Abstract
Insulin-like growth factor binding protein 6 (IGFBP6) is a key factor in regulating the effects of insulin-like growth factor 2 (IGF2) on the animal growth and development, but the mechanism is far from known. In this study, the 5'-terminal sequence of the IGFBP6 gene (from −920 to −1 bp) which may regulate gene expression was sequenced in the Bama mini-pigs, the Tibetan mini-pigs, the Landrace pigs, the Large White pigs and the Northeast wild boars to screen for the single nucleotide polymorphisms (SNPs) and analyze the relations with the body size traits of swine through a chi-square test analysis. The genotype frequencies of the SNPs in the 5'-terminal sequence have shown that c.-726C > T, c.-722T > C, c.-535C > A, c.-488T > C, c.-403A > G, and c.-378T > C may be related to the dwarf traits of the Bama mini-pig and the Tibetan mini-pigs (P < .05). A haplotype analysis of the 5'-terminal sequence of the IGFBP6 gene in the Landrace pigs, the Large White pigs and the Northeast wild boars found the two SNPs at −403 nt and −378 nt were in linkage and formed three kinds of haplotypes; AT was the dominant haplotype and the haplotype block was not formed in the Bama mini-pigs and Tibetan mini-pigs. Above all, the SNPs and haplotypes of the 5'-terminal sequence of the IGFBP6 gene may be involved in the regulation of swine body size.
Introduction
Insulin-like growth factor binding protein 6 (IGFBP6) is a member of the insulin-like growth factor binding protein (IGFBP) family and functions through both insulin-like growth factor (IGF)-dependent and IGF-independent systems. IGFBP6 differs from the other IGFBPs in that it binds insulin-like growth factor 2 (IGF2) with marked preferential affinity over IGF-1. Therefore, it is able to regulate the growth, development, cell adhesion and other functions mediated by IGF2 (Murphy Citation1998). The swine IGFBP6 gene includes four exons and three introns on chromosome 5 and spans 4.7 kb. Four clustered transcription initiation sites (TIS) were found in the 5'-flanking regions of the human IGFBP6 gene, which does not include TATA box but has Sp1 sites that are clustered near the TIS of the IGFBP6 promoter, which is also the case for other TATA-less promoters (Dusing & Wiginton Citation1994; Huber et al. Citation1998). The finding from genetic markers located the IGFBP6 gene and 5' upstream sequences in the quantitative trait locus for birth weight of swine. In serum, the IGFBP6 content was consistent with that of IGFBP-2, higher than that of IGFBP-1 and lower than 5% that of IGFBP-3 (Rechler & Nissley Citation1990). Mice that overexpressed human IGFBP6 suffered weight loss (Bienvenu et al. Citation2004, Citation2005). Furthermore, downregulation of IGFBP6 has been found to lead to premature entry into cellular senescence. Because the overexpression of IGFBP6 increases the cellular lifespan, the data have suggested that IGFBP6, in contrast to other IGFBPs, is a negative regulator of cellular senescence in human fibroblasts (Micutkova et al. Citation2011).
Miniature pigs are small in body size, and their physiology and genetics are closest to that of humans; thus, they are an ideal animal model for biological and medical research. The Tibetan mini-pigs and the Bama mini-pigs are valuable germplasm resources of miniature pigs. The study of Chinese miniature pigs is still in its infancy, and the growth mechanisms of the Chinese miniature pigs are unclear.
Our previous studies detected the differentiated expression rules of IGFBP6 in various swine varieties with different body sizes. In studies of the molecular mechanisms of the animal growth and development, it is important to understand how the initiation and expression of related genes are regulated. However, the impact of the single nucleotide polymorphisms (SNPs) at the 5' upstream DNA sequences of the IGFBP6 gene on the animal growth and body size traits has not been previously reported. In this experiment, Tibetan mini-pigs and Bama mini-pigs were used (with Northeast wild boars, Large White pigs and Landrace pigs as controls) to screen and analyze the linkage of the SNPs of IGFBP6 (upstream sequence from −920 to −1 bp) with the body size traits of pig.
Materials and methods
Animal resources
The Tibetan mini-pigs were provided by Beijing Tongheshengtai Institute of Comparative Medicine. The Bama mini-pigs were provided by the Yunfu Zhaoqing Pig Farm. The Northeast wild boars were supplied by the Yezhulin Wild Boar Breeding Farm of Jiang Yuan County, Jilin Province. The Landrace pigs and Large White pigs were provided by the Pig Breeds Farm of Jilin University. The five breeds pigs were grouped by body size to the mini-pigs (<45 kg for adult; BamaXiang pigs and Tibetan mini-pigs) and large-sized pigs (>200 kg for adult; Daibai Pigs, Northeast Wild pigs and Junmu No.1 White pigs). All pigs are chosen by random sampling and all breeds are with equal numbers of male and female.
