ABSTRACT
TRIM46 is a RING finger E3 ligase which belongs to TRIM (tripartite motif-containing) protein family. TRIM46 is required for neuronal polarity and axon specification by driving the formation of parallel microtubule arrays, whereas its embryological functions remain to be determined yet. Expression patterns and biological functions of trim46a, a zebrafish homologue of TRIM46, were studied in zebrafish embryo. First, maternal transcripts of trim46a were present at 1 cell stage whereas zygotic messages were abundant in the eyes, MHB (Midbrain-Hindbrain Boundary) and hindbrain at 24 hpf (hours post fertilization). Second, transcriptional regulatory region of trim46a contains cis-acting elements binding a transcriptional factor Foxa2. Transcription of foxa2 is positively regulated by Sonic Hedgehog (SHH), and treatment of cyclopamine, an SHH inhibitor, represses transcription of foxa2 in 4 hpf through 24 hpf embryos. Third, the transcriptional repression of foxa2 inhibited transcription of trim46a to cause developmental defects in the midbrain and MHB. Finally, spatiotemporal expression patterns of a midbrain marker otx2b in the developmental defects confirmed inhibition of SHH by cyclopamine caused underdevelopment of the midbrain and MHB at 24 hpf. We propose a signaling network where trim46a contributes to development of the midbrain and MHB via Foxa2, a downstream element of SHH signaling in zebrafish embryogenesis.
Introduction
Eukaryotic cells are equipped to degrade proteins via the ubiquitin-proteasome system (UPS). Proteins become degraded upon their conjugation to chains of ubiquitin where they are directed to the 26S proteasome, a macromolecular protease (Deshaies and Joazeiro Citation2009; Kleiger and Mayor Citation2014). The post-translational attachment of ubiquitin, a highly conserved 76-amino-acid polypeptide, also directs myriad eukaryotic proteins to a variety of fates and functions, such as internalization and lysosomal targeting, modulation of protein interactions, alteration of subcellular distribution, regulation of transcription, DNA repair and propagation of transmembrane signaling, most notably in the nuclear factor kappa B (NF-κB) pathway (Metzger et al. Citation2012). Derangements of the UPS can lead to the dysregulation of cellular homeostasis and the development of multiple disorders including neurodegenerative and systemic autoimmune diseases (Nalepa et al. Citation2006).
TRIM46 (tripartite motif-containing 46) belongs to the large family of tripartite motif proteins that have been implicated in many biological processes including UPS, cell differentiation, transcriptional regulation and signaling pathway (McNab et al. Citation2011; Versteeg et al. Citation2013). TRIM proteins are characterized by the presence of a highly conserved N-terminal RBCC motif that consists of a RING finger domain, one or two B-boxes, and a coiled-coil domain but differ in their C-terminal regions that confer functional specificity (Reymond et al. Citation2001; Meroni and Diez-Roux Citation2005; Short and Cox Citation2006). Based on their C-terminal domain composition, TRIM46 has been classified into C11 subgroups (Ozato et al. Citation2008).
Ubiquitin ligase TRIM46 has been revealed as an essential element in neurogenesis as well as in the initiation and maintenance of axonal transport. TRIM46 is localized in the proximal axon, where it specifies neuronal polarity of microtubule bundles oriented with their plus-end pointing outward (Curcio and Bradke Citation2015), defining an axonal cytoskeletal compartment for microtubule organization during neuronal development (van Beuningen et al. Citation2015). TRIM46 regulates microtubule fasciculation in the axon initial segment (AIS) (Harterink et al. Citation2019) and is transported by microtubule-based motors KIF3A/B/KAP3 under a MARK2 (microtubule affinity-regulating kinases 2) phosphorylation cascade in axonal cargo trafficking (Fréal et al. Citation2019; Ichinose et al. Citation2019). TRIM46 has been known to be involved in inflammatory response through ubiquitination. It ubiquitinates DUSP1 (Dual-specificity phosphatase 1), which in turn induces activation of NF-κB and MAPK (Mitogen-activated protein kinases) in colonic inflammation (Li et al. Citation2020). On the other hand, TRIM46 acts as an oncogene in osteosarcoma by interacting with and ubiquitinating peroxisome proliferator-activated receptor alpha (PPARα), resulting in the activation of the NF-κB signaling pathway (Jiang et al. Citation2020). Despite the many reports describing the various molecular functions of TRIM46 in mammalian cells, its embryological functions have yet to be determined. We thus investigated expression patterns as well as signaling networks governing the transcription of trim46a in zebrafish embryos. We propose a signaling pathway including Sonic Hedgehog (SHH) and Foxa2, which controls transcription of trim46a.
