ABSTRACT
The genetic investigation of the archeological or museum samples, including endangered species, provides vital information necessary to plan, implement, and revisit conservation strategies. In South Korea, the Asian black bear went almost extinct in wild by 2002, without leaving any authentic specimens representing the native population. Recently researchers found a set of animal bones in a natural cave in Mt. Taebaek (South Korea), suspected to be of a bear. In the present study, we undertook a molecular investigation and radiocarbon dating to establish the species’ identity, phylogenetic position, and approximate age of the recovered specimen. The genetic investigation (CytB, COI, D-loop, SRY, and ZFX-ZFY) identified the sample as a male Asian black bear with close phylogenetic affinity with Northeast Asian bears. Radiocarbon dating estimated the bones to be aged 1800–1942 calAD. These findings indicate that the bone specimens found in the natural cave in Mt. Taebaek were from an individual that naturally inhabited South Korea long before the importing of farm bears (the 1980s) and initiation of wild population restoration (2004). The present study provides the first genetic information record of the native South Korean black bear. Our findings reaffirm the appropriateness of the ongoing bear restoration program in South Korea, with the reintroduction of individuals from North Korea and Russia.
Introduction
Ancient or old DNA-based studies are useful in understanding the phylogenetic relationship between extinct and extant species. Museum sample genetic analysis was found to be useful in establishing a phylogenetic relationship between extinct quagga and extant mountain zebra (Higuchi et al. Citation1984). Based on the mitochondrial DNA analysis of Holocene sub-fossil giant panda and modern panda, Sheng et al. (Citation2018) concluded that extant panda populations do not suffer any immediate threat from the perspective of genetic evolutionary potential. Ancient DNA analysis was also found useful to elucidate the evolutionary history of Yakutian brown bears and Japanese otters (Park et al. Citation2019; Rey-Iglesia et al. Citation2019). Archeological genetic studies are difficult to perform as old fossilized samples yield low-quality DNA with high chances of environmental contamination (Rohland and Hofreiter Citation2007). These studies require dedicated laboratory facilities, skilled researchers, and efficient protocols for DNA extraction and PCR.
Asian black bear is included in the endangered species list of the South Korean Ministry of Environment. In South Korea, Asian black bears can be categorized into three groups – native, farmed, and reintroduced. Bear farms were established in the 1980s to support local livelihood (MOE Citation2005; Lee et al. Citation2013; Jang Citation2014). The genetic ancestry of farmed black bears is ambiguous and unknown (Kim et al. Citation2012). The native Asian black bear population of South Korea was predicted less than 5 by the early twenty-first century due to excessive persecution and habitat loss. The South Korean government initiated the Asian black bear population restoration project (Mt. Jiri), in 2004, using individuals imported from North Korea, China, and Russia (Lee and Jeong Citation2009; Kim et al. Citation2012). According to the International Union for Conservation of Nature (IUCN) guidelines for re-introductions, the population restoration of endangered species should be preceded by a phylogenetic examination of the native population to ensure the selection of candidates from a genetically similar population or subspecies (IUCN Citation1998). Hong (Citation2005) and Kim et al. (Citation2011) conducted a phylogenetic study on Asian black bears, including the Ussuri subspecies, and found that candidate populations, for the South Korean Asian black bear restoration project, were appropriate. However, their examination does not include any native South Korean Asian black bear specimen.
Asian black bears are divided into seven subspecies based on their geographical distribution (Won Citation1968; Smith and Xie Citation2008; Ohdachi et al. Citation2015) and are distributed in a wide area of East Asia, Southeast Asia, and South Asia. Among these, the Asian black bear populations in the Korean Peninsula-Northeast China–Russia Far East Asian region are considered and classified as subspecies U. t. ussuricus (Won Citation1968; Smith and Xie Citation2008; Ohdachi et al. Citation2015). In South Korea, there exists confusion about the subspecies nomenclature of native Asian black bears with the use of the two scientific names – U. t. ussuricus (major use) and U. t. wulsini (rare use) (Jo et al. Citation2018). This confusion might have arisen because of the similar pronunciation between Chihli of Hebei (China), the type locality of U. t. ussuricus, and Mt. Jiri (South Korea) (Cultural Heritage Administration Citation2016; Jo et al. Citation2018). Thus, a re-evaluation of the phylogenetic position of the native South Korean black bears is required to clarify the controversy.
