ABSTRACT
Natural products (NPs) have greatly contributed to the development of novel treatments for human diseases such as cancer, metabolic disorders, and infections. Compared to synthetic chemical compounds, primary and secondary metabolites from medicinal plants, fungi, microorganisms, and our bodies are promising resources with immense chemical diversity and favorable properties for drug development. In addition to the well-validated significance of secondary metabolites, endogenous small molecules derived from central metabolism and signaling events have shown great potential as drug candidates due to their unique metabolite-protein interactions. In this short review, we highlight the values of NPs, discuss recent scientific and technological advances including metabolomics tools, chemoproteomics approaches, and artificial intelligence-based computation platforms, and explore potential strategies to overcome the current challenges in NP-driven drug discovery.
Introduction
Humans has long relied on natural products (NPs) to treat and manage various diseases including infection, cancer, and metabolic disorders (e.g. obesity and type 2 diabetes) (Koehn and Carter Citation2005; Baker et al. Citation2007; Cragg and Newman Citation2013; Atanasov et al. Citation2021). Historical records from Mesopotamia dating back to 2,600 BC clearly demonstrate that early civilizations were aware of the medicinal properties of approximately 1,000 plants. Traditional Asian medicine is also based on the knowledge obtained from thousands of years of disease management and treatment using NPs. Since the nineteenth century, the concept of rational drug discovery began to flourish with the successful isolation of analgesic chemicals from opium and many bioactive compounds such as quinine, nicotine, and rapamycin from various natural sources. Driven by the industrial need to maximize production yields and quality, natural compounds such as salicylic acid have been successfully synthesized through chemical procedures. The discovery of penicillin further broadened our interests by recognizing the potent activities of microbial NPs. Crude and semi-pure extracts from medicinal plants, animals, and microbes including fungi provided the best available medications in this early period. However, with the introduction and validation of the receptor theory of drug action in pharmacology, specific chemical compounds in crude NP extracts were identified as the primary factors mediating the biological and pharmacological properties of these extracts. Well-known examples of approved drugs derived from NPs include penicillin (antibacterial), morphine (analgesic), artemisinin (antimalarial), fingolimod (immunosuppressor), rapamycin (immunosuppressor, anticancer), and paclitaxel (anticancer). Small bioactive molecules from NPs are also widely used to improve biological activity and pharmaceutical properties, as demonstrated by the use of salicylic acid for aspirin production.
Our intense efforts to investigate NPs as sources of novel human therapeutics became fruitful between the 1970s and 1980s, leading to pharmaceutical development influenced by non-synthetic molecules. While classical and combinatorial chemical compound libraries are competitively emerging, mainly due to their favorable physicochemical properties and lower toxicity, NPs have been continuously considered an attractive source of compounds for drug discovery (). Nearly 25% of new drugs approved worldwide in the past four decades are NPs and their derivatives, whereas another 25% are synthetic drugs with an NP pharmacophore or drugs that mimic the structure and properties of an NP (Newman and Cragg Citation2020). These novel drugs have been used to treat a wide variety of disorders including infectious (bacterial, fungal, parasitic, and viral), immunological, cardiovascular, neurological, inflammatory, and related diseases, as well as cancer.
Compared to synthetic small molecules, the most valuable feature of NPs as a resource for drug discovery is their structural scaffold complexity (Rodrigues et al. Citation2016; Yñigez-Gutierrez and Bachmann Citation2019; Lautié et al. Citation2020). All small molecules found in nature are metabolites, which are defined as intermediate or final products of metabolism (e.g. synthesis and breakdown of carbohydrates, proteins, fats, and nucleic acids) catalyzed by various arrays of cellular enzymes. Primary metabolites are the chemical compounds produced from central metabolic processes, thus contributing to the regulation of energy homeostasis, growth, and reproduction. Secondary metabolites derived from central metabolic pathways in plants and fungi are not required for homeostatic metabolic processes. However, they offer a wide array of new chemical structures, many of which have numerous biological and pharmacological properties against virtually every existing disease including cancer (Seca and Pinto Citation2018; Keller Citation2019). In this short review, we summarize how our knowledge of the biological activities of primary metabolites can be used for drug discovery. We then highlight the importance of secondary metabolite activities and finally focus on current/future strategies to harness their therapeutic properties and promote drug discovery and development.
Emerging roles of primary metabolites as signaling factors
Primary metabolites have long been viewed as a simple fuel source for energy metabolism or as fundamental substrates required for the degradation and biosynthesis of macromolecules. However, several recent discoveries have demonstrated that certain primary metabolites can trigger or mediate potent biological activities () (Fang et al. Citation2001; Schreiber Citation2005; Keller et al. Citation2012; Shimazu et al. Citation2013; Lee et al. Citation2015; Mills et al. Citation2018; Bae et al. Citation2020; Gomes et al. Citation2020; Harayama and Shimizu Citation2020; Lee et al. Citation2020c; Martínez-Reyes and Chandel Citation2020; Shyer et al. Citation2020; Lee et al. Citation2021). Particularly, these metabolites can control cellular signaling pathways, which are the main targets for drug development (Li and Snyder Citation2011; Wang and Lei Citation2018; Milanesi et al. Citation2020). These findings suggest that our cellular and physiological systems have evolved to utilize specialized endogenous small molecules as primary or secondary signaling messengers to fine-tune various biological events. Therefore, identifying specific cellular metabolite-protein interactions provides insights into the development of synthetic drug molecules with enhanced properties such as efficacy and stability (Piazza et al. Citation2018). Endogenous small molecules can thus be classified depending on their modes of interaction ().
Figure 1. Biological actions of endogenous small molecules in the control of cellular signaling and metabolism via metabolite-protein interactions.
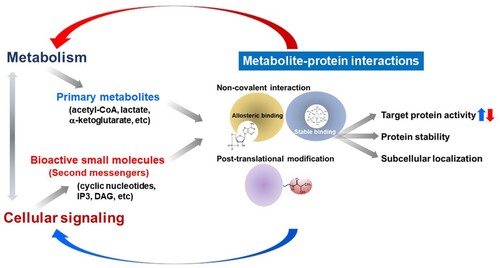
First, small molecules in our body can act as ligands toward specific receptor molecules. In addition to the well-known involvement of hydrophobic steroid hormones in the activation of nuclear receptor proteins (Mangelsdorf et al. Citation1995), primary metabolites (e.g. adenosine, sphingosine-1-phosphate, free fatty acids, amino acid neurotransmitters) also stimulate key signaling pathways for cellular responses such as cell growth, differentiation, immunity, neuronal activation, and morphological changes by directly binding and stimulating their cognate cellular receptors (Schreiber Citation2005; Jacobson and Gao Citation2006; Traynelis et al. Citation2010; Blaho and Hla Citation2014; Milligan et al. Citation2017).
