ABSTRACT
The gut microbiome plays a crucial role in maintaining health in a variety of organisms, from insects to humans. Further, beneficial symbiotic microbes are believed to contribute to improving the quality of life of the host. Drosophila is an optimal model for studying host–commensal microbe interactions because it allows for convenient manipulation of intestinal microbial composition. Fly microbiota has a simple taxonomic composition and can be cultivated and genetically tracked. This permits functional studies and analyses of the molecular mechanisms underlying their effects on host physiological processes. In this context, we briefly introduce the principle of juvenile developmental growth in Drosophila. Then, we discuss the current understanding of the molecular mechanisms underlying the effects of gut commensal bacteria, such as Lactiplantibacillus plantarum and Acetobacter pomorum, in the fly gut microbiome on Drosophila juvenile growth, including specific actions of gut hormones and metabolites in conserved cellular signaling systems, such as the insulin/insulin-like (IIS) and the target of rapamycin (TOR) pathways. Given the similarities in tissue function/structure, as well as the high conservation of physiological systems between Drosophila and mammals, findings from the Drosophila model system will have significant implications for understanding the mechanisms underlying the interaction between the host and the gut microbiome in metazoans.
Introduction
Ellie Metchnikoff first recognized that certain bacteria are host-friendly and could improve health and delay aging. Since then, numerous researchers have attempted to classify the bacterial species that constitute the gut microbiome and determine how specific species or combinations of species affect host physiology in vertebrates. Despite the use of advanced techniques, such as next-generation sequencing and meta-transcriptomic analysis, reports on the effects of particular bacterial species or gut microbiota complexes on host physiology are inconsistent. This can be attributed to the extreme variability in the gut microbiome, which depends on environmental factors as well as the difficulty in artificially changing intestinal microbial communities. The advantages of the Drosophila model system have been emphasized for studying host–gut microbe interactions, specifically to overcome the abovementioned limitations. In contrast to mammals, the gut microbiome of Drosophila is relatively simple as it is composed of approximately 30 species (Cox and Gilmore Citation2007; Chandler et al. Citation2011; Fink et al. Citation2013; Han et al. Citation2017). This provides a more accurate method for assessing the effects of different combinations of microbial communities on the host. Additionally, it also provides the perfect opportunity for artificial colonization of single bacterial taxa, co-cultures of multiple taxa, and undefined microbiota in the feces or dissected guts of conventionally reared (CR) flies.
Furthermore, Drosophila shares many similarities with mammals in various aspects such as functional organs and physiology (Hyun Citation2013; Buchon et al. Citation2014; Lee SH and Kim Citation2021). For example, insulin/insulin-like (IIS) and target of rapamycin (TOR) signaling pathway are a highly conserved signaling pathway that plays a major role in metazoan growth and development in response to the nutritional status of the host. Thus, Drosophila research has been helpful in the current understanding of the role of microbial communities and their functions in host physiology. In this review, we discuss the regulation of body growth and juvenile development and its association with the IIS/TOR pathway and summarize the important findings on how gut commensal bacteria regulate appropriate body growth and developmental processes through the IIS/TOR pathway in Drosophila.
IIS/TOR Drosophila nutrient-dependent pathway and juvenile growth
Organisms must modify their metabolism to adapt to the changing conditions for developmental growth and survival. To achieve this adaptation, they must be able to sense changes in the external environmental conditions and adjust their internal states accordingly. Metabolic adaptability in response to changes in nutritional status is necessary to maintain energy balance and developmental progress (Leopold and Perrimon Citation2007; Owusu-Ansah and Perrimon Citation2014). The IIS/TOR signaling pathway is an evolutionarily conserved signaling cascade across metazoans that coordinates organismal growth and development by sensing the internal nutritional status (Wullschleger et al. Citation2006; Hyun Citation2013).