Materials and methods
Adult zebrafish care and embryos
Wild types of adult zebrafish were maintained at 28.5°C with 10 h dark/14 h light cycles. Embryos were obtained through natural spawning and raised, and staged as described previously (Kimmel et al. Citation1995; Westerfield Citation2000). The pigmentation of embryos for whole-mount in situ hybridization (WISH) was blocked by treating them with 0.002% phenylthiourea (PTU) after onset of somitogenesis.
Sequence analysis
The sequences and additional information of zebrafish trim46a and other orthologs were obtained by searching the database of National Center for Biotechnology Information (NCBI) (http://www.ncbi.nim.nih.gov/). Amino acid sequences of trim46a and other orthologs were aligned as previously described (Choe et al. Citation2020) to identify the similarity among them.
RNA isolation, cDNA synthesis and reverse transcription-PCR (RT-PCR)
Total RNA isolation, cDNA synthesis and reverse transcription polymerase chain reaction (RT-PCR) were performed as described in (Kang et al. Citation2014). The primers were used for PCR to analyze the zebrafish trim46a specific template (228 bp) in different stages of embryos, forward primer; 5′-GGACTGCGTTAAATTGGACCTCTCAG-3′ and reverse primer; 5′-ATCGCTGAAACTGTGCCGTTGTTC-3′, and for β-actin (328 bp), as an internal control for this experiment, forward primer; 5′-GCCCATCTATGAGGGTTACG-3′ and reverse primer; 5′-GCAAGATTCCATACCCAGGA-3′. Upon completion of the PCR reaction, every set of reaction was confirmed with running 1% agarose gel in TE buffer using gel electrophoresis. After cloning in pGEM®T-easy vector and confirmation with the digestion process, plasmid construct was sent for sequencing to SolGent Co. Ltd. The protocols were established and applied as previously in our laboratory (Kumar et al. Citation2017).
Whole-mount in situ hybridization
Whole-mount in situ hybridization (WISH) was performed following a standard procedure with minor modifications as described in (Thisse et al. Citation1993; Jung et al. Citation2019). After confirming the sequences in the plasmid construct, cloned trim46a construct was linearized with restriction enzyme. Then DIG-labeled antisense and sense probe of trim46a were synthesized using the DIG RNA Labeling Kit (SP6/T7) (Roche, USA). Embryos were fixed in 4% paraformaldehyde (PFA) overnight, and dehydrated in 100% methanol. All images and their magnification were captured with Leica MZ16 microscopy systems.
Bioinformatic analysis of putative promoter regions
Promoter region sequences (3899 bp) of trim46a were obtained from National Center for Biotechnology Information (NCBI) (http://www.ncbi.nim.nih.gov/) and searched for the occurrences of binding motifs for transcription factors (TF) as defined by Transfac database (Wingender et al. Citation1996).
Treatment of cyclopamine to zebrafish embryos
Embryos were staged and synchronized at 4 hpf by discarding any embryos not at sphere stage, and then incubated beginning at 4 hpf until 24 hpf in embryo medium containing 20 μM and 60 μM cyclopamine respectively (LC Laboratories), diluted from a 10 mM stock in ethanol, at 28.5°C. Control embryos were treated with an equivalent concentration of ethanol. Embryos were raised at low density (no more than 50 embryos/100 mm Petri dish) to ensure developmental synchrony across individuals (Kay et al. Citation2005), and manually dechorionated at 24 hpf. Phenylthiourea (PTU), normally used to block the pigmentation of embryos for WISH, was not treated to avoid interference in cyclopamine treatment. To stop the treatment, embryos at 24 hpf were rinsed three times in fresh embryo medium, and fixed for WISH analysis.