Furthermore, our metabolism produces second messenger molecules such as cyclic nucleotides, inositol polyphosphates, and bioactive lipids upon activation of target cells (Berridge and Irvine Citation1984; Conti Citation2000; Murad Citation2006; Newton et al. Citation2016). The production and degradation of these second messengers are tightly regulated by the dynamic changes of cellular and physiological programs such as differentiation, cellular death, development, and tissue regeneration. Recent studies on cellular nutrient and energy sensing continue to uncover unexpected metabolite-protein interactions (e.g. leucine-sestrin, inositol pyrophosphate-Akt) (Alvarez et al. Citation2010; Chakraborty et al. Citation2010; Lee et al. Citation2016; Lee et al. Citation2020c; Wolfson et al. Citation2016; Li et al. Citation2017). Moreover, druggable targets can be further developed based on our knowledge of the allosteric interaction between endogenous small molecules and their effector proteins. Some primary metabolites are also known to mediate stable interactions with target proteins, suggesting their pivotal roles in coordinating protein stability. For example, inositol polyphosphate is known as an essential factor to mediate capsid formation for HIV (human immunodeficiency virus) viral particle assembly, suggesting that IP6 (inositol hexakisphosphate) metabolism is a promising target for the development of new anti-HIV therapeutics (Dick et al. Citation2018). The stabilization of the molecular complexes (e.g. RNA editing enzyme ADAR2, Integrator) by inositol polyphosphates is another example demonstrating the significance of stable metabolite-protein interactions for the development of novel therapeutics (Macbeth et al. Citation2005; Lin et al. Citation2022).
Besides the above-mentioned non-covalent metabolite-protein interactions of cellular metabolites, some metabolic biomolecules can also lead to various post-translational modifications (PTMs) (Figlia et al. Citation2020). In addition to ATP, which is an essential substrate for protein phosphorylation, many other metabolites can also modify and modulate target protein functions. For example, nitric oxide (NO), a product derived from arginine metabolism, is the primary source for protein S-nitrosylation, a process that leads to changes in the activities, subcellular localization, and stability of target proteins (Jaffrey et al. Citation2001; Stomberski et al. Citation2019). S-adenosyl methionine, acetyl-CoA, and other metabolites (e.g. alpha-ketoglutarate) can indeed control many signaling pathways and mediate epigenetic changes through unique PTMs (e.g. methylation, acetylation, succinylation) (Kaelin and McKnight Citation2013). The identification of other PTMs and their responsible metabolites could provide specific molecular targets and metabolism for novel drug discovery and development.
Recent advances in the characterization of primary metabolites and endogenous small molecules as signaling factors have greatly expanded our understanding of the vast potential of NPs as sources of therapeutic compounds. Fundamental structural and biological information on the specific covalent and/or non-covalent interactions among bioactive primary metabolites and target proteins provide a valuable resource for the development of efficient small molecules and therapeutic compounds based on known druggable targets. In addition to targeting metabolite-protein interactions, approaches to modulate selective metabolites will also become an important strategy to control serious diseases such as cancer (Wang and Lei Citation2018; Milanesi et al. Citation2020; Stine et al. Citation2022). As exemplified by recent studies, increasing the levels of toxic metabolites can be selectively and efficiently kill tumor cells (Kim et al. Citation2015; Lee et al. Citation2020b). In summary, the primary metabolites in our cells and tissues are less structurally complex compared to secondary metabolites from other organisms, but endogenous small molecules and their control of cellular signaling should be further elucidated and applied for the development of next-generation drugs.
Pharmacologically-active secondary metabolites as major resources for the discovery of potent drugs
Many medicinal NPs derived from plants, fungi, and microorganisms have long been used to alleviate and cure a wide range of medical conditions such as infectious diseases, inflammatory reactions, obesity/diabetes, cardiovascular disease, and cancer, as well as psychiatric disorders (Dias et al. Citation2012; Atanasov et al. Citation2021). Our information on these potent effects of extracts and/or single compounds derived from NPs have been continuously updated ( and ). The potent activities of these NPs largely originate from their secondary metabolites, which are organic compounds produced through the modification of primary metabolites. Unlike primary metabolites, secondary metabolites and related metabolic reactions are involved in ecological functions (e.g. pathogen sensing and defense mechanisms) (Li et al. Citation2020).
Table 1. Selected examples of bioactive NP extracts.
Table 2. Selected list of bioactive secondary metabolites derived from plant NPs.
Structural diversity and complexity are the most important features of secondary metabolites as an essential resource for drug in competition with synthetic chemical libraries (Hong Citation2011; Lautié et al. Citation2020). Approximately 40% of the chemical scaffolds from NPs are not found in commercial synthetic compounds (Henkel et al. Citation1999). Furthermore, 83% of the ring scaffolds in NPs are not present in synthetic molecules (Hert et al. Citation2009). Complex molecules can more readily interact with a greater variety of chemical structures and modifications, suggesting a superior potential for secondary metabolites to complement the spatial characteristics of target proteins. Although many secondary metabolites do not fit into drug-likeness standards such as Lipinski's ‘rule of five’ (e.g. logP ≤ 5, molecular weight ≤ 500 Da, number of hydrogen bond acceptors ≤ 10), they exhibit more favorable metabolic and pharmacokinetic properties such as absorption, distribution, metabolism, and excretion/toxicity than synthetic molecules (Müller-Kuhrt Citation2003; Atanasov et al. Citation2021).
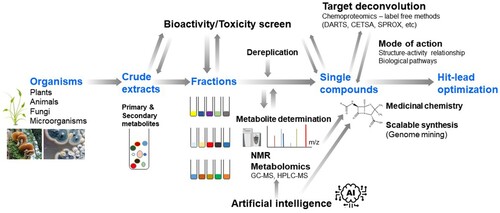
The journey from the identification of promising bioactive secondary metabolites to drug discovery indeed entails a series of demanding processes (). The therapeutic potential of secondary metabolites depends on the quality and quantity of the bioactive chemicals in medicinal organisms, which in turn is influenced by various environmental factors (e.g. growth conditions, age, climate changes). The purification of bioactive metabolites involves various strategies such as combinatorial chemistry, isolation assays, and efficacy-based high-quality fractionation. It is also critical to avoid the replication of previous efforts by correctly identifying known compounds. The determination of de novo structure of novel compounds has greatly benefited from recent advances in spectroscopic techniques such as high-resolution nuclear magnetic resonance (NMR) technologies. When the biological activity profile of a therapeutic candidate meets the optimal criteria for potency and selectivity, structure–activity relationship (SAR) studies are then conducted and large-scale purification processes are developed. Once synthetic modification methods become feasible, hit-to-lead optimization is further accelerated by conventional medicinal chemistry approaches.
Strategies for accelerating natural metabolite-driven drug discovery
To continue the long and successful history of NPs in drug discovery and their unique structural diversity, several challenges must be overcome (Koehn and Carter Citation2005; Lam Citation2007; Atanasov et al. Citation2021). NP-derived lead compounds typically exhibit low solubility or chemical instability, which impedes further drug development. Many NPs also exhibit high molecular weight and complex structures, which often results in poor absorption and complicates the development of oral formulations. Although naturally active substances usually make well-qualified lead compounds, many of them can hardly fulfill the criteria for druggability. Therefore, the most important step during NP-based drug discovery is the efficient and accurate selection of natural sources for the extraction and isolation of bioactive metabolites with desired biological activities and structural properties ().