Reagents
LA Taq, DL2000 DNA Marker, dNTPs (TaKaRa Bio. Co., Ltd., Dalian, China) and 2 × Power Taq PCR Master Mix (Bio Teck Biotechnology Co., Ltd., Beijing, China) were used. The DNA purification kit and animal tissue DNA extraction kit were purchased from Axygene.
DNA extraction and determination
The genomic DNA was extracted from the liver or tail tissue of individual pigs of the five swine varieties according to the manufacturer's instructions, and the purity and concentration of the genomic DNA were determined with a NanoDrop 2000 UV spectrophotometer. The DNA preparations in an appropriate amount were used for the gel electrophoresis on 1% agarose gel.
Primers and Polymerase Chain Reaction (PCR) amplification of the 5'-terminal sequences of the IGFBP6 gene
The 5'-terminal sequences of the IGFBP6 gene (NC_010447.4) was amplified by PCR of the genomic DNA pool of the each pig breeds (Bama mini-pigs, Tibetan mini-pigs, Landrace pigs, Northeast wild boars and Large White pigs) and the individuals in five pig breeds with the primers (the forward: 5'-GCACAACACAGCCTCAATAGT-3' and the reverse: 5'-AGCGAGCAACAGAGTTAGCA-3') listed above. The PCR reactions of the IGFBP6 gene 5' terminus included 2.5 μL 10 × LA Taq buffer (Mg2+ Free), 2.0 μL dNTPs (2.5 mM), 0.5 μL forward and reverse primers (10 μM), 2.0 μL template DNA (25 ng/μL), 0.5 μL LA Taq Polymerase (5 U/μL) and 17.0 μL ultra-pure water. The PCR reaction program for amplifications of the 5'-terminal sequence: 95°C for 5 min followed by 30 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 1 min, 10 min extension at 72°C and then held at 4°C. The PCR products (4.0 μL) were used in the gel electrophoresis (1% agarose gel) to detect the PCR amplifications.
SNP genotyping and predicting of transcription factor binding sites
The PCR products were purified according to the manufacturer's instructions and submitted to GENEWIZ, Inc. for DNA Sanger dideoxy sequencing. The sequence alignments were performed using DNASTAR Lasergene (DNAStar, Inc., USA) on the Genebank sequencing data (NC_010447.4) to screen for inter-race mutations in the five swine varieties. The target peaks in the sequencing chromatographs from the five swine varieties were labeled and analyzed using Chormas (Technelysium Pty Ltd, Australia) to screen for the mutations. The SNPs were positioned, and a genotyping statistics and analysis with the PCR products of individuals in each breed were performed. The transcription factor binding sites of sequence −1 to −920 bp in the IGFBP6 gene were predicted on the website (http://www.cbrc.jp/research/db/TFSEARCH.html).
Statistical analysis
The five breeds pigs were grouped by body size to the mini-pigs (<45 kg for adult) and large-sized pigs (>200 kg for adult). A chi-square test analysis was performed on each SNP locus using Graphpad prism 6.0 (Graphpad software, Inc., USA). P < .05 was defined as statistically significant. The gene heterozygosity (He) and gene homozygosity (Ho) were used to measure the genic variation of a population and methods were calculated according to Nei (Citation1973). The polymorphism information content (PIC) was calculated according to Botstein's (Citation1980) methods. A linkage analysis on the haplotype was conducted using HaploView 4.2 software (Daly Lab, USA), and the SNPs with an allele frequency (Minor Allele Frequency) < 1% were excluded from the haplotype analysis. All of the remaining SNPs had a Hardy–Weinberg Equilibrium P > .001. The haplotype block was constructed at and inside the 95% confidence interval of the D' value.
Results
PCR amplification of 5'-terminal sequences of the IGFBP6 gene
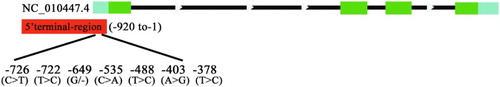
The gel electrophoresis result of the PCR amplification of the 5'-terminal sequences of the IGFBP6 gene is shown in . Distinct bands of amplification products (1134 bp, from −920 to 51 bp of exon 1) were observed.