Quantification of expression level of foxa2 and trim46a
To provide objective and quantitative assessments of foxa2 and trim46a expression, the total area of WISH staining was examined and analyzed by ImageJ software (National Institutes of Health, Bethesda, MD). First, the images were converted to 8-bit grayscale and the area to be measured was outlined, and the sum of intensity (reaction product plus background) was determined in this region of interest (ROI). Next, the outline was moved to an adjacent region of the embryo that showed no reaction product and the background intensity was determined. This was then subtracted from the ROI. Using the sum intensity and dividing by the ROI pixel area, the mean intensity of an embryo was calculated. The average mean intensity of each gene was obtained from 5 to 15 embryos in each group.
Statistical analysis
Statistically significant differences between the two groups were determined using the unpaired t tests. P < 0.05 was considered to indicate a statistically significant difference. GraphPad Prism 9 software was used for data analysis.
Results
Identification of trim46a and structural analysis of Trim46a
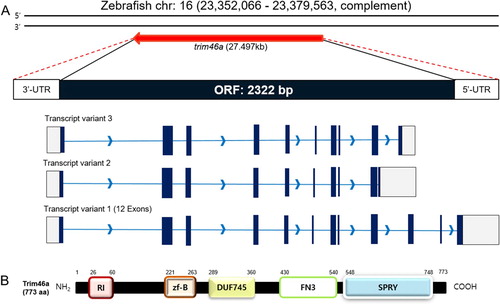
To identify trim46a, an ortholog of TRIM46 in zebrafish, NCBI database was used as reference. Zebrafish trim46a (Gene ID;569905) is mapped to chromosome 16 and located in NC_007127.7 (23,352,066, 23,379,563, complement) which spans about 27.497 kb of nucleotides. trim46a transcript consists of 4136 bp containing the open reading frame (ORF) between 340 bp and 2661 bp, which encodes 773 amino acids ((A)). Protein structure of Trim46a analyzed by NCBI Conserved Domain Search with the default parameters found that Trim46a consists of the RING_Ubox superfamily domain (105 a. a), zf-B box domain (43 a. a), DUF745 superfamily domain (72 a. a), FN3 domain (111 a. a) and SPRY superfamily domain (201 a. a) ((B)). Comparative analysis of Trim46 orthologs among zebrafish, mouse (M. musculus), and human (H. sapiens) using CLUSTAL OMEGA under default parameters for multiple sequence alignment demonstrated that zebrafish Trim46a shares homology with mouse and human by 58.96% and 59.36%, respectively (data not shown).
Figure 1. Schematic diagrams of the ORF of trim46a and protein structures of Trim46a. (A) Zebrafish trim46a is present on chromosome 16 and spans 27,494 bp on the genome. trim46a includes 2322 bp of open reading frame (ORF) encoding 773 amino acids long Trim46a. (B) Trim46a contains RING_Ubox superfamily domain (amino acids 26–60), zf-B box domain (amino acids 221–263), DUF745 superfamily domain (amino acids 289–360), FN3 domain (amino acids 430–540), and SPRY superfamily domain (amino acids 548–748).