The Drosophila nervous system plays a vital role in sensing internal nutrient conditions, coordinating metabolic processes, and regulating energy metabolism. When glucose levels rise, insulin-producing cells (IPCs) in the brain release Drosophila insulin-like peptides (Dilps) into the bloodstream, mainly Dilp2, Dilp3, and Dilp5 (Hyun Citation2013; Owusu-Ansah and Perrimon Citation2014). These Dilps bind to insulin receptors (InR) and trigger downstream signaling. This signaling cascade involves the phosphorylation of Chico, which activates phosphoinositide-3-kinase (PI3 K). PI3 K converts phosphatidylinositol 4,5-diphosphate (PIP2) to phosphatidylinositol 3,4,5-triphosphate (PIP3) (Auger et al. Citation1989; Lee JO et al. Citation1999), leading to the recruitment and activation of phosphoinositide-dependent kinase 1 (PDK1) and Akt (Oldham and Hafen Citation2003; Mora et al. Citation2004; Lee JE et al. Citation2021). Akt suppresses the activity of Drosophila forkhead box O (dFOXO), a transcription factor that regulates stress responses and cell growth (Arden Citation2008). Imaginal morphogenesis protein-late 2 (Imp-L2) acts similarly to human IGF-binding protein 7 (IGFBP-7) by binding directly to Dilps and inhibiting their activity, reducing systemic IIS levels (Honegger et al. Citation2008; Alic et al. Citation2011). When amino acid levels, especially branched-chain amino acids, increase in the hemolymph, they enter cells through Slimfast amino acid transporters and activate the TOR pathway, which mediates cellular nutrient responses (Colombani et al. Citation2003). TOR exists in two complexes: TOR complex 1 (TORC1) and TOR complex 2 (TORC2). TORC1 is sensitive to amino acid changes and diet, promoting cell growth by enhancing protein translation and ribosome biogenesis through phosphorylation of the translation initiation factor 4E-binding protein (4EBP) and ribosomal protein S6 kinase (S6 K), respectively (Hara et al. Citation2002; Yang et al. Citation2013). On the other hand, TORC2, which does not respond to amino acids or rapamycin, is more involved in insulin signaling and cell regulation (Jacinto et al. Citation2004; Hietakangas and Cohen Citation2007; Yang et al. Citation2013; Jevtov et al. Citation2015).
During the juvenile growth period, the nutritional status significantly influences the growth and final adult size. In Drosophila, the fat body acts as an endocrine organ, much like the mammalian liver and adipose tissues, and plays a pivotal role in regulating body size through the insulin/TOR signaling pathway. Depletion of Slimfast in the fat body leads to systemic growth defects by reducing S6 K phosphorylation, indicating that the fat body can slow down growth in response to decreased amino acids, closely tied to the TOR pathway (Colombani et al. Citation2003). Circulating Dilps in Drosophila promote body growth by binding to InR in the larval fat body, activating the PI3 K signaling pathway and repressing dFOXO. Undernourishment reduces InR-dependent PI3 K activity, causing dFOXO to inhibit cell proliferation and reduce the final body size (Britton et al. Citation2002; Kramer et al. Citation2003; Puig et al. Citation2003).
In humans, the cessation of growth coincides with increased circulating steroid hormones. High-fat and high-protein imbalances in nutrition can lead to early and excessive sex hormone production, causing precocious puberty and a shortened growth period (Tanner and Davies Citation1985; Calcaterra et al. Citation2021; Tang et al. Citation2022), highlighting the close link between nutritional status-driven insulin/TOR signaling and steroid hormone production. In Drosophila, ecdysone, a major steroid hormone, is produced in the prothoracic glands through the IIS/TOR pathway (Colombani et al. Citation2005; Layalle et al. Citation2008). Ecdysone affects body size by regulating peripheral IIS directly or indirectly. miR-8 in Drosophila plays a role in ecdysone-dependent growth regulation by targeting U-shaped (Ush), the repressor of PI3 K, and the upregulation of miR-8 increases peripheral IIS activity and final body size (Hyun et al. Citation2009; Jin et al. Citation2012). Additionally, ecdysone-induced Imp-L2 production in the larval fat body suppresses peripheral IIS activity and body growth (Lee GJ et al. Citation2018).
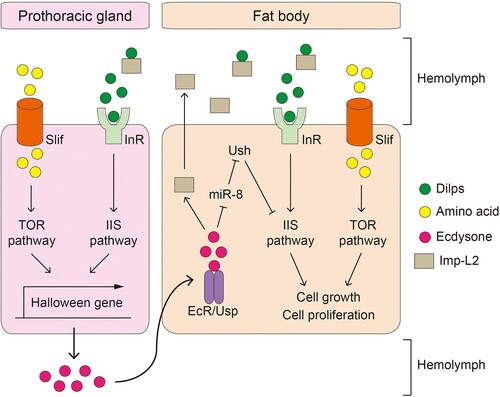
In summary, these studies suggest that the IIS/TOR pathway, body growth rate, and sex hormone secretion are interconnected, with the final size determined by the length of the juvenile growth period and the rate of body growth (Hyun Citation2013, Citation2018) ().