Screening of SNPs on the 5'-terminal sequences of the IGFBP6 gene
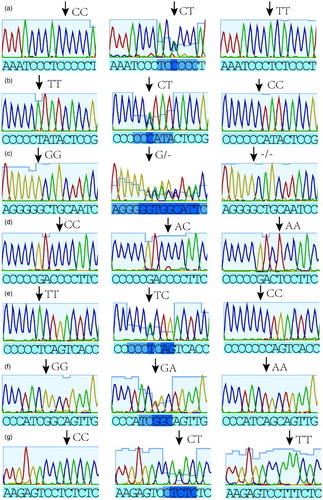
From the sequencing results and sequencing chromatographs of the PCR products, seven SNPs were found in the 5'-terminal sequences of the IGFBP6 gene ().
The detected 5'-terminus SNPs included the following: C.-726C > T, C.-722T > C, C.-649delG, C.-535C > A, C.-488T > C, C.-403A > G and C.-378T > C, as noted on the chromatographs (); C.-403A > G and C.-378T > C have been already reported in the National Center for Biotechnology Information (NCBI) (rs324366597 and rs329035010).
Prediction of the potential transcription factor binding sites in the 5'-terminal sequences of IGFBP6
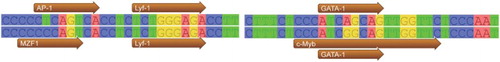
Through predictions, it was found that the C.-488T > C in the 5'-terminal sequence of the swine IGFBP6 gene led to the elimination of the transcription factor binding site AP-1 in the region from −489 to −481 bp and the addition of a new Myeloid zinc finger 1 (MZF1) transcription factor binding site in the region from −491 to −484 bp; however, C.-403A > G in the region from −409 to −392 bp led to the addition of a c-Myb transcription factor binding site, which are shown in .
SNPs loci analysis
The analysis results for each locus of SNPs of the IGFBP6 gene 5'-terminal sequence in the mini-pigs and large-sized pigs are shown in . The differences in genotype distribution of the all the seven SNP sites between the large pigs and mini-pigs at the locus were significant (P < .05 or .01).The values of Ho, He approached 0.5 about most of SNPs in five breeds, and the minimum and maximum PIC values were 0.0761 and 0.5925. According to the classification of the PIC value (PIC value < 0.25, low polymorphism; 0.25 < PIC value < 0.5, intermediate polymorphism; and PIC value > 0.5, high polymorphism), these pig populations mainly belonged to intermediate polymorphism in these SNPs.
Table 1. The frequencies and chi-square test of SNP genotypes in the 5'-terminus of the IGFBP6 gene.
For C.-726C > T, only the TT genotype was observed in the large pigs, whereas the CC, CT and TT genotypes were observed in the mini-pigs, with the TT genotype showing dominance in the Tibetan mini-pigs and the three genotypes showing an equivalent gene frequency in the Bama mini-pigs.
At −649bp, the G/- genotype was dominant in the Large White pigs, but not observed in the Northeast wild boars and Landrace pigs, whereas the GG genotype was exhibited in both the Tibetan mini-pigs and Bama mini-pigs. The allele of G was the dominant allele in the mini-pigs.
For C.-722T > C and C.-488T > C, the TT, TC and CC genotypes were detected only in the Northeast wild boars and no mutations were detected in the remaining four swine varieties at the locus.
C.-535C > A was only detected in the Tibetan mini-pigs, and CA was the dominant genotype; for the remaining four swine varieties, they all exhibited the CC genotype.
C.-403A > G exhibited the AA, AG and GG genotypes in all five swine varieties, and GA was heterozygous dominant in the Landrace pigs, AA was dominant in the Large White pigs, GA was dominant in the Northeast wild boars, GG was dominant in the Bama mini-pigs and GA was dominant in the Tibetan mini-pigs.
C.-378T > C showed TC as the dominant genotype in both the Landrace pigs and Large White pigs, TT as the dominant genotype in the Northeast wild boars and CC as the dominant genotype type in the Bama mini-pigs and Tibetan mini-pigs.
Linkage analysis of the SNPs loci in the haplotype
The results of the linkage analysis of the SNPs in the 5'-terminus of IGFBP6 gene are shown in .
Figure 5. Linkage disequilibrium of the SNPs of the 5'-terminal sequences of the IGFBP-6 gene. Large: large size pigs; Mini: mini-pigs.