Technical advancements in the field of metabolomics — the sensitive, unbiased, and high-throughput study of complex metabolites — have enabled the characterization and quantification of bioactive metabolites from complex mixtures derived from NPs (Liu and Locasale Citation2017). Metabolomics can thus be widely applied to the analyses of pharmaceutically relevant NP resources, as well as for the discovery of bioactive metabolites (Wishart Citation2016; Wolfender et al. Citation2019; Stuart et al. Citation2020). Coupling metabolomics with NMR further facilitates the acquisition of structural information of potent metabolites, which saves a substantial amount of time and labor when extracting or isolating metabolites (Lin et al. Citation2008; Gathungu et al. Citation2020). Efforts have been recently made to establish a comprehensive experimental tandem mass spectrometry (MS/MS) database of NPs. The Global Natural Products Social (GNPS) molecular networking platform contains thousands of MS datasets from NP extracts (Wang et al. Citation2016). Moreover, this platform clusters structurally related metabolites and provides insights regarding their relationships. In addition to the GNPS platform, the METLIN (Guijas et al. Citation2018) and CSI:FingerID (Dührkop et al. Citation2015) databases provide useful information to expedite metabolite identification by combining fragmentation tree computation and machine learning.
The subsequent process of identifying the molecular targets of bioactive NP-derived hits, which is also known as ‘target deconvolution,’ is essential for underpinning the mechanisms of drug action, as well as for the application of the identified hits to fully elucidate the biological processes modulated by a drug candidate (Terstappen et al. Citation2007). Recent advancements in chemical proteomics have led to the development of efficient and sensitive methods analyzing the proteins that the bioactive metabolite of interest binds. Based on the assumption that a molecule binding to a protein target alters the target’s stability, two major methods have been widely applied. Drug affinity-responsive target stability (DARTS) assesses the changes in the stability of a protein to proteolysis upon binding with the ligand (Lomenick et al. Citation2009). The cellular thermal shift assay (CETSA) and the stability of proteins from rates of oxidation (SPROX) rely on the thermal stabilization of a protein bound to a ligand (Molina et al. Citation2013; Strickland et al. Citation2013). Label-free metabolites of interest can be used in these platforms, thus foregoing the need for laborious chemical modifications such as biotinylation. Combined with MS proteomics, the CETSA-MS and DARTS-MS platforms become more powerful, thus enabling the acquisition of protein–ligand interactome data, as well as the accompanying physiological changes in biological samples such as cell lysates, intact cells, as well as tissues (Savitski et al. Citation2014; Pai et al. Citation2015).
Small molecule drug candidates derived from NPs can be applied by conjugating them with other bioactive molecules, thus expanding the use of NP metabolites for the development of novel, bifunctional, and more effective drugs for disease treatment. The immense range of bifunctional conjugates used for the development of NP hybrid drugs includes antibody–drug conjugates and aptamer drug conjugates, PROteolysis TArgeting Chimeras (PROTAC), and AUTOphagy-TArgeting Chimeras (AUTOTAC) (Yoon et al. Citation2017; Newman Citation2021; He et al. Citation2022; Ji et al. Citation2022). For example, the PROTAC approach is based on the development of bifunctional hybrid molecules comprised of a ligand for an E3 ligase and a ligand for the target protein joined by a linker, thus leading to the ubiquitination of the target protein and proteasomal degradation (Nalawansha and Crews Citation2020; Li and Crews Citation2022). For example, the PROTAC approach, which is based on the NP apigenin (i.e. a low estrogenic flavonoid with anticancer activity), was developed to specifically target the aryl hydrocarbon receptor for degradation (Puppala et al. Citation2008). Wogonin-based PROTACs were also used for the synthesis of CDK9-targeting PROTACs capable of selectively degrading CDK9 (Bian et al. Citation2018). Since the first study with PROTACs was performed with a natural polyketide ovalicin-derived molecule (Sakamoto et al. Citation2001), the use of NP-mediated target protein degradation has emerged as a promising strategy to treat diseases such as cancer and metabolic disease (Li et al. Citation2022).
Artificial intelligence (AI) has garnered increasing attention in various academic fields as well as industrial decision-making and processing applications because it allows for fast and efficient analysis and reduces human errors. Therefore, AI has recently been applied in drug discovery to analyze molecular properties, identify synthetic routes, and predict bioactive metabolites. By using various machine learning algorithms coupled with cloud computing technologies, big data accumulated from drug discovery and development can be processed to facilitate the identification of therapeutic candidates. For example, machine learning software (e.g. ACD/structure elucidator, Mestrelab Mnova) has been used for structure determination and dereplication (Claridge Citation2009; Elyashberg and Williams Citation2021). An AI-based structure prediction tool (DP4-AI) has been also developed to predict metabolite structures as well as MS2DeepScore, a machine learning-based mass spectral similarity-predicting algorithm, to identify metabolites based on clustering analysis (Howarth et al. Citation2020; Huber et al. Citation2021). Machine learning can also be used to identify drug targets. For example, BANDIT, a Bayesian machine-learning algorithm, is used to integrate multiple data types and predict the targets of nearly 4,000 compounds with a 90% accuracy (Madhukar et al. Citation2019). Other AI platforms include DEcRyPT (Drug–Target Relationship Predictor) (Rodrigues et al. Citation2018), SuperPred (Gallo et al. Citation2022), and NPClassifier (Kim et al. Citation2021a). The integration and curation of different forms of NP-derived databases (taxonomic, structural, genomic, transcriptomic, proteomic, and metabolomics databases) should be systematically pursued to overcome the common drawbacks of AI-powered technology for drug discovery. In turn, this approach is highly expected to reduce errors and increase predictability.
Conclusions
The quest for the discovery of new drugs derived from natural metabolites has led to many breakthroughs and achievements (e.g. taxol, artemisinin, rapamycin, and penicillin). In addition to the discovery of several potent bioactive secondary metabolites, recent findings on the signaling activities of endogenous primary metabolites have greatly contributed to the identification of novel metabolite-protein interactions (), which are critical for disease control. Recent technological improvements and systems biology approaches coupled with the application of available omics technologies and AI-powered computational strategies will potentially pave the way for the discovery of new NP-derived drug candidates (). In turn, this strategic integration of various technologies enables the design of a new generation of first- and best-in-class drugs. Less than 1% of Earth’s vast biodiversity has been investigated as a potential source of drug candidates. However, the discovery of novel therapeutic compounds is being threatened by the massive destruction of ecosystems (e.g. deforestation) and the consequent loss of species diversity and habitats. Therefore, promoting NP research through the construction of metabolite databases and the use of integrative drug discovery platforms could develop new and more effective NP-based therapeutics, sooner than expected.
Acknowledgments
We feel sorry for not citing the work of many other NP-based drug discovery investigators and related experts owing to space constraints. We thank all the members of the Kim lab for discussion.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Ahmed MB, Islam SU, Lee YS. 2020. Decursin negatively regulates LPS-induced upregulation of the TLR4 and JNK signaling stimulated by the expression of PRP4 in vitro. Anim Cells Syst (Seoul). 24(1):44–52.
- Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, et al. 2010. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 465(7301):1084–1088.
- Atanasov AG, Zotchev SB, Dirsch VM. 2021. International natural product sciences taskforce, Supuran CT. 2021. Natural products in drug discovery: advances and opportunities. Nat Rev Drug Discov. 20(3):200–216.
- Austin C, Stewart D, Allwood JW, McDougall GJ. 2018. Extracts from the edible seaweed, Ascophyllum nodosum, inhibit lipase activity in vitro: contributions of phenolic and polysaccharide components. Food Funct. 9(1):502–510.
