus. Insects. 2:354–368. doi: 10.3390/insects2030354
- Nalepa CA. 1984. Colony composition, protozoan transfer and some life history characteristics of the woodroach Cryptocercus punctulatus Scudder (Dictyoptera: Cryptocercidae). Behav Ecol Sociobiol. 14:273–279. doi: 10.1007/BF00299498
- Park YC, Choe JC. 2008. Evolution of social life in wood-eating cockroaches (Cryptocercus spp.): effects of the winter climate on the evolution of subsociality. J Ecol Field Biol. 31:97–105. doi: 10.5141/JEFB.2008.31.2.097
- Park YC, Grandcolas P, Choe JC. 2002. Colony composition, social behavior and some ecological characteristics of the Korean wood-feeding cockroach (Cryptocercus kyebangensis). Zool Sci. 19:1133–1139. doi: 10.2108/zsj.19.1133
- Pellens R, D'Haese CA, Bellés X, Piulachs MD, Legendre F, Wheeler WC, Grandcolas P. 2007. The evolutionary transition from subsocial to eusocial behaviour in Dictyoptera: phylogenetic evidence for modification of the “shift-in-dependent-care” hypothesis with a new subsocial cockroach. Mol Phylogenet Evol. 43:616–626. doi: 10.1016/j.ympev.2006.12.017
- Rand DM. 1993. Endotherms, ectotherms, and mitochondrial genome-size variation. J Mol Evol. 37:281–295. doi: 10.1007/BF00175505
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. doi: 10.1093/sysbio/sys029
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. doi: 10.1093/molbev/msr121
- Xiao B, Chen AH, Zhang YY, Jiang GF, Hu CC, Zhu CD. 2012. Complete mitochondrial genomes of two cockroaches, Blattella germanica and Periplaneta americana, and the phylogenetic position of termites. Curr Genet. 58:65–77. doi: 10.1007/s00294-012-0365-7
- Yamauchi MM, Miya MU, Nishida M. 2004. Use of a PCR-based approach for sequencing whole mitochondrial genomes of insects: two examples (cockroach and dragonfly) based on the method developed for decapod crustaceans. Insect Mol Biol. 13:435–442. doi: 10.1111/j.0962-1075.2004.00505.x
- Zhang YY, Xuan WJ, Zhao JL, Zhu CD, Jiang GF. 2010. The complete mitochondrial genome of the cockroach Eupolyphaga sinensis (Blattaria: Polyphagidae) and the phylogenetic relationships within the Dictyoptera. Mol Biol Rep. 37:3509–3516. doi: 10.1007/s11033-009-9944-1