Zygotic transcripts of trim46a are restricted to the neural plate, telencephalon, mesencephalon, and hindbrain in embryos at 24 hpf
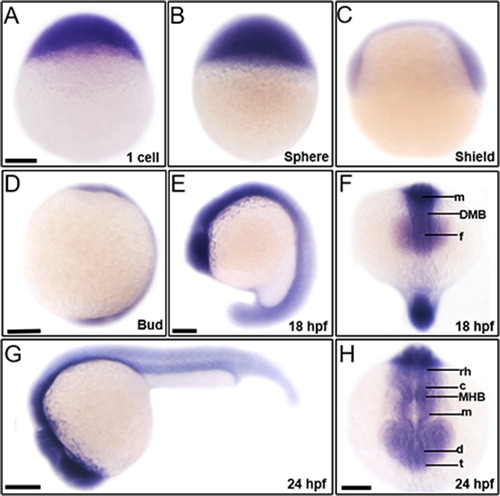
To elucidate contribution of trim46a to vertebrate embryogenesis, we initially examined spatiotemporal expression of trim46a in zebrafish embryos at various developmental stages. Analysis of RT-PCR (Reverse Transcription Polymerase Chain Reaction) exhibited that trim46a transcripts were abundant at 1-cell stage, and the expression persisted until 24 hpf, indicating that it is maternally and zygotically expressed (data not shown). Spatiotemporal expression patterns of trim46a were analyzed with whole-mount in situ hybridization (WISH) using trim46a-specific digoxigenin-labeled RNA probe in zebrafish embryos at 1-cell ((A)), sphere ((B)), and shield ((C)) stages. trim46a transcripts were present as maternal messages while the zygotic transcripts were localized to the central nervous system at bud stage and 18 hpf ((D, E)). trim46a transcripts were restricted to the neural plate, forebrain, midbrain, and hindbrain at 18 hpf ((F)) and further to the eyes, telencephalon, diencephalon, midbrain, MHB (Midbrain-Hindbrain Boundary), cerebellum and rhombomeres ((G,H)).
Figure 2. Spatiotemporal expression patterns of trim46a in zebrafish embryos at 1 cell, sphere, shield, bud, 18 hpf and 24 hpf. trim46a transcripts were distributed in the precursor region of brain along the central nervous system. (A) Maternal messages of trim46a were present at 1-cell stage. (B) trim46a transcripts were abundant in the deep cell layer (DEL), enveloping layer (EVL) and I-YSL (yolk syncytial layer) at sphere stage. (C) trim46a transcripts were localized to the ventral & dorsal region at shield stage. (D) trim46a transcripts were abundant in the central nervous system at bud stage. (E and F) trim46a transcripts were evenly distributed in the brain region. (G and H) Lateral (G) and anterior (H) view of zebrafish embryos at 24 hpf; trim46a messages were restricted to the forebrain through the telencephalon, diencephalon, midbrain, MHB, cerebellum and rhombomere. All embryos were collected synchronously from WT zebrafish for WISH analysis at the corresponding stages. MHB, midbrain-hindbrain boundary; DMB, diencephalic-mesencephalic boundary; c, cerebellum; rh, rhombomere; t, telencephalon; m, midbrain; d, diencephalon; f, forebrain. (A–H) Scale bars: 50 μm.
Trim46a promoter contains cis-acting elements preferentially binding to Foxa2
To identify potential regulatory elements of trim46a transcription, 5’-upstream (3,000 bp long) region from the transcriptional start site and 500bp of the first intron were analyzed by Transfac database (Wingender et al. Citation1996). Various cis-acting elements were identified; Ikaros family zinc finger protein 1 (IKZF1), LIM homeobox gene 3 (LHX3), Hepatocyte nuclear factor 4 alpha 1 (HNF-4alpha1), Forkhead box A2 (FOXA2), Forkhead box D3 (FOXD3), Octamer transcription factor 1 (OCT-1), TATA-binding protein (TBP), E2F, Meis1A:Hoxa9 and Runt-related transcription factor 1 (RUNX1) ((B)). Among the candidate transcriptional factors, Foxa2 was selected as a potential regulator of the transcription of trim46a because its DNA recognition domain is well conserved in zebrafish (-1244 bp: AAATGTTTGCTTT), mouse (-1618 bp: AGTCAATAGTGCG) and human (-458 bp: GAGGAAATAATGCG) (data not shown). Forkhead box (FOX) proteins comprise a large family of transcription factors (TFs), members of which display functional diversity and participate in cellular processes ranging from development to immunity and metabolism (Hannenhalli and Kaestner Citation2009; Lam et al. Citation2013).
Figure 3. Putative cis-acting elements in the promoter region (3899 bp) of trim46a. Putative cis-acting elements defined by the Transfac database (Wingender et al. Citation1996). (A) Schematic representation of zebrafish trim46a genomic region. Twelve exons (E1 to E12) and eleven introns are depicted; the translation initiation site is indicated with an arrow. (B) List of the sixteen transcription factors which might bind to their corresponding response elements within the trim46a promoter (3899 bp). (C) The putative transcription factors are highlighted in bold and blue color represents exon1 (Cut-off p-value = 0).