In 2016, a set of bones was found in a cave in South Korea, presumed to be Ursidae based on morphology. In Korea, most of the bear bones excavated from natural caves or prehistoric sites belong to brown bears, and the remains and fossils of Asian black bears have rarely been found (Won Citation1967; Jeong Citation2018; Jo et al. Citation2018). The discovery of these bones could offset the lack of sample availability, and using these samples, we developed the following hypotheses: (1) Recovered bones belong to the Asian black bear (Ursus thibetanus), (2) Asian black bear in hypothesis-1 is an animal that naturally lived in the wild in South Korea and is not a bear (or descendant) of a domestic farm, introduced from abroad in the 1980s. (3) The South Korean Asian black bear population had an active genetic exchange with other populations in Northeast Asia (North Korea, Northeastern China, and Far East Russia) and represent the same clade in a phylogenetic tree – Northeast Asia clade or Ursus thibetanus ussuricus. We undertook a scientific study using molecular techniques and radiocarbon dating to test the above hypothesis. We examined the recovered sample to determine – (1) species of origin (black or brown bear), (2) approximate age, and (3) phylogenetic position.
Material and methods
Samples and DNA extraction
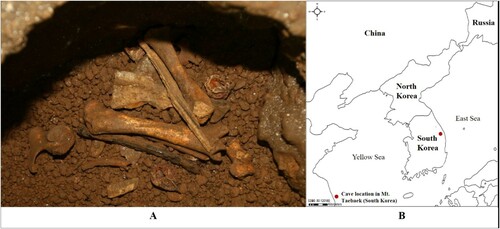
Samples were obtained from a set of animal bones found in a natural limestone cave in the Mt. Taebaek area, Samcheok City, Gangwon Province, Korea (N37.0451, E129.0936, 700 m) (). These bones were initially assumed to be of a carnivore (Jeong Citation2016) and were later identified as bear bones based on the morphological characterization (bone size and shape) by an expert (Swenson et al. Citation2007; Huber and van Manen Citation2019). The bear bone specimens have been stored in the Natural Heritage Center (Geology) of Korea and registered with specimen number NHCG a10953.
Figure 1. The animal bone specimens from a natural limestone cave in Mt. Taebaek in South Korea that we used in this study – (A) bone specimen and (B) cave location (marked with a red dot).
The DNA extraction was performed in an isolated, sterile facility dedicated to ancient and museum samples at the Seoul National University College of Veterinary Medicine (South Korea). We selected the thigh bone for DNA extraction. The bone was cleaned, sterilized, and punctured using a drill, and bone powder was collected from the bone marrow region (Rohland and Hofreiter Citation2007). DNA was extracted from the bone powder using the QIAamp DNA Micro Kit (Qiagen, Germany) following the supplied protocol for bone DNA extraction. Negative controls were included in every step to monitor contamination.
Polymerase chain reaction (PCR)
For the molecular analysis of the bone samples from the cave, fragments of the D-loop region (620 bp), cytochrome b gene (CytB, 697 bp), and cytochrome oxidase subunit I gene (COI, 631 bp) in mtDNA, ZFX-ZFY, and SRY gene in nDNA were PCR amplified. CytB and COI genes are commonly used for species identification and mtDNA D-loop for phylogenetic analysis of mammals (Tobe et al. Citation2009; Yasukochi et al. Citation2009; Kartavtsev Citation2011; Kartavtsev Citation2013; Shen et al. Citation2013; Lee et al. Citation2016). According to the morphological characteristics of the bones, we used bear-specific primers. For efficient PCR amplification of archaeological DNA, we used primers that amplified a short target sequence. All primers except for the URL2/URH2 primer pair (Uchiyama Citation1998) against the D-loop region were newly constructed using the published Asian black bear DNA sequence (GenBank Accession No. EF681884.1, AM748310, AM941050) (). Primers were designed using Primer3 software (Untergasser et al. Citation2012).