Figure 1. The molecular pathways involved in Drosophila juvenile development and growth. Amino acids in the hemolymph enter cells through Slimfast (Slif) and thereafter activate TOR signaling. Drosophila insulin like peptides (Dilps) bind to insulin receptor (InR), thereby activating the insulin/insulin-like growth factor signaling (IIS) pathway. Both signaling pathways stimulate transcription of Halloween genes in the prothoracic gland, which facilitates the synthesis of ecdysone. The release of ecdysone into the hemolymph promotes fly development. Additionally, Dilps and amino acids stimulate IIS and TOR signaling in the larval fat body, respectively, to promote cell growth and proliferation. Activation of IIS/TOR signaling in the larval fat body is thought to contribute to body growth rate through regulation of non-autonomous tissue growth; however, the underlying mechanisms are still elusive. Ecdysone in the hemolymph penetrates the larval fat body and immediately binds to the ecdysone receptor (EcR). When EcR is activated, it secretes imaginal morphogenesis protein-Late 2, which inhibits the activity of Dilps and prevents the activation of the systemic IIS pathway.
Anatomy and functions of the Drosophila gut
The gut is one of the largest organs in the body and plays a key role in the digestion and absorption of nutrients. Additionally, it forms the largest immune epithelial barrier and serves as the first line of defense against ingested pathogens. The structure and functions of the Drosophila gut are analogous to those of mammals, which is why it is a widely used alternative model for research on the human gastrointestinal system. The Drosophila gut can be divided into the midgut and hindgut, which correspond to the small and large intestines, respectively. Similar to mammal, the distribution of predominant bacteria may vary depending on the intestine tract (Donaldson et al. Citation2016; Chiang et al. Citation2022). After ingestion, food passes through the esophagus to the midgut, where it is broken down into small units by digestive enzymes, including proteases, carbohydrates, and lipases. Subsequently, the hindgut selectively absorbs nutrients, water, and electrolytes. The Drosophila gut comprised four cell types: intestinal stem cells (ISCs), enteroblasts (EBs), differentiated enterocytes (ECs), and enteroendocrine cells (EEs). ISCs and EBs are undifferentiated and differentiated ISC daughter cells, respectively, and the latter can differentiate into two types of functional cells: ECs and EEs. Similar to mammalian goblet cells, ECs secrete digestive enzymes, absorb and transport nutrients, and maintain epithelial barriers by secreting mucus. EEs are scattered throughout both Drosophila and mammalian guts, and sense various environmental stimuli and secrete hormonal peptides to regulate the host’s nutritional status. For detailed information on the structure and function of the Drosophila gut, readers can refer to other excellent reviews (Wang and Hou Citation2010; Apidianakis and Rahme Citation2011; Miguel-Aliaga et al. Citation2018).
The microbiome composition and effective bacteria species involved in Drosophila juvenile growth
For many years, numerous studies have investigated the Drosophila intestinal microbiome using 16S rRNA pyrosequencing. These studies have found that flies generally support relatively simple microbial communities, composed of approximately 30 species, represented by two phyla, Firmicutes and Proteobacteria, and further dominated by two major families, Acetobacteraceae and Lactobacillaceae, and two minor families, Enterococceae and Enterobacteriaceae (Cox and Gilmore Citation2007; Chandler et al. Citation2011; Fink et al. Citation2013; Han et al. Citation2017). Interestingly, Drosophila reared on complex polysaccharide diets, such as cornmeal or soy flour, have a high abundance of Lactiplantibacillus species, whereas Drosophila reared on sugar-rich diets have microbiomes dominated by Acetobacteraceae, particularly Acetobacter and Gluconobacter species (Chaston et al. Citation2014; Huang and Douglas Citation2015). According to the nutrient niche theory, ecological niches in the gut can be defined by available nutrients, and only a few bacterial species that efficiently use partially limiting nutrients can be colonized (Pereira and Berry Citation2017). Several analyses of fly gut microbiota collected from various geographic locations and laboratory sources have shown that bacterial community abundance is dependent on food preferences, implying that gut microbial composition is primarily determined by the host's diet and nutritional environment (Staubach et al. Citation2013; Wong AC et al. Citation2013). Despite considerable variation at the species level, Lactiplantibacillus and Acetobacter mostly inhabit the gut and both genera likely play important roles in the physiology of their hosts. Researchers have attempted to understand the specific commensal bacteria-dependent effects on host traits, and focused on how specific commensal bacteria affect body growth and development during the juvenile period in Drosophila. Germ-free (GF) animals can be easily created using the Drosophila model system and numerous tools are available for temporal expression, down-regulation, and tissue-specific gene expression. Moreover, Drosophila dynamically exhibits changes in body size and developmental metamorphosis during its short life cycle, thereby facilitating quick and easy observation of bacteria-dependent effects.