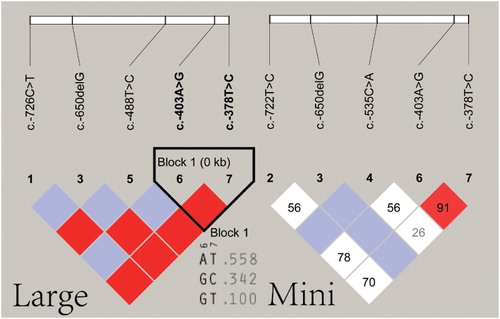
In the five breeds, strong linkages existed between the SNPs at −726 and −488 nt (D' = 1.0, r2 = 1.0), at −726 and −378 nt (D' = 1.0, r2 = 0.066), at −649 and −403 nt (D' = 1.0, r2 = 0.152), at −649 and −378 nt (D' = 1.0, r2 = 0.184) and −488 and −378 nt (D' = 1.0, r2 = 0.066). The remaining SNPs showed weak linkages.
In the Large White pigs, Landrace pigs and Northeast wild boars, a strong linkage was observed between the SNPs at −726 and −488 nt (D' = 1.0, r2 = 1.0), at −726 and −378 nt (D' = 1.0, r2 = 0.058), at −649 and −403 nt (D' = 1.0, r2 = 0.209), at −649 and −378 nt (D' = 1.0, r2 = 0.318), at −488 and −378 nt (D' = 1.0, r2 = 0.058) and −403 and −378 nt (D' = 1.0, r2 = 0.656). A linkage block was formed by the SNPs at −403 and −378 nt, which exhibited three haplotypes: AT (0.558), GC (0.342) and GT (0.100). The remaining SNPs showed weak linkages.
In the Tibetan mini-pigs and Bama mini-pigs, the linkage between the SNPs was weak and no linkage block was formed.
Discussion
IGFBPs include six mutually related peptides and exhibit high-affinity binding to IGFs family proteins (Hwa et al. Citation1999). IGFBPs and IGF receptors compete in the binding of IGF, and the outcome influences cell proliferation and animal growth and development. Although IGFBPs have the same characteristics when interacting with IGF, the expression of each IGFBP is time- and tissue-specific, highly regulated and functions distinctively. The main biological function of IGFBP6 is to inhibit IGF2, and it is also potentially involved in the regulations that are independent of the IGF system (Bach Citation2005).
To understand the molecular mechanisms of animal growth and development, the methods by which related genes are turned on and their expression is regulated must be determined. The mutations at the 5' terminus of a gene can introduce changes in the transcription factor binding sites, thus leading to the changes of the gene transcription efficiency and transcriptional defects that impact the expression and biological functions. In this study, the predicted transcription factor binding sites at the 5' terminus (from −920 to −1 nt) of IGFBP6 had a mutation of C.-403A > G conversion, which led to the addition of a c-Myb transcription factor binding site in the five swine varieties that had the G allele which has a higher frequency in the mini-pigs than the large-sized pigs. The c-Myb transcription factor belongs to the MYB family and plays a key role in the regulation of blood cell formation and may also be involved in tumorigenesis. The newly added c-Myb transcription factor binding site may induce the changes in the expression level of IGFBP6 in the process of the leukocyte differentiation and tumorigenesis (Sala Citation2005). In the 5'-terminal sequences of IGFBP6 in the Northeast wild boars, the C.-488T > C mutation was detected only in the Northeast wild boars which led to the elimination of an AP-1 transcription factor binding site in the region from −489 to −481 bp and addition a MZF1 transcription factor binding site from −491 to −484 bp. Many cytokines and growth factors genes have the AP-1 transcription factor binding site in the gene regulatory region, and certain sites are AP-1 dependent indicating the importance of AP-1 for the cell proliferation and the differentiation transformation (Ameyar et al. Citation2003). The elimination of the AP-1 transcription factor binding site might result in the decline of the expression levels of IGFBP6. MZF1 is a transcription factor belonging to the Krüppel family of zinc finger proteins (Hromas et al. Citation1991) which could increase cell proliferation and tumorigenesis in MZF1 knockout mice (Gaboli et al. Citation2001). And MZF1 was previously reported as the most important factor for follistatin which could tightly regulated during growth and development in pigs. What is more, added a MZF1 transcription factor binding site may play an important role in the Northeast wild boars growth and development by regulating expression of IGFBP6 (Sun et al. Citation2015).
An analysis of the seven SNPs in the 5' terminus of the IGFBP6 gene showed that in the Large White pigs, Landrace pigs and Northeast wild boars, the linkage between two SNPs at −403 and −378 nt formed a haplotype block that exhibited three haplotypes: AT, GC and GT, with AT as the dominant haplotype. In the Tibetan mini-pigs and Bama mini-pigs, however, no such block was formed in this region, indicating that the selective pressure on the SNPs in the DNA region in the large size pigs and mini-pigs was different. In the mini-pigs, the linkages between each SNP were weak, which suggested that the selection pressure leading to the formation of the SNPs in the 5' terminal sequence of the IGFBP6 gene was different between the Tibetan mini-pigs and Bama mini-pigs, which was probably caused by the two mini-pig varieties living in the close geographical environments. The SNPs at −403 and −378 nt were correlated with the dwarfism, suggesting that the haplotype exerted a degree of influence on the swine body size traits. In addition, the haplotype can be used as a potential molecular marker in swine breeding programs.