- Bae S, Kim MK, Kim HS, Moon YA. 2020. Arachidonic acid induces ER stress and apoptosis in HT-29 human colon cancer cells. Anim Cells Syst (Seoul). 24(5):260–266.
- Baker DD, Chu M, Oza U, Rajgarhia V. 2007. The value of natural products to future pharmaceutical discovery. Nat Prod Rep. 24(6):1225–1244.
- Bao YY, Zhou SH, Fan J, Wang QY. 2013. Anticancer mechanism of apigenin and the implications of GLUT-1 expression in head and neck cancers. Future Oncol. 9(9):1353–1364.
- Berridge MJ, Irvine RF. 1984. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature. 312(5992):315–321.
- Bian J, Ren J, Li Y, Wang J, Xu X, Feng Y, Tang H, Wang Y, Li Z. 2018. Discovery of Wogonin-based PROTACs against CDK9 and capable of achieving antitumor activity. Bioorg Chem. 81:373–381.
- Blaho VA, Hla T. 2014. An update on the biology of sphingosine 1-phosphate receptors. J Lipid Res. 55(8):1596–1608.
- Burlec AF, Pecio Ł, Kozachok S, Mircea C, Corciovă A, Vereștiuc L, Cioancă O, Oleszek W, Hăncianu M. 2021. Phytochemical profile, antioxidant activity, and cytotoxicity assessment of tagetes erecta L. flowers. Molecules. 26(5):1201.
- Cai Z, He S, Li T, Zhao L, Zhang K. 2020. Plumbagin inhibits proliferation and promotes apoptosis of ovarian granulosa cells in polycystic ovary syndrome by inactivating PI3K/Akt/mTOR pathway. Anim Cells Syst (Seoul). 24(4):197–204.
- Chakraborty A, Koldobskiy MA, Bello NT, Maxwell M, Potter JJ, Juluri KR, Maag D, Kim S, Huang AS, Dailey MJ, et al. 2010. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 143(6):897–910.
- Chandrashekar N, Pandi A. 2022. Baicalein: a review on its anti-cancer effects and mechanisms in lung carcinoma. J Food Biochem. 46(9):e14230.
- Choi HJ, Choi HJ, Park MJ, Lee JY, Jeong SI, Lee S, Kim KH, Joo M, Jeong HS, Kim JE, et al. 2015. The inhibitory effects of geranium thunbergii on interferon-γ- and LPS-induced inflammatory responses are mediated by Nrf2 activation. Int J Mol Med. 35(5):1237–1245.
- Choi Y, Seo H, Cho M, Kim J, Chung HS, Lee I, Kim MJ. 2021. Rutin inhibits DRP1-mediated mitochondrial fission and prevents ethanol-induced hepatotoxicity in HepG2 cells and zebrafish. Anim Cells Syst (Seoul). 25(1):74–81.
- Claridge T. 2009. MNova: NMR data processing, analysis, and prediction software. J Chem Inf Model. 49(4):1136–1137.
- Collins FW. 1989. Oat phenolics: avenanthramides, novel substituted N-cinnamoylanthranilate alkaloids from oat groats and hulls. J Agric Food Chem. 37(1):60–66.
- Colombo R, Ferron L, Papetti A. 2021. Colored corn: an up-date on metabolites extraction, health implication, and potential use. Molecules. 26(1):199.
- Conti M. 2000. Phosphodiesterases and cyclic nucleotide signaling in endocrine cells. Mol Endocrinol. 14(9):1317–1327.
- Couladis M, Tzakou O, Verykokidou E, Harvala C. 2003. Screening of some Greek aromatic plants for antioxidant activity. Phytother Res. 17(2):194–195.
- Cragg GM, Newman DJ. 2013. Natural products: a continuing source of novel drug leads. Biochim Biophys Acta. 1830(6):3670–3695.
- Cui G, Wei F, Wei M, Xie L, Lin Z, Feng X. 2021. Modulatory effect of tagetes erecta flowers essential oils via Nrf2/HO-1/NF-κB/p65 axis mediated suppression of N-methyl-N'nitro-N-nitroguanidine (MNNG) induced gastric cancer in rats. Mol Cell Biochem. 476(3):1541–1554.
- Cui H, Bashar MAE, Rady I, El-Naggar HA, El-Maoula LMA, Mehany ABM. 2022. Antiproliferative activity, proapoptotic effect, and cell cycle arrest in human cancer cells of some marine natural product extract. Oxid Med Cell Longev. 2020:7948705.
- Deepika MP. 2022. Health benefits of quercetin in age-related diseases. Molecules. 27(8):2498.
- Dias DA, Urban S, Roessner U. 2012. A historical overview of natural products in drug discovery. Metabolites. 2(2):303–336.
- Dick RA, Zadrozny KK, Xu C, Schur FKM, Lyddon TD, Ricana CL, Wagner JM, Perilla JR, Ganser-Pornillos BK, Johnson MC, et al. 2018. Inositol phosphates are assembly co-factors for HIV-1. Nature. 560(7719):509–512.
- Dührkop K, Shen H, Meusel M, Rousu J, Böcker S. 2015. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc Natl Acad Sci USA. 112(41):12580–12585.
- Elyashberg M, Williams A. 2021. ACD/structure elucidator: 20 years in the history of development. Molecules. 26(21):6623.
- Enogieru AB, Haylett W, Hiss DC, Bardien S, Ekpo OE. 2018. Rutin as a potent antioxidant: implications for neurodegenerative disorders. Oxid Med Cell Longev. 2018:6241017.
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 294(5548):1942–1945.
- Félix-Silva J, Giordani RB, SilvaJrA, Zucolotto SM, Fernandes-Pedrosa M. 2014. Jatropha gossypiifolia L. (Euphorbiaceae): a review of traditional uses, phytochemistry, pharmacology, and toxicology of this medicinal plant. Evid Based Complement Alternat Med. 2014:369204.
- Feng JH, Jung JS, Hwang SH, Lee SK, Lee SY, Kwak YG, Kim DH, Song CY, Kim MJ, Suh HW, et al. 2022. The mixture of Agrimonia pilosa Ledeb. and Salvia miltiorrhiza Bunge. extract produces analgesic and anti-inflammatory effects in a collagen-induced arthritis mouse model. Anim Cells Syst (Seoul). 26(4):166–173.
- Figlia G, Willnow P, Teleman AA. 2020. Metabolites regulate cell signaling and growth via covalent modification of proteins. Dev Cell. 54(2):156–170.
- Fu X, Li M, Tang C, Huang Z, Najafi M. 2021. Targeting of cancer cell death mechanisms by resveratrol: a review. Apoptosis. 26(11–12):561–573.
- Gabbia D, Martin SD. 2020. Brown seaweeds for the management of metabolic syndrome and associated diseases. Molecules. 25(18):4182.
- Gallo K, Goede A, Preissner R, Gohlke BO. 2022. Superpred 3.0: drug classification and target prediction-a machine learning approach. Nucleic Acids Res. 50(W1):W726–W731.
- Garg A, Garg S, Zaneveld LJ, Singla AK. 2001. Chemistry and pharmacology of the citrus bioflavonoid hesperidin. Phytother Res. 15(8):655–669.