Inhibition of Sonic Hedgehog (SHH) signaling caused developmental defects in the midbrain at 24 hpf
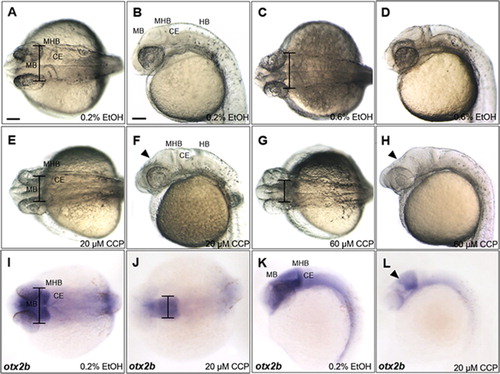
Organizing midbrain signaling centers is known to be tightly associated with SHH signaling in chick embryos (Bayly et al. Citation2012) while Foxa2 is a direct downstream target of SHH pathway and pharmacologically blocked by cyclopamine (Chen et al. Citation2002; Schäfer et al. Citation2007). Because the upstream promoter region of trim46a contains a binding site for Foxa2 (), it is conceivable that SHH signaling might control transcription of trim46a via Foxa2. To test this possibility, an inhibitor of SHH, cyclopamine was treated to zebrafish embryos at 4 hpf through 24 hpf. Embryos treated with cyclopamine at 20 μM and 60 μM demonstrated severe shrinkage in the midbrain along the dorso-ventral axis, accompanying notable reduction in width of the midbrain at 24 hpf ((A–H)). Developmental defects of the midbrain in the cyclopamine-treated embryos were further analyzed with a midbrain marker otx2b (Li et al. Citation1994). otx2b transcripts detected in the midbrain were outstandingly lower than those in the ethanol-treated controls at 24 hpf ((I–L)), suggesting that inhibition of SHH signaling with cyclopamine hinders proper development of the midbrain. Considering the presence of Foxa2 binding site in the upstream promoter region of trim46a and SHH as an upstream regulator of Foxa2, it is highly probable that trim46a defines development of the midbrain via SHH-Foxa2 signaling pathway in zebrafish embryos.
Figure 4. Inhibition of SHH signaling with cyclopamine resulted in developmental defects in the midbrain of zebrafish embryos at 24 hpf. (A–D) Zebrafish embryos at 24 hpf which were treated 0.2% and 0.6% EtOH from 4 hpf. 0.6% EtOH-exposed embryos showed similar phenotypes of midbrain and MHB in comparison to those of 0.2% EtOH-exposed embryos. (E and F) Cyclopamine-treated embryos (20 μM) showed reduction in width of the midbrain and dorsally shrunken midbrain in comparison to those of EtOH controls. (G and H) 60 μM cyclopamine-treated embryos exhibited severe defects in the midbrain. (I–L) Spatiotemporal expression patterns of otx2b in the midbrain of cyclopamine-treated embryos at 24 hpf. CCP, cyclopamine. (A–L) Scale bars: 50 μm.