Table 1. Primer information in this study.
The PCR (Takara TP600) cycle conditions for mtDNA fragments were as follows: 5 min initial denaturation at 94°C; 50 cycles of 45 s at 94°C, 45 s at 54°C, and 45 s at 72°C; and a final extension for 5 min at 72°C. Similarly, PCR amplifications were performed for SRY and ZFX-ZFY genes using the following conditions: 5 min initial denaturation at 94°C; 40 cycles of 45 s at 94°C, 45 s at 58°C for SRY and at 56°C for ZFX-ZFY, and 45 s at 72 °C; and a final extension for 10 min at 72°C. PCR negative was included in each experiment to monitor contamination. Post amplification, amplified products were resolved on 1.5% agarose gel.
DNA sequencing
The amplified PCR products were sequenced using a Big Dye version 3.1 cycle sequencing kit and ABI Prism 3730XL DNA analyzer (Applied Biosystems, Foster City, CA, USA). The sequences determined in this study were used to identify the species by comparing their similarity with sequences reported in the NCBI nucleotide database through BLAST searches (Supplementary information 1).
For sex identification, the DNA sequences of the bone specimens were compared to those of male and female Asian black bears as controls. Sex discrimination was performed based on the presence or absence of the SRY gene and homozygosity/heterozygosity of the ZFX-ZFY gene.
Phylogenetic analysis
The 620 bp sequence of the D-loop region was used for phylogenetic analysis as in previous studies (Kim et al. Citation2011). For the phylogenetic analysis, the corresponding sequence of Asian black bears from China, Russia, Japan, and Southeast Asia (Accession Nos.: HM135178-135193 and EU264496-264527) were obtained from NCBI GenBank. The American black bear (Ursus americanus) (Accession Nos.: HE657194, HE657196) was used as the outgroup. Geneious 5.0.4 (Kearse et al. Citation2012) was used for multiple sequence alignment, and a phylogenetic tree was constructed using IQtree 1.5.6 with an ultrafast bootstrap of 1000 replicates (Minh et al. Citation2013; Hoang et al. Citation2018; Kalyaanamoorthy et al. Citation2017). The best-fit model was chosen based on the Bayesian information criterion (BIC) (Nguyen et al. Citation2015; Hoang et al. Citation2018; Kalyaanamoorthy et al. Citation2017). The maximum-likelihood (ML) consensus trees were illustrated using FigTree v 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Radiocarbon dating
The age of the bone specimens was determined using the radiocarbon dating method proposed by the Korea Institute of Geoscience and Mineral Resources (KIGAM), Daejeon, South Korea. The estimated age calculations used Libby’s half-life (5568 years) (Arnold and Libby Citation1949; Stuiver and Polach Citation1977), which is relative to the year 1950.
The bone powder was used for radiocarbon dating. The foreign particles were removed from the bone and collagen was extracted and gelatinized (Kim et al. Citation2010). The whole process includes the following steps – (1) the sample was added to 0.5 M HCl for 1 h at room temperature and washed to be neutralized, (2) the sample was added to 0.1 M NaOH solution for 1 h at room temperature and washed to neutral pH, (3) the sample was added to 0.5 M HCl for 30 min at room temperature and neutralized, (4) the sample was added to HCl solution (pH 3) for 12 h at 70° to proceed with gelatinization. Then, the sample was collected that passed through the 2.7 μm filter. Finally, using a Centriprep ultrafilter, gelatin with a molecular weight of ≥30,000 Da was collected (Kim et al. Citation2010) and freeze-dried using OPR-FDG-120 lyophilizer (OPERON). In the processed samples, 12C, 13C, 14C, 13C/12C, and 14C/12C were measured using AMS (Accelerator Mass Spectrometer). The radiocarbon age and δ13C were calculated according to the method proposed by Stuiver and Polach (Citation1977). Then the calendar age was analyzed using calibration curves of IntCal13 (Reimer et al. Citation2013) and Korean Tree Ring (Hong et al. Citation2013) with Bayesian statistics (Ramsey Citation2009).