- Gathungu RM, Kautz R, Kristal BS, Bird SS, Vouros P. 2020. The integration of LC-MS and NMR for the analysis of low molecular weight trace analytes in complex matrices. Mass Spectrom Rev. 39(1–2):35–54.
- Gomes AP, Ilter D, Low V, Endress JE, Fernández-García J, Rosenzweig A, Schild T, Broekaert D, Ahmed A, Planque M, et al. 2020. Age-induced accumulation of methylmalonic acid promotes tumour progression. Nature. 585(7824):283–287.
- Gomes CL, de Albuquerque Wanderley Sales V, de Melo CG, da Silva RMF, Nishimura RHV, Rolim LA, Neto PJR. 2021. Beta-lapachone: natural occurrence, physicochemical properties, biological activities, toxicity and synthesis. Phytochemistry. 186:112713.
- Guijas C, Montenegro-Burke R, Domingo-Almenara X, Palermo A, Warth B, Hermann G, Koellensperger G, Huan T, Uritboonthai W, Aisporna AE, et al. 2018. METLIN: a technology platform for identifying knowns and unknowns. Anal Chem. 90(5):3156–3164.
- Gupta SP, Siddiqi NJ, Khan HA, Alrokayan SH, Alhomida AS, Singh RK, Verma PK, Kumar S, Acharya A, Sharma B. 2022. Phytochemical profiling of microalgae Euglena tuba and its anticancer activity in Dalton's lymphoma cells. Front Biosci (Landmark Ed). 27(4):120.
- Hakkou Z, Maciuk A, Leblais V, Bouanani NE, Mekhfi H, Bnouham M, Aziz M, Ziyyat A, Rauf A, Hadda TB, et al. 2017. Antihypertensive and vasodilator effects of methanolic extract of inula viscosa: biological evaluation and POM analysis of cynarin, chlorogenic acid as potential hypertensive. Biomed Pharmacother. 93:62–69.
- Harayama T, Shimizu T. 2020. Roles of polyunsaturated fatty acids, from mediators to membranes. J Lipid Res. 61(8):1150–1160.
- He M, Cao C, Ni Z, Liu Y, Song P, Hao S, He Y, Sun X, Rao Y. 2022. PROTACs: great opportunities for academia and industry (an update from 2020 to 2021). Signal Transduct Target Ther. 7:181.
- Henkel T, Brunne RM, Muller H, Reichel F. 1999. Statistical investigation into the structural complementarity of natural products and synthetic compounds. Angew Chem Int Ed. 38(5):643–647.
- Hert J, Irwin JJ, Laggner C, Keiser MJ, Shoichet BK. 2009. Quantifying biogenic bias in screening libraries. Nat Chem Biol. 5(7):479–483.
- Hong J. 2011. Role of natural product diversity in chemical biology. Curr Opin Chem Biol. 15(3):350–354.
- Hong JS, Feng JH, Park JS, Lee HJ, Lee JY, Lim SS, Suh HW. 2020. Antinociceptive effect of chrysin in diabetic neuropathy and formalin-induced pain models. Anim Cells Syst (Seoul). 24(3):143–150.
- Hou J, Jeon B, Baek J, Yun Y, Kim D, Chang B, Kim S, Kim S. 2022. High fat diet-induced brain damaging effects through autophagy-mediated senescence, inflammation and apoptosis mitigated by ginsenoside F1-enhanced mixture. J Ginseng Res. 46(1):79–90.
- Howarth A, Ermanis K, Goodman JM. 2020. DP4-AI automated NMR data analysis: straight from spectrometer to structure. Chem Sci. 11(17):4351–4359.
- Huber F, van der Burg S, van der Hooft JJJ, Ridder L. 2021. MS2DeepScore: a novel deep learning similarity measure to compare tandem mass spectra. J Cheminform. 13(1):84.
- Hwang GH, Ryu JM, Jeon YJ, Choi J, Han HJ, Lee Y-M, Lee S, Bae J-S, Jung J-W, et al. 2015. The role of thioredoxin reductase and glutathione reductase in plumbagin-induced, reactive oxygen species-mediated apoptosis in cancer cell lines. Eur J Pharmacol. 765:384–393.
- Hwang M, Kim JN, Kim BJ. 2020. Hesperidin depolarizes the pacemaker potentials through 5-HT4 receptor in murine small intestinal interstitial cells of Cajal. Anim Cells Syst (Seoul). 24(2):84–90.
- Ior LD, Otimenyin SO, Okwuasaba FK. 2021. Antipsychotic-like effect of ethyl acetate fraction of Terminalia macroptera leaf in mice. IBRO Neurosci Rep. 10:83–89.
- Jacobson KA, Gao ZG. 2006. Adenosine receptors as therapeutic targets. Nat Rev Drug Discov. 5(3):247–264.
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. 2001. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 3(2):193–197.
- Jan R, Khan M, Asaf S, Lubna AS, Kim KM. 2022. Bioactivity and therapeutic potential of kaempferol and quercetin: new insights for plant and human health. Plants (Basel). 11(19):2623.
- Jang MG, Oh JM, Ko HC, Kim JW, Baek S, Jin YJ, Hur SP, Kim SJ. 2021. Clerodendrum trichotomum extract improves metabolic derangements in high fructose diet-fed rats. Anim Cells Syst (Seoul). 25(6):396–404.
- Jeong MJ, Lim DS, Kim SO, Park C, Choi YH, Jeong SJ. 2021. Effect of rosmarinic acid on differentiation and mineralization of MC3T3-E1 osteoblastic cells on titanium surface. Anim Cells Syst (Seoul). 25(1):46–55.
- Ji CH, Kim HY, Lee MJ, Heo AJ, Park DY, Lim S, Shin S, Ganipisetti S, Yang WS, Jung CA, et al. 2022. The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system. Nat Commun. 13(1):904.
- Jing W, Xiaolan C, Yu C, Feng Q, Haifeng Y. 2022. Pharmacological effects and mechanisms of tannic acid. Biomed Pharmacother. 154:113561.
- John OD, Mushunje AT, Surugau N, Guad RM. 2022. The metabolic and molecular mechanisms of α mangostin in cardiometabolic disorders. Int J Mol Med. 50(3):120.
- Jung H, Jung DM, Lee SS, Kim EM, Yoon K, Kim KK. 2022. Mangifera Indica leaf extracts promote hair growth via activation of Wnt signaling pathway in human dermal papilla cells. Anim Cells Syst (Seoul). 26(3):129–136.
- Kaelin WG Jr, McKnight SL. 2013. Influence of metabolism on epigenetics and disease. Cell. 153(1):56–69.
- Karali D, Georgescu L, Pirintsos S, Athanassakis I. 2016. T cell regulation by Phlomis lanata protein extracts in mice. Pharm Biol. 54(2):207–214.
- Keller KE, Tan IS, Lee YS. 2012. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science. 338(6110):1069–1072.
- Keller NP. 2019. Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol. 17(3):167–180.
- Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato RL, Chudnovsky Y, Pacold ME, Chen WW, Cantor JR, et al. 2015. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 520(7547):363–367.
- Kim DB, Unenkhuu B, Kim GJ, Kim SW, Kim HS. 2022a. Cynarin attenuates LPS-induced endothelial inflammation via upregulation of the negative regulator MKP-3. Anim Cells Syst (Seoul). 26(3):119–128.