Inhibition of SHH reduced the level of transcripts of foxa2 and trim46a in the embryos at 24 hpf
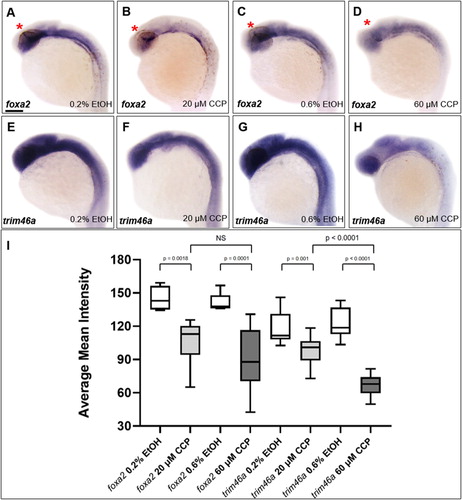
To determine if the inhibition of SHH regulates transcription of trim46a, spatiotemporal expression patterns of trim46a were analyzed in zebrafish embryos treated with cyclopamine. Spatiotemporal expression patterns of foxa2 were initially studied to ask if Foxa2, a downstream target of SHH signaling regulates transcription of trim46a via the putative Foxa2 binding site in the promoter. Upon the treatment of cyclopamine to zebrafish embryos, level of foxa2 transcripts was significantly reduced in the ZLI ((B,D)) compared to that of the control at 24 hpf ((A,C)). In order to examine if the reduced transcripts of foxa2 caused any changes in transcription trim46a, spatiotemporal expression patterns of trim46a in the cyclopamine-treated embryos were analyzed with WISH. Level of trim46a transcripts were decreased in the hypothalamus, midbrain and MHB in dose-dependent manners ((F,H)). Furthermore, staining intensity of the transcripts from trim46a and foxa2 were measured for validation of the anatomical territories in the brain by employing ImageJ (National Institutes of Health, Bethesda, MD) analysis software. Region of interest (ROI) consists of the forebrain, midbrain and cerebellum except for the rhombomeres. Calculation of the mean intensity indicated that inhibition of SHH signaling by cyclopamine reduced level of trim46a transcripts more significantly than that of foxa2 in the ROI of the zebrafish embryos at 24 hpf ((I)). Based upon all the observations, it is highly probable that proper expression of trim46a via SHH signaling is essential to development of the midbrain in zebrafish embryos.
Figure 5. Inhibition of SHH signaling by cyclopamine decreased the level of trim46a transcripts in dose-dependent manners. (A–D) WISH analysis using foxa2 specific probe detected foxa2 transcripts in the midbrain at 24 hpf. Level of foxa2 transcripts decreased in the embryos treated with cyclopamine versus the control. (E–H) WISH analysis of cyclopamine-treated embryos using trim46a as a probe. Cyclopamine caused reduction in the level of trim46a transcripts in comparison to those of EtOH control in dose-dependent manners. (I) Quantification of foxa2 transcripts in the ZLI, and trim46a transcripts in the forebrain, midbrain and cerebellum at 24 hpf using ImageJ software. CCP, cyclopamine. (A–H) Scale bars: 50 μm.
Discussion
SHH signaling is required for trim46a to develop the ZLI in midbrain in zebrafish embryogenesis
Cyclopamine led to the repression of genes specifically associated with hedgehog signaling (Incardona et al. Citation1998; Chen et al. Citation2002). Formation of the ZLI in midbrain is closely related to temporal adaptation of gradient SHH for the pre-thalamic and thalamic gene expression domains during diencephalic patterning (Krauss et al. Citation1993; Balaskas et al. Citation2012). Our data in this report provide evidences that the cyclopamine-treated embryos exhibited dorsally reduced transcripts of foxa2 in the ZLI ((B,D)). This, in turn, implies that acquisition of trim46a expression is dependent on direct SHH signaling for proper neurogenesis of the diencephalon, which requires the ZLI-derived SHH signaling (Kiecker and Lumsden Citation2004).