Results
Species identification: cave bones belong to a male Asian black bear
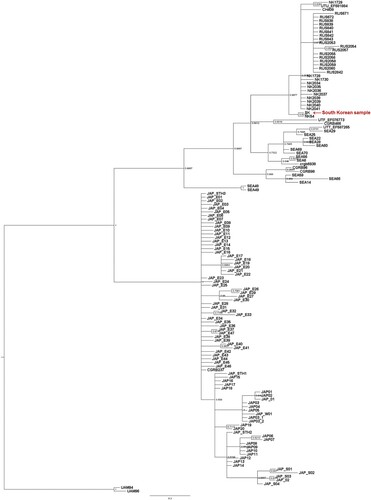
The partial nucleotide sequences of the CytB gene (697 bp), and COI gene (631 bp) were determined from the mtDNA fragments amplified from the thigh bone. Results of the BLAST search revealed that the partial mtDNA gene sequences from the unknown animal bone were of the Asian black bear, Ursus thibetanus (Supplementary information 1). The COI and CytB sequences were 100% identical to GenBank Accession Nos. EF6667005 and EU573174, respectively (Supplementary information 1). All mitochondrial gene sequences used in this study matched 100% to the sequences of Asian black bears (i.e. CGRB54, Ducksung17, Jangkang21, and Songwon), which had been reintroduced to Mt. Jiri from North Korea. Therefore, the investigated animal bones belong to the Asian black bear, U. t. ussuricus.
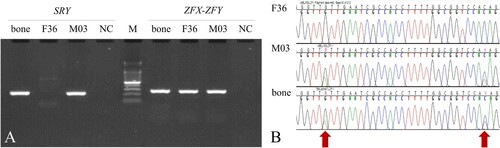
In molecular sex determination tests, the fragment of the SRY gene (196 bp) was successfully amplified () from the DNA extracted from the bear bone, and the nucleotide sequence was consistent with the sequences of the SRY gene previously reported for Ursidae. In addition, the amplified ZFX-ZFY (288 bp) gene sequence showed heterozygosity (), suggesting that this investigated sample was a male Asian black bear.
Figure 2. Molecular sexing for the animal bone DNA. (A) PCR amplification for male-specific SRY and sex-chromosome-linked ZFX-ZFY genes. (B) Multiple alignments for DNA sequences of ZFX-ZFY amplified in the PCR test (A). Bone indicates the DNA sample of animal bone found in the Taebaeksan area. F36 and M03 are female and male bears reintroduced from China and Russia, respectively. NC is the negative control for PCR amplification and M is the DNA size marker, 100 bp DNA Ladder.