- Kim HW, Wang M, Leber CA, Nothias LF, Reher R, Kang KB, van der Hooft JJJ, Dorrestein PC, Gerwick WH, Cottrell GW. 2021a. NPClassifier: a deep neural network-based structural classification tool for natural products. J Nat Prod. 84(11):2795–2807.
- Kim KH, Kim S, Jung MY, Ham IH, Whang WK. 2009. Anti-inflammatory phenylpropanoid glycosides from Clerodendron trichotomum leaves. Arch Pharm Res. 32(1):7–13.
- Kim SW, Kim DB, Kim HS. 2022b. Neuroprotective effects of tannic acid in the postischemic brain via direct chelation of Zn2+. Anim Cells Syst (Seoul). 26(4):183–191.
- Kim WS, Kim CH, Lee JM, Jeon JH, Kang BG, Warkad MS, Inci G, Suh HW, Lim SS, Kim SC, et al. 2021b. Purple corn extract (PCE) alleviates cigarette smoke (CS)-induced DNA damage in rodent blood cells by activation of AMPK/Foxo3a/MnSOD pathway. Anim Cells Syst (Seoul). 25(1):65–73.
- Kimura S, Tung YC, Pan MH, Su NW, Lai YJ, Cheng KC. 2017. Black garlic: a critical review of its production, bioactivity, and application. J Food Drug Anal. 25(1):62–70.
- Koehn FE, Carter GT. 2005. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 4(3):206–220.
- Koenig RT, Dickman JR, Kang CH, Zhang T, Chu YF, Ji LL. 2016. Avenanthramide supplementation attenuates eccentric exercise-inflicted blood inflammatory markers in women. Eur J Appl Physiol. 116(1):67–76.
- Lajter I, Pan SP, Nikles S, Ortmann S, Vasas A, Csupor-Löffler B, Forgó P, Hohmann J, Baueret R. 2015. Inhibition of COX-2 and NF-kB1 gene expression, NO production, 5-LOX, and COX-1 and COX-2 enzymes by extracts and constituents of Onopordum acanthium. Planta Med. 81(14):1270–1276.
- Lam KS. 2007. New aspects of natural products in drug discovery. Trends Microbiol. 15(6):279–289.
- Lautié E, Russo O, Ducrot P, Boutin JA. 2020. Unraveling plant natural chemical diversity for drug discovery purposes. Front Pharmacol. 11:397.
- Lee DC, Sohn HA, Park ZY, Oh S, Kang YK, Lee KM, Kang M, Jang YJ, Yang SJ, Hong YK, et al. 2015. A lactate-induced response to hypoxia. Cell. 161(3):595–609.
- Lee H, Jang JH, Kim SJ. 2021. Malonic acid suppresses lipopolysaccharide-induced BV2 microglia cell activation by inhibiting the p38 MAPK/NF-κB pathway. Anim Cells Syst (Seoul). 25(2):110–118.
- Lee H, Kim W, Kang HG, Kim WJ, Lee SC, Kim SJ. 2020a. Geranium thunbergii extract-induced G1 phase cell cycle arrest and apoptosis in gastric cancer cells. Anim Cells Syst (Seoul). 24(1):26–33.
- Lee N, Spears ME, Carlisle AE, Kim D. 2020b. Endogenous toxic metabolites and implications in cancer therapy. Oncogene. 39(35):5709–5720.
- Lee S, Kim MG, Ahn H, Kim S. 2020c. Inositol pyrophosphates: signaling molecules with pleiotropic actions in mammals. Molecules. 25(9):2208.
- Lee TS, Lee JY, Kyung JW, Yang Y, Park SJ, Lee S, Pavlovic I, Kong B, Jho YS, Jessen HJ, et al. 2016. Inositol pyrophosphates inhibit synaptotagmin-dependent exocytosis. Proc Natl Acad Sci USA. 113(29):8314–8319.
- Lee Y, Kwon J, Jeong JH, Ryu JH, Kim KI. 2022. Kazinol C from Broussonetia kazinoki stimulates autophagy via endoplasmic reticulum stress-mediated signaling. Anim Cells Syst (Seoul). 26(1):28–36.
- Li J, Bonkowski MS, Moniot S, Zhang D, Hubbard BP, Ling AJY, Rajman LA, Qin B, Lou Z, Gorbunova V. 2017. A conserved NAD+ binding pocket that regulates protein-protein interactions during aging. Science. 355(6331):1312–1317.
- Li J, Cai Z, Li XW, Zhuang C. 2022. Natural product-inspired targeted protein degraders: advances and perspectives. J Med Chem. 65(20):13533–13560.
- Li K, Crews CM. 2022. PROTACs: past, present and future. Chem Soc Rev. 51(12):5214–5236.
- Li X, Snyder M. 2011. Metabolites as global regulators: a new view of protein regulation: systematic investigation of metabolite-protein interactions may help bridge the gap between genome-wide association studies and small molecule screening studies. Bioessays. 33(7):485–489.
- Li Y, Kong D, Fu Y, Sussman MR, Wu H. 2020. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol Biochem. 148:80–89.
- Lim W, Kang C. 2020. Avenanthramide C suppresses hypoxia-induced cyclooxygenase-2 expression through sirtuin1 activation in non-small-cell lung cancer cells. Anim Cells Syst (Seoul). 24(2):79–83.
- Lin MH, Jensen MK, Elrod ND, Huang KL, Welle KA, Wagner EJ, Tong L. 2022. Inositol hexakisphosphate is required for integrator function. Nat Commun. 13(1):5742.
- Lin Y, Schiavo S, Orjala J, Vouros P, Kautz R. 2008. Microscale LC-MS-NMR platform applied to the identification of active cyanobacterial metabolites. Anal Chem. 80(21):8045–8054.
- Liu X, Locasale JW. 2017. Metabolomics: a primer. Trends Biochem Sci. 42(4):274–284.
- Lomenick B, Hao R, Jonai N, Huang J. 2009. Target identification using drug affinity responsive target stability (DARTS). Proc Natl Acad Sci USA. 106(51):21984–21989.
- Luduvico KP, Spohr L, Soares MSP, Teixeira FC, de Farias AS, Bona NP, Pedra NS, de Oliveira Campello Felix A, Spanevello RM, Stefanello FM. 2020. Antidepressant effect and modulation of the redox system mediated by tannic acid on lipopolysaccharide-induced depressive and inflammatory changes in mice. Neurochem Res. 45(9):2032–2043.
- Luo C, Zou L, Sun H, Peng J, Gao C, Bao L, Ji R, Jin Y, Sun S. 2020. A review of the anti-inflammatory effects of rosmarinic acid on inflammatory diseases. Front Pharmacol. 11:153.
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. 2005. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 309(5740):1534–1539.
- Machana S, Weerapreeyakul N, Barusrux S. 2012. Anticancer effect of the extracts from polyalthia evecta against human hepatoma cell line (HepG2). Asian Pac J Trop Biomed. 2(5):368–374.
- Madhukar NS, Khade PK, Huang L, Gayvert K, Galletti G, Stogniew M, Allen JE, Giannakakou P, Elemento O. 2019. A Bayesian machine learning approach for drug target identification using diverse data types. Nat Comm. 10(1):5221.