Foxa2 is a putative transcriptional regulator of trim46a in response to SHH signaling
TRIM46 promoted cell growth and inhibited apoptosis of osteosarcoma cells by activating NF-κB signaling pathway through the ubiquitination of PPAR (Peroxisome proliferator-activated receptor). TRIM46 induces ubiquitination of PPAR-α and regulates cell viability, cell cycle, and apoptosis of OS (Jiang et al. Citation2020). PPARs regulate many cellular processes in the CNS, for instance, PPARγ regulates neural cell differentiation, death, and inflammation, in addition to playing a neuroprotective role in human neural stem cells (Chiang et al. Citation2016). Two duplicate paralogs of mammalian PPARA namely, Pparαa and Pparαb are present in the zebrafish genome. Both Pparαa and Pparαb are essential regulators of the proliferation of neuronal and glial precursors in zebrafish embryos (Hsieh et al. Citation2018). Based on our findings that trim46a is implicated in zebrafish neurogenesis, further study of its relevance to Pparαa and Pparαb would be an interesting topic. In addition, bioinformatics studies in this report discovered a putative Foxa2 binding site in the upstream region of trim46a (). Furthermore, the repression of Foxa2 by cyclopamine caused the transcriptional repression of trim46a (), which resulted in the developmental defects of the ZLI of zebrafish embryos (). Most paralogous FOX proteins bind to cis-acing element 5′-RYAAAYA-3′ (R = A or G, Y = C or T) (Koh et al. Citation2009; Chen et al. Citation2016). The conserved C-terminus of FOXA proteins interact with histones H3 and H4 and overcome physical barrier of nucleosomes (Clark et al. Citation1993). FOXA2, which belongs to pioneer transcription factors (PTFs) has ability to engage their target sequence on nucleosomal DNA and initiate gene expression that confers cell identities (Soufi et al. Citation2015). Foxa2 is critical to continued survival of the mesodiencephalic dopaminergic neurons (Kittappa et al. Citation2007) and cooperates with Lmx1a and Lmx1b to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon (Nakatani et al. Citation2010). In particular, it is most likely that Foxa2 contributes to the specification of dopaminergic neurons and the control of floor plate cell differentiation in the developing mesencephalon by regulating the transcription of trim46a in the zebrafish embryos.
Acknowledgements
This work was supported by research fund of Chungnam National University. Jaehun Kim is grateful for financial support from Hyundai Motor Chung Mong-Koo Foundation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, Ribes V. 2012. Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell. 148:273–284.
- Bayly RD, Brown CY, Agarwala S. 2012. A novel role for FOXA2 and SHH in organizing midbrain signaling centers. Dev Biol. 369:32–42.
- Chen X, Ji Z, Webber A, Sharrocks AD. 2016. Genome-wide binding studies reveal DNA binding specificity mechanisms and functional interplay amongst Forkhead transcription factors. Nucleic Acids Res. 44:1566–1578.
- Chen JK, Taipale J, Cooper MK, Beachy PA. 2002. Inhibition of Hedgehog signaling by direct binding of cyclopamine to smoothened. Genes Dev. 16:2743–2748.
- Chiang MC, Nicol CJ, Cheng YC, Lin KH, Yen CH, Lin CH. 2016. Rosiglitazone activation of PPARγ-dependent pathways is neuroprotective in human neural stem cells against amyloid-beta-induced mitochondrial dysfunction and oxidative stress. Neurobiol Aging. 40:181–190.
- Choe S, Huh TL, Rhee M. 2020. Trim45 is essential to the development of the diencephalon and eye in zebrafish embryos. Anim Cells Syst. 24:99–106.
- Clark KL, Halay ED, Lai E, Burley SK. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 364:412–420.
- Curcio M, Bradke F. 2015. Microtubule organization in the axon: TRIM46 determines the orientation. Neuron. 88:1072–1074.
- Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 78:399–434.
- Fréal A, Rai D, Tas RP, Pan X, Katrukha EA, van de Willige D, Stucchi R, Aher A, Yang C, Altelaar AFM, et al. 2019. Feedback-driven assembly of the axon initial segment. Neuron. 104:305–321.
- Hannenhalli S, Kaestner KH. 2009. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 10:233–240.
- Harterink M, Vocking K, Pan X, Soriano Jerez EM, Slenders L, Fréal A, Tas RP, van de Wetering WJ, Timmer K, Motshagen J, et al. 2019. TRIM46 organizes microtubule fasciculation in the axon initial segment. J Neurosci. 39:4864–4873.
- Hsieh YC, Chiang MC, Huang YC, Yeh TH, Shih HY, Liu HF, Chen HY, Wang CP, Cheng YC. 2018. Pparα deficiency inhibits the proliferation of neuronal and glial precursors in the zebrafish central nervous system. Dev Dyn. 247:1264–1275.
- Ichinose S, Ogawa T, Jiang X, Hirokawa N. 2019. The spatiotemporal construction of the axon initial segment via KIF3/KAP3/TRIM46 transport under MARK2 signaling. Cell Rep. 28:2413–2426.
- Incardona JP, Gaffield W, Kapur RP, Roelink H. 1998. The teratogenic veratrum alkaloid cyclopamine inhibits sonic hedgehog signal transduction. Development. 125:3553–3562.