Radiocarbon dating: identified male Asian black bear that lived before the Korean War
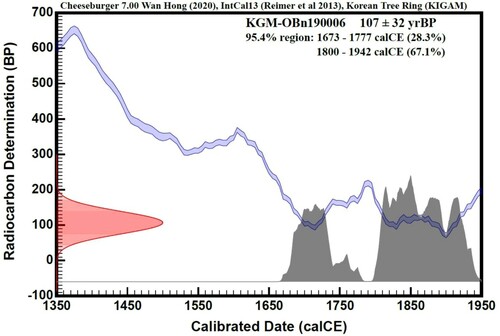
Radiocarbon dating based on the ratio of the carbon isotopes 12C and 14C produced estimates of 1843 ± 32 yrsBP. After the carbon date was adjusted to calibrated date using IntCal13 and Korean Tree Ring calibration curves, a range of 1800–1942 calCE (median 1871) was indicated at a 67.1% probability (). Radiocarbon dating was highly reliable as our inferred sample age range (1800–1942) lies within the 95% accuracy range (). Within the age accuracy range of 95.4%, two distinct age ranges were observed 1673–1777 (28.3% probability) and 1800–1942 (67.1% probability), thus age was determined 1800–1942 calCE. In other words, the bones found in the cave in the Mt. Taebaek area were estimated to be from a bear that lived on the Korean peninsula 80–220 years ago. The results of the age analysis methods suggested that the age of this sample was estimated to be 1800–1942 AD (Mid-point to late 1800s), i.e. approximately 150 years ago. This period corresponds to the late Joseon Dynasty (nineteenth century) or early twentieth century, before the Korean War in 1950.
Figure 3. Age dating graph. Radiocarbon determination dates (14C age) are calculated by Libby’s half-life, based on the ratio of 14C/12C (ordinate). The red color indicates the radiocarbon determination (BP) results of the bone sample. The Abscissa shows calibrated date (calCE = calAD) which is adjusted data of radiocarbon determination using calibration curves (blue line) of IntCal13 and Korean tree ring. The Grey color indicates calibrated age dating of bear specimens in the 95.4% region.
Phylogenetic position and conservation of the Asian black bear native to South Korea
Analysis of the phylogenetic relationships using the D-loop region of mtDNA showed that this South Korean bear was grouped with Asian black bears from North Korea, China, and Russia and that it was distinct from black bears from Southeast Asia and Japan (). Due to a single representative sample for each northeast China and South Korea in our study, we recommend having further detailed bear phylogeny in the future with additional representative samples for under-sampled northeast China and South Korea.
Discussion
Asian Black bears inhabited much of the Korean peninsula until the early decades of twentieth century and their population plummeted due to habitat loss, capture, and hunting. A wild bear was reportedly poached in Mt. Seorak in 1983, since then, there had been no formal reports of wild bears in South Korea until 2000. In 2000, the activities of a black bear in Mt. Jiri were filmed by a broadcaster's cameras, but the number of Asian black bears in the wild has been very low, and these bears have been considered to be almost extirpated (KNPS Citation2004; Yang Citation2008). Since 2004, the reintroduction of Asian black bears from Russia, China, and North Korea has been carried out in Mt. Jiri to restore the Asian black bear population (MOE Citation2005; Lee and Jeong Citation2009; SRTI Citation2018). Additionally, Asian black bears were recently confirmed to inhabit the demilitarized zone (DMZ) via the use of unmanned sensor cameras (MOE Citation2016; MOE Citation2019).
Understanding of molecular affinity (species status and phylogenetic position) of archeological samples is essential to maintain the utility in species conservation and restoration. Black bears represented the Korean Peninsula in all previous genetic studies were from North Korea due to a lack of authentic native South Korean specimens. Phylogenetic analysis confirmed the species (Asian black bear) and subspecies identity (Ussuri spp) of recovered bones. The radiocarbon dating of specimens suggested that the bear bones found in the cave did not originate from bears imported into South Korea between 1981 and 1985 for bear farming. This confirms that the bones found and investigated in the present study originated from a bear belonging to the native South Korean population of Asian black bears. The finding has academic significance as no prior specimen has been found derived from an Asian black bear that lived in South Korea before the 1950s.
The phylogenetic results presented here indicate that the native South Korean, North Korean, and Russian Asian black bears belong to the same genetic group. Therefore, it would be the best strategy to reintroduce individuals from Northeast Asia, including regions such as North Korea, Russia, and northeast China. The present study is supported by previous studies (Hong Citation2005; Kim et al. Citation2011; Wu et al. Citation2015) suggesting that the conservation unit of the Asian black bear group is composed of black bear populations of the Korean peninsula (with North Korean samples), Russian Primorskiy region, and northeastern China. Furthermore, the results of our study indicate that the selection of individuals from Far Eastern Russia, northeast China, and North Korea for the ongoing bear reintroduction project (Lee and Jeong Citation2009; Kim et al. Citation2011; SRTI Citation2018) was the appropriate choice, in that they are from Asian black bear subspecies that share the same genetic group with the native South Korean black bears.