Based on the results, it is suggested that the IGFBP6 gene may be one of the candidate genes that affects the swine body size traits. The specific mechanism of the IGFBP6 action requires further study. This study provided the molecular evidence of IGFBP6's influence on the body size of mini-pigs and provided a good foundation for the investigations of the growth mechanism of mini-pigs. The study also provided a theoretical basis for the development and application of mini-pig resources.
Funding
This work was supported by the National Natural Science Foundation of China [31101781, 31072102].
Disclosure
No potential conflict of interest was reported by the authors.
References
- Ameyar M, Wisniewska M, Weitzman JB. 2003. A role for AP-1 in apoptosis: the case for and against. Biochimie. 85:747–752. doi: 10.1016/j.biochi.2003.09.006
- Bach LA. 2005. IGFBP-6 five years on; not so ‘forgotten’? Growth Horm IGF Res. 15:185–192. doi: 10.1016/j.ghir.2005.04.001
- Bienvenu G, Seurin D, Grellier P, Froment P, Baudrimont M, Monget P, Le Bouc Y, Babajko S. 2004. Insulin-like growth factor binding protein-6 transgenic mice: postnatal growth, brain development, and reproduction abnormalities. Endocrinology. 145:2412–2420. doi: 10.1210/en.2003-1196
- Bienvenu G, Seurin D, Le Bouc Y, Even P, Babajko S, Magnan C. 2005. Dysregulation of energy homeostasis in mice overexpressing insulin-like growth factor-binding protein 6 in the brain. Diabetologia. 48:1189–1197. doi: 10.1007/s00125-005-1767-6
- Botstein D, White RL, Skolnick M, Davis RW. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Human Genetics. 32:314–331.
- Dusing MR, Wiginton DA. 1994. Sp1 is essential for both enhancer-mediated and basal activation of the TATA-less human adenosine deaminase promoter. Nucleic Acids Res. 22:669–677. doi: 10.1093/nar/22.4.669
- Gaboli M, Kotsi PA, Gurrieri C, Cattoretti G, Ronchetti S, Cordon-Cardo C, Broxmeyer HE, Hromas R, Pandolfi PP. 2001. Mzf1 controls cell proliferation and tumorigenesis. Genes Develop. 15:1625–1630. doi: 10.1101/gad.902301
- Hromas R, Collins SJ, Hickstein D, Raskind W, Deaven LL, O'Hara P, Hagen FS, Kaushansky K. 1991. A retinoic acid-responsive human zinc finger gene, mzf-1, preferentially expressed in myeloid cells. J Biol Chem. 266:14183–14187.
- Huber R, Schlessinger D, Pilia G. 1998. Multiple Sp1 sites efficiently drive transcription of the TATA-less promoter of the human glypican 3 (GPC3) gene. Gene. 214:35–44. doi: 10.1016/S0378-1119(98)00233-9
- Hwa V, Oh Y, Rosenfeld RG. 1999. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev. 20:761–787.
- Micutkova L, Diener T, Li C, Rogowska-Wrzesinska A, Mueck C, Huetter E, Weinberger B, Grubeck-Loebenstein B, Roepstorff P, Zeng R, Jansen-Duerr P. 2011. Insulin-like growth factor binding protein-6 delays replicative senescence of human fibroblasts. Mech Ageing Dev. 132:468–479. doi: 10.1016/j.mad.2011.07.005
- Murphy LJ. 1998. Insulin-like growth factor-binding proteins: functional diversity or redundancy? J Mol Endocrinol. 21:97–107. doi: 10.1677/jme.0.0210097
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proc Nat Acad Sci U S A. 70:3321–3323. doi: 10.1073/pnas.70.12.3321
- Rechler MM, Nissley SP. 1990. Insulin-like growth factors: peptide growth factors and their receptors I. Sporn M & Roberts A. New York: Springer.
- Sala A. 2005. B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur J Cancer. 41:2479–2484. doi: 10.1016/j.ejca.2005.08.004
- Sun YM, Wang L, Yang XQ, Zhang DJ, Liu D. 2015. Myeloid zinc finger 1 (mzf1) is the most important transcriptional factor for porcine follistatin promoter. J Int Agri. 14:1383–1389. doi: 10.1016/S2095-3119(14)60893-5