- Jiang W, Cai X, Xu T, Liu K, Yang D, Fan L, Li G, Yu X. 2020. Tripartite motif-containing 46 promotes viability and inhibits apoptosis of osteosarcoma cells by activating NF-B signaling through ubiquitination of PPAR. Oncol Res. 28:409–421.
- Jung J, Udhaya Kumar S, Choi I, Huh TL, Rhee M. 2019. Znf76 is associated with development of the eyes, midbrain, MHB, and hindbrain in zebrafish embryos. Anim Cells Syst. 23:26–31.
- Kang N, Won M, Rhee M, Ro H. 2014. Siah ubiquitin ligases modulate nodal signaling during zebrafish embryonic development. Mol Cells. 37:389–398.
- Kay JN, Link BA, Baier H. 2005. Staggered cell-intrinsic timing of ath5 expression underlies the wave of ganglion cell neurogenesis in the zebrafish retina. Development. 132:2573–2585.
- Kiecker C, Lumsden A. 2004. Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat Neurosci. 7:1242–1249.
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn. 203:253–310.
- Kittappa R, Chang WW, Awatramani RB, McKay RD. 2007. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol. 5:e325.
- Kleiger G, Mayor T. 2014. Perilous journey: a tour of the ubiquitin-proteasome system. Trends Cell Biol. 24:352–359.
- Koh KP, Sundrud MS, Rao A. 2009. Domain requirements and sequence specificity of DNA binding for the forkhead transcription factor FOXP3. PLoS One. 4:e8109.
- Krauss S, Concordet JP, Ingham PW. 1993. A functionally conserved homolog of the drosophila segment polarity gene HH is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 75:1431–1444.
- Kumar A, Huh TL, Choe J, Rhee M. 2017. Rnf152 is essential for NeuroD expression and delta-notch signaling in the zebrafish embryos. Mol Cells. 40:945–953.
- Lam EW, Brosens JJ, Gomes AR, Koo CY. 2013. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 13:482–495.
- Li Y, Allende ML, Finkelstein R, Weinberg ES. 1994. Expression of two zebrafish orthodenticle-related genes in the embryonic brain. Mech Dev. 48(3):229–244.
- Li Y, Xu S, Xu Q, Chen Y. 2020. Clostridium difficile toxin B induces colonic inflammation through the TRIM46/DUSP1/MAPKs and NF-κB signalling pathway. Artif Cells Nanomed Biotechnol. 48:452–462.
- McNab FW, Rajsbaum R, Stoye JP, O’Garra A. 2011. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol. 23:46–56.
- Meroni G, Diez-Roux G. 2005. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 27:1147–1157.
- Metzger MB, Hristova VA, Weissman AM. 2012. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 125:531–537.
- Nakatani T, Kumai M, Mizuhara E, Minaki Y, Ono Y. 2010. Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev Biol. 339:101–113.
- Nalepa G, Rolfe M, Harper JW. 2006. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 5:596–613.
- Ozato K, Shin DM, Chang TH, Morse HC. 2008. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 8:849–860.
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140–2151.
- Schäfer M, Kinzel D, Winkler C. 2007. Discontinuous organization and specification of the lateral floor plate in zebrafish. Dev Biol. 301:117–129.
- Short KM, Cox TC. 2006. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem. 281:8970–8980.
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. 2015. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 161:555–568.
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. 1993. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 119:1203–1215.
- van Beuningen SFB, Will L, Harterink M, Chazeau A, van Battum EY, Frias CP, Franker MAM, Katrukha EA, Stucchi R, Vocking K, et al. 2015. TRIM46 controls neuronal polarity and axon specification by driving the formation of parallel microtubule arrays. Neuron. 88:1208–1226.
- Versteeg GA, Rajsbaum R, Sánchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, García-Sastre A. 2013. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 38:384–398.
- Westerfield M. 2000. The zebrafish book: A guide for the laboratory use of zebrafish (Danio rerio). Eugene: Univ. of Oregon Press.
- Wingender E, Dietze P, Karas H, Knüppel R. 1996. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 24:238–241.