Conclusion
The unknown animal bones found in the Mt. Taebaek limestone cave were identified to be the remains of a wild male Asian black bear (Ussuri subspecies) that inhabited the Korean Peninsula in the nineteenth century or early twentieth century. Moreover, the findings reaffirm the management decision of selecting Northeast Asian black bears as the source population for the reintroduction project in South Korea.
In wildlife conservation, it is important to preserve a sufficient population size to ensure that the species does not become endangered, but it is also critical to maintaining the unique biological characteristics of the population that has accumulated for an extended period in one area (Lee et al. Citation2021). The bear bones identified in this study are precious specimens that can clarify the biological features inherent to the Asian black bears that inhabited South Korea. In the future, information on the osteological, morphological, and genetic characteristics of these samples will be used not only to describe the features of the native population of Asian black bears in South Korea but also to contribute to understanding the evolution and regional differences in the Northeast Asian black bear populations, including those from the Korean peninsula.
Supplemental Material
Download MS Word (590.8 KB)Acknowledgements
We gratefully acknowledge Dr. Gyujun Park for his valuable comments on this manuscript. The authors would like to thank Daecheol Jeong for his assistance about non technical work. This study was partially supported by the Research Institute for Veterinary Science, Seoul National University. The supporting had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Arnold JR, Libby WF. 1949. Age determinations by radiocarbon content; checks with samples of known age. Science. 110(2869):678–680. doi: 10.1126/science.110.2869.678
- Cultural Heritage Administration. 2016. Research service to prepare a plan for the protection and management of natural monument animal species; p. 18.
- Higuchi R, Bowman B, Freiberger M, Ryder OA, Wilson AC. 1984. DNA sequences from the quagga, an extinct member of the horse family. Nature. 312:282–284. doi: 10.1038/312282a0
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. doi: 10.1093/molbev/msx281
- Hong W, Park JH, Park G, Sung KS, Park WK, Lee JG. 2013. Regional offset of radiocarbon concentration and its variation in the Korean atmosphere from AD 1650 – AD 1850. Radiocarbon. 55(2-3):753–762. doi: 10.1017/S003382220005791X
- Hong Y-J. 2005. Molecular phylogenetic study on asiatic black bears (Ursus thibetanus) in Korea: defining the conservation unit of Korean black bears [master’s thesis]. Seoul: Seoul National University.
- Huber Đ, van Manen FT. 2019. Bear morphology. In: Vonk Jennifer et Shackelford Todd, editor. Encyclopedia of animal cognition and behavior [en ligne]. Cham: Springer International Publishing; p. 1–11.
- IUCN. 1998. IUCN guidelines for Re-introductions. Gland: IUCN.
- Jang S-H. 2014. A study on policy-making process about ending bear farming – based on Kingdon’s policy stream model [master’s thesis]. Seoul: Seoul National University.
- Jeong H-K. 2016. Are there really wild tigers and wild leopards in Korea? J MICE Tour Res. 16(4):77–95.
- Jeong JS. 2018. A study of the carnivore fossils from the chommal cave site in Jecheon, Korea [master’s thesis]. Seoul: Yonsei University.
- Jo Y-S, Baccus JT, Koprowski JL, Ji Y-H. 2018. Mammals of Korea. Incheon: National Institute of Korea.