- Mahdiani S, Omidkhoda N, Heidari S, Hayes AW, Karimi G. 2022. Protective effect of luteolin against chemical and natural toxicants by targeting NF-κB pathway. Biofactors. 48(4):744–762.
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. 1995. The nuclear receptor superfamily: the second decade. Cell. 83(6):835–839.
- Mani R, Natesan V. 2018. Chrysin: sources, beneficial pharmacological activities, and molecular mechanism of action. Phytochemistry. 145:187–196.
- Martínez-Reyes I, Chandel NS. 2020. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commnun. 11(1):102.
- Milanesi R, Coccetti P, Tripodi F. 2020. The regulatory role of key metabolites in the control of cell signaling. Biomolecules. 10(6):862.
- Milligan G, Shimpukade B, Ulven T, Hudson BD. 2017. Complex pharmacology of free fatty acid receptors. Chem Rev. 117(1):67–110.
- Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, Jedrychowski MP, Costa ASH, Higgins M, Hams E, et al. 2018. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 556(7699):113–117.
- Molina DM, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, Nordlund P. 2013. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 341(6141):84–87.
- Moon SB, Choi NR, Kim JN, Kwon MJ, Kim BS, Ha KT, Lim EY, Kim YT, Kim BJ. 2022. Effects of black garlic on the pacemaker potentials of interstitial cells of Cajal in murine small intestine in vitro and on gastrointestinal motility in vivo. Anim Cells Syst (Seoul). 26(1):37–44.
- Moore J, Yousef M, Tsiani E. 2016. Anticancer effects of rosemary (Rosmarinus officinalis L.) extract and rosemary extract polyphenols. Nutrients. 8(11):731.
- Müller-Kuhrt L. 2003. Putting nature back into drug discovery. Nat Biotechnol. 21(6):602.
- Murad F. 2006. Shattuck lecture. Nitric oxide and cyclic GMP in cell signaling and drug development. N Engl J Med. 355(19):2003–2011.
- Nalawansha DA, Crews CM. 2020. PROTACs: an emerging therapeutic modality in precision medicine. Cell Chem Biol. 27(8):998–1014.
- Newman DJ. 2021. Natural product based antibody drug conjugates: clinical status as of November 9, 2020. J Nat Prod. 84(3):917–931.
- Newman DJ, Cragg GM. 2020. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 83(3):770–803.
- Newton AC, Bootman MD, Scott JD. 2016. Second messengers. Cold Spring Harb Perspect Biol. 8(8):a005926.
- No H, Nam SH, Seo HW, Seo JH, Park SH, Kim SB, Jung JS, Park J, Choi J, Lee JY, et al. 2021. Purple corn extract alleviates 2,4-dinitrochlorobenzene-induced atopic dermatitis-like phenotypes in BALB/c mice. Anim Cells Syst (Seoul). 25(5):272–282.
- Noratto GD, Bertoldi MC, Krenek K, Talcott ST, Stringheta PC, Mertens-Talcott SU. 2010. Anticarcinogenic effects of polyphenolics from mango (Mangifera indica) varieties. J Agric Food Chem. 58(7):4104–4112.
- Nugroho A, Harahap IA, Ardiansyah A, Bayu A, Rasyid A, Murniasih T, Setyastuti A, Putra MY. 2022. Antioxidant and antibacterial activities in 21 species of Indonesian sea cucumbers. J Food Sci Technol. 59(1):239–248.
- Nursid M, Marraskuranto E, Chasanah E. 2019. Cytotoxicity and apoptosis induction of sea cucumber Holothuria atra extracts. Pharmacogn Res. 11(1):41–46.
- Pai MY, Lomenick B, Hwang H, Schiestl R, McBride W, Loo JA, Huang J. 2015. Drug affinity responsive target stability (DARTS) for small molecule target identification. Methods Mol Biol. 1263:287–298.
- Park C, Choi SH, Jeong JW, Han MH, Lee H, Hong SH, Kim GY, Moon SK, Kim WJ, Choi YH. 2020. Honokiol ameliorates oxidative stress-induced DNA damage and apoptosis of c2c12 myoblasts by ROS generation and mitochondrial pathway. Anim Cells Syst (Seoul). 24(1):60–68.
- Park C, Jeong JW, Han MH, Lee H, Kim GY, Jin S, Park JH, Kwon HJ, Kim BW, Choi YH. 2021. The anti-cancer effect of betulinic acid in u937 human leukemia cells is mediated through ROS-dependent cell cycle arrest and apoptosis. Anim Cells Syst (Seoul). 25(2):119–127.
- Park H, Park H, Chung TW, Choi HJ, Jung YS, Lee SO, Ha KT. 2017. Effect of Sorbus commixta on the invasion and migration of human hepatocellular carcinoma Hep3B cells. Int J Mol Med. 40(2):483–490.
- Pham AT, Malterud KE, Paulsen BS, Diallo D, Wangensteen H. 2014. alpha-Glucosidase inhibition, 15-lipoxygenase inhibition, and brine shrimp toxicity of extracts and isolated compounds from Terminalia macroptera leaves. Pharm. Biol. 52(9):1166–1169.
- Piazza I, Kochanowski K, Cappelletti V, Fuhrer T, Noor E, Sauer U, Picotti P. 2018. A map of protein-metabolite interactions reveals principles of chemical communication. Cell. 172(1–2):358–372.e23.
- Puppala D, Lee H, Kim KB, Swanson HI. 2008. Development of an aryl hydrocarbon receptor antagonist using the proteolysis-targeting chimeric molecules approach: a potential tool for chemoprevention. Mol Pharmacol. 73(4):1064–1071.
- Rahmani AH, Alsahli MA, Almatroudi A, Almogbel MA, Khan AA, Anwar S, Almatroodi SA. 2022. The potential role of apigenin in cancer prevention and treatment. Molecules. 27(18):6051.
- Ratan ZA, Haidere MF, Hong YH, Park SH, Lee JO, Lee J, Cho JY. 2021. Pharmacological potential of ginseng and its major component ginsenosides. J Ginseng Res. 45(2):199–210.
- Rauf A, Olatunde A, Imran M, Alhumaydhi FA, Aljohani ASM, Khan SA, Uddin S, Mitra S, Emran TB, Khayrullin M. 2021. Honokiol: a review of its pharmacological potential and therapeutic insights. Phytomedicine. 90:153647.
- Ríos JL, Máñez S. 2018. New pharmacological opportunities for betulinic acid. Planta Med. 84(1):8–19.
- Robertovna GE, Alexeevich KD, Alexeevich SA, Petrovna GM, Kenzhebaevna OK. 2019. A traditional medicine plant, oOnopordum acanthium L. (Asteraceae): chemical composition and pharmacological research. Plants (Basel). 8(2):40.
- Rodrigues T, Reker D, Schneider P, Schneider G. 2016. Counting on natural products for drug design. Nat Chem. 8(6):531–541.
- Rodrigues T, Werner M, Roth J, da Cruz EHG, Marques MC, Akkapeddi P. 2018. Machine intelligence decrypts β-lapachone as an allosteric 5-lipoxygenase inhibitor. Chem Sci. 9(34):6899–6903.
- Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. 2001. Protacs: chimeric molecules that target proteins to the Skp1-cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci USA. 98(15):8554–8559.
- Savitski MM, Reinhard FBM, Franken H, Werner T, Savitski MF, Eberhard D, Molina DM, Jafari R, Dovega RB, Klaeger S, et al. 2014. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science. 346(6205):1255784.
- Schreiber SL. 2005. Small molecules: the missing link in the central dogma. Nat Chem Biol. 1(2):64–66.
- Seca AML, Pinto DCGA. 2018. Plant secondary metabolites as anticancer agents: successes in clinical trials and therapeutic application. Int J Mol Sci. 19(1):263.
- Shanab SMM, Mostafa SSM, Shalaby EA, Mahmoud GI. 2012. Aqueous extracts of microalgae exhibit antioxidant and anticancer activities. Asian Pac J Trop Biomed. 2(8):608–615.
- Sharma P, McAlinden KD, Ghavami S, Deshpande DA. 2021. Chloroquine: autophagy inhibitor, antimalarial, bitter taste receptor agonist in fight against COVID-19, a reality check? Eur J Pharmacol. 15(897):173928.
- Shehzad A, Parveen S, Qureshi M, Subhan F, Lee YS. 2018. Decursin and decursinol angelate: molecular mechanism and therapeutic potential in inflammatory diseases. Inflamm Res. 67(3):209–218.
- Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Moan NL, Grueter CA, Lim H, Saunders LR, Stevens RD, et al. 2013. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 339(6116):211–214.
- Shyer JA, Flavell RA, Bailis W. 2020. Metabolic signaling in T cells. Cell Res. 30(8):649–659.
- Stine ZE, Schug ZT, Salvino JM, Dang CV. 2022. Targeting cancer metabolism in the era of precision oncology. Nat Rev Drug Discov. 21(2):141–162.
- Stomberski CT, Hess DT, Stamler JS. 2019. Protein S-nitrosylation: determinants of specificity and enzymatic regulation of S-nitrosothiol-based signaling. Antioxid Redox Signal. 30(10):1331–1351.
- Stompor-Gorący M, Bajek-Bil A, Machaczka M. 2021. Chrysin: perspectives on contemporary status and future possibilities as pro-health agent. Nutrients. 13(6):2038.
- St-Pierre A, Blondeau D, Lajeunesse A, Bley J, Bourdeau N, DesgagnéPenix I. 2018. Phytochemical screening of quaking aspen (Populus tremuloides) extracts by UPLC-QTOF-MS and evaluation of their antimicrobial activity. Molecules. 23(7):1739.
- Strickland EC, Geer MA, Tran DT, Adhikari J, West GM, DeArmond PD, Xu Y, Fitzgerald MC. 2013. Thermodynamic analysis of protein-ligand binding interactions in complex biological mixtures using the stability of proteins from rates of oxidation. Nat Protoc. 8(1):148–161.
- Stuart KA, Welsh K, Walker MC, Edrada-Ebel RA. 2020. Metabolomic tools used in marine natural product drug discovery. Expert Opin Drug Discov. 15(4):499–522.
- Sung YY, Yoon T, Yang WK, Kim SJ, Kim HK. 2011. Anti-obesity effects of geranium thunbergii extract via improvement of lipid metabolism in high-fat diet-induced obese mice. Mol Med Rep. 4(6):1107–1113.
- Sur B, Lee B. 2022. Luteolin reduces fear, anxiety, and depression in rats with post-traumatic stress disorder. Anim Cells Syst (Seoul). 26(4):174–182.
- Terstappen GC, Schlüpen C, Raggiaschi R, Gaviraghi G. 2007. Target deconvolution strategies in drug discovery. Nat Rev Drug Discov. 6(11):891–903.
- Topal M, Gocer H, Topal F, Kalin P, Kose LP, Gulcin I, Cakmak KC, Kucuk M, Durmaz L, Goren AC, et al. 2016. Antioxidant, antiradical, and anticholinergic properties of cynarin purified from the illyrian thistle (onopordum illyricumL.). J Enzyme Inhib Med Chem. 31(2):266–275.
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. 2010. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 62(3):405–496.
- Turumtay H, Midilli A, Turumtay EA, Demir A, Selvi EK, Budak EE, Er H, Kocaimamoglu F, Baykal H, Belduz AO, et al. 2017. Gram (-) microorganisms DNA polymerase inhibition, antibacterial and chemical properties of fruit and leaf extracts of Sorbus acuparia and Sorbus caucasica var. yaltirikii. Biomed Chromatogr. 31(6):e3901.
- Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, et al. 2016. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat Biotechnol. 34(8):828–837.
- Wang YP, Lei QY. 2018. Metabolite sensing and signaling in cell metabolism. Signal Transduct Target Ther. 3:30.
- Wen S, Zhang X, Wu Y, Yu S, Zhang W, Liu D, Yang K, Sun J. 2022. Agrimonia pilosa Ledeb.: a review of its traditional uses, botany, phytochemistry, pharmacology, and toxicology. Heliyon. 8(8):e09972.
- Wishart DS. 2016. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 15(7):473–484.
- Wolfender JL, Nuzillard JM, van der Hooft JJJ, Renault JH, Bertrand S. 2019. Accelerating metabolite identification in natural product research: toward an ideal combination of liquid chromatography–high-resolution tandem mass spectrometry and NMR profiling, in silico databases, and chemometrics. Anal Chem. 91(1):704–742.
- Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, Sabatini DM. 2016. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 351(6268):43–48.
- Yen YH, Farooqi AA, Li KT, Butt G, Tang JY, Wu CY, Cheng YB, Hou MF, Chang HW. 2014. Methanolic extracts of Solieria robusta inhibits proliferation of oral cancer Ca9-22 cells via apoptosis and oxidative stress. Molecules. 19(11):18721–18732.
- Yim D, Singh RP, Agarwal C, Lee S, Chi H, Agarwal R. 2005. A novel anticancer agent, decursin, induces G1 arrest and apoptosis in human prostate carcinoma cells. Cancer Res. 65(3):1035–1044.
- Yñigez-Gutierrez AE, Bachmann BO. 2019. Fixing the unfixable: the art of optimizing natural products for human medicine. J Med Chem. 62(18):8412–8428.
- Yoon S, Huang KW, Reebye V, Spalding D, Przytycka TM, Wang Y, Swiderski P, Li L, Armstrong B, Reccia I, et al. 2017. Aptamer-drug conjugates of active metabolites of nucleoside analogs and cytotoxic agents inhibit pancreatic tumor cell growth. Mol Ther Nucleic Acids. 6:80–88.
- Zhang KJ, Gu QL, Yang K, Ming XJ, Wang JX. 2017. Anticarcinogenic effects of α-Mangostin: a review. Planta Med. 83(3–04):188–202.
- Zhang LX, Li CX, Kakar MU, Khan MS, Wu PF, Amir RM, Dai DF, Naveed M, Li QY, Saeed M, et al. 2021. Resveratrol (RV): a pharmacological review and call for further research. Biomed Pharmacother. 143:112164.
- Zhou W, Wang H, Yang Y, Chen ZS, Zou C, Zhang J. 2020. Chloroquine against malaria, cancers and viral diseases. Drug Discov Today. 25(11):2012–2022.