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature methods. 14(6):587–589. doi: 10.1038/nmeth.4285
- Kartavtsev YH. 2011. Divergence at Cyt-b and Co-1 mtDNA genes on different taxonomic levels and genetics of speciation in animals. Mitochondrial DNA. 22(3):55–65. doi: 10.3109/19401736.2011.588215
- Kartavtsev YH. 2013. Sequence diversity at Cyt-b and Co-1 mtDNA genes in animal taxa proved neo-darwinism. J Phylogen Evolution Biol. 1:4. doi: 10.4172/2329-9002.1000120
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. doi:10.1093/bioinformatics/bts199.
- Kim KJ, Hong W, Park JH, Woo HJ, Hodgins G, Jull AJT. 2010. Development of radiocarbon dating methods for modern bone collagenization. Radiocarbon. 52:1657–1659. doi: 10.1017/S0033822200056381
- Kim M, Shin N-S, Youn H-Y, Cho J-K, Seo K-W, Na K-J, Youn H-J, Lee I-B, Lee J-W, Yoo J. 2012. Research on the status of breeding bears and establishment of management plans (in Korean). Gwacheon: Ministry of Environment. https://library.me.go.kr/#/search/detail/5549766.
- Kim YK, Hong YJ, Min MS, Kim KS, Kim YJ, Voloshina I, Myslenkov A, Smith GJ, Cuong ND, Tho HH, et al. 2011. Genetic status of asiatic black bear (Ursus thibetanus) reintroduced into South Korea based on mitochondrial DNA and microsatellite loci analysis. J Hered. 102(2):165–174. doi: 10.1093/jhered/esq121
- KNPS, Korean National Park Service. 2004. Report of asiatic black bear pre-release.
- Lee BG, Jeong DH. 2009. Restoration of asiatic black bears through reintroduction on Mt. Jiri, South Korea. Newslett Int Assoc Bear Res Manag (IBA) and IUCN/SSC Bear Spec Group. 18:8–10.
- Lee C, Fong JJ, Jiang J-P, Li P-P, Waldman B, Chong JR, Lee H, Min M-S. 2021. Phylogeographic study of the Bufo gargarizans species complex, with emphasis on northeast Asia. Animal Cells Syst (Seoul). 25(6):434–444. doi: 10.1080/19768354.2021.2015438
- Lee J-H, Lee G-G, Lee M-J, Cha J-Y. 2013. Improvement disciplines for relief policy of breeding bears at the perspective of animal welfare in the Republic of Korea. J Korean Env Res Tec. 16(6):31–48.
- Lee M-Y, Lee S-M, Song E-G, An JH, Voloshina I, Chong JR, Johnson WE, Min M-S, Lee H. 2016. Phylogenetic relationships and genetic diversity of badgers from the Korean peninsula: implications for the taxonomic status of the Korean badger. Biochem Syst Ecol. 69:18–26. doi: 10.1016/j.bse.2016.07.015
- Minh B, Nguyen M-A, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30:1188–1195. doi:10.1093/molbev/mst024.
- Ministry of Environment (MOE). 2005. Breeding bear management guidelines. Press release of Division of Natural Resources of MOE.
- MOE. 2016. A comprehensive report on the biodiversity around the Demilitarized Zone.
- MOE. 2019. Wild asiatic black bear captures vivid images in the DMZ. Available from: http://www.me.go.kr/home/web/board/read.do?menuId=286&boardMasterId=1&boardCategoryId=39&boardId=981435.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. doi:10.1093/molbev/msu300.
- Ohdachi SD, Ishidashi Y, Iwasa MA, Fukui D, Saitoh T. 2015. The wild mammals of Japan. Kyoto: Shoukadoh Book Sellers and the Mammal Society of Japan.
- Park H-C, Kurihara N, Kim KS, Min M-S, Han SY, Lee H, Kimura J. 2019. What is the taxonomic status of east Asian otter species based on molecular evidence?: focus on the position of the Japanese otter holotype specimen from museum. Animal Cells Syst (Seoul). 23(3):228–234. doi:10.1080/19768354.2019.1601133.
- Ramsey CB. 2009. Bayesian analysis of radiocarbon dates. Radiocarbon. 51(1):337–360. doi: 10.1017/S0033822200033865
- Reimer PJ, Bard E, Bayliss A, Beck JW, Blackwell PG, Bronk Ramsey C, Buck CE, Cheng H, Edwards RL, Friedrich M, et al. 2013. Intcal13 and marine13 radiocarbon age calibration curves 0–50,000 yearscal BP. Radiocarbon. 55(4):1869–1887. doi: 10.2458/azu_js_rc.55.16947
- Rey-Iglesia A, García-Vázquez A, Treadaway EC, van der Plicht J, Baryshnikov GF, Szpak P, Bocherens H, Boeskorov GG, Lorenzen ED. 2019. Evolutionary history and palaeoecology of brown bear in north-east Siberia re-examined using ancient DNA and stable isotopes from skeletal remains. Sci Rep. 9(1):4462. doi: 10.1038/s41598-019-40168-7
- Rohland N, Hofreiter M. 2007. Ancient DNA extraction from bones and teeth. Nat Protoc. 2:1756–1762. doi: 10.1038/nprot.2007.247
- Shen YY, Chen X, Murphy RW. 2013. Assessing DNA barcoding as a tool for species identification and data quality control. PLoS One. 8(2):e57125. doi: 10.1371/journal.pone.0057125
- Sheng GL, Barlow A, Cooper A, Hou XD, Ji XP, Jablonski NG, Zhong BJ, Liu H, Flynn LJ, Yuan JX, et al. 2018. Ancient DNA from giant panda (Ailuropoda melanoleuca) of south-western China reveals genetic diversity loss during the holocene. Genes (Basel). 9(4):E198. doi: 10.3390/genes9040198
- Smith AT, Xie Y. 2008. A guide to the mammals of China. Princeton (NJ): Princeton University Press.
- SRTI, Species Restoration Technology Institute. 2018. Annual report 2017. SRTI, Korea National Park Service; p. 21–24.
- Stuiver M, Polach HA. 1977. Discussion: reporting of 14C data. Radiocarbon. 19(3):355–363. doi: 10.1017/S0033822200003672
- Swenson JE, Adamič M, Huber Đ, Stokke S. 2007. Brown bear body mass and growth in northern and Southern Europe. Oecologia. 153:37–47. doi: 10.1007/s00442-007-0715-1
- Tobe SS, Andrew K, Adrian L. 2009. Cytochrome b or cytochrome c oxidase subunit I for mammalian species identification – an answer to the debate. Forensic Sci Int Genet Suppl Ser. 2(1):306–307. doi: 10.1016/j.fsigss.2009.08.053
- Uchiyama. 1998. DNA analysis for the asiatic black bear (Ursus thibetanus) [master’s thesis]. Fukuoka: University of Kyushu.
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3 – new capabilities and interfaces. Nucleic Acids Res. 40(15):e115. doi: 10.1093/nar/gks596
- Won HK. 1968. The mammals of Korea. Pyeongyang: Institute of Science Press.
- Won PH. 1967. Illustrated encyclopedia of fauna and flora of Korea. Vol. 7, Mammals. Seoul: Ministry of Education; p. 127–130.
- Wu J, Kohno N, Mano S, Fukumoto Y, Tanabe H, Hasegawa M, Yonezawa T. 2015. Phylogenetic and demographic analysis of the Asian black bear (Ursus thibetanus) based on mitochondrial DNA. PLoS One. 10(9):e0136398. doi: 10.1371/journal.pone.0136398
- Yang DH. 2008. Ecological characteristics of asiatic black bear (Ursus thibetanus ussuricus) released in Jirisan National Park [doctor’s thesis]. Changwon: Kyungnam University.
- Yasukochi Y, Nishida S, Han SH, Kurosaki T, Yoneda M, Koike H. 2009. Genetic structure of the asiatic black bear in Japan using mitochondrial DNA analysis. J Hered. 100(3):297–308. doi: 10.1093/jhered/esn097