Lactiplantibacillus species
Lactiplantibacillus species (formerly known as Lactobacillus) are found in a wide range of ecological niches including the mucosal surfaces of several animals. Although the probiotic effects of diverse Lactiplantibacillus species have been explored in detail, there is a limited understanding of the processes through which Lactiplantibacillus species alter animal physiology (Oelschlaeger Citation2010). Members of the genus Lactiplantibacillus, including Lactiplantibacillus plantarum, Lactiplantibacillus fructivorans, and Lactiplantibacillus brevis are dominant in the Drosophila gut and are gram-positive, rod-shaped, lactic acid-producing microaerophilic bacteria from the Firmicutes phylum. Among these, L. plantarumWJL (hereinafter referred to as Lp) was reported to promote Drosophila larval development (Shin et al. Citation2011; Storelli et al. Citation2011; Lee J et al. Citation2020). Under nutritional scarcity, GF larvae showed developmental delays and lower body growth rates than CR larvae, which was reversed when GF larvae were mono-associated with Lp (Storelli et al. Citation2011). The body growth and development-promoting effects of Lp were lost when the larvae were fed a nutrient-rich diet but were noticeable when the host had nutritional deficits. Further, inactivation of the amino acid transporter Slimfast in the larval fat body and PG was sufficient to disrupt Lp-induced body growth and juvenile development, respectively (Storelli et al. Citation2011). Therefore, Lp is hypothesized to promote both body growth and juvenile development by providing the host with essential amino acids for proper TOR activity ().
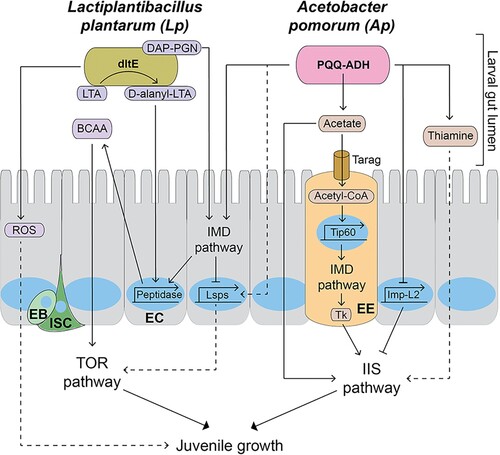
Figure 2. The molecular pathways mediating the effects of Lactiplantibacillus plantarum (Lp) and Acetobacter pomorum (Ap) on Drosophila juvenile development and growth. The IMD pathway recognizes the diaminopimelic acid-type peptidoglycan in cell walls of Lp and Ap, and promotes peptidase gene transcription in enterocytes (ECs). D-alanylated-lipoteichoic acid in Lp is modified from lipoteichoic acid by dltE and induces peptidase transcription in ECs, independently of the IMD pathway. The pyrroquinoline quinone-dependent alcohol dehydrogenase enzyme in Ap produces acetate, which stimulates the IIS pathway, promoting body growth and development. Acetate in the gut lumen enters enteroendocrine cells (EEs) via a monocarboxylic acid transporter called Tarag and is converted into acetyl-CoA. Acetyl-CoA pools stimulate the Tip60 histone acetylase complex, leading to an increase in the IMD pathway at the transcriptional level. The IMD pathway in EE activates the systemic IIS pathway by increasing the expression of Tk. Ap promotes the expression of larval serum proteins (Lsps) the IMD pathway in ECs, which is suppressed by IMD pathway. Lsps in the hemolymph are likely up taken in nutrition-sensing tissues such as fat bodies by endocytosis and stimulate the TOR pathway and larval development. Ap suppresses the expression of Imp-L2 via ecdysone signaling in ECs, which in turn stimulates the IIS pathway. Thiamine secreted from Ap can regulate larval development; however, it’s the underlying mechanism is not clear. DAP-PGN: diaminopimelic acid-type peptidoglycan. Nucleus is described as a blue circle or an oval. Dotted lines indicate putative pathways. LTA: Lipoteichoic acid. D-alanyl-LTA: D-alanylated-lipoteichoic acid. ROS: Reactive oxygen species. BCAA: Branched-chain amino acids. Lsps: Larval serum proteins. PQQ-ADH: Pyrroquinoline quinone-dependent alcohol dehydrogenase. Tk: Tachykinin. EC: Enterocyte. EE: Enteroendocrine cell. EB: Enteroblast. ISC: Intestinal stem cell.
Upon microbial infection, Drosophila rapidly induces the expression of a series of antimicrobial peptides (AMPs) including diptericins, drosomycin, and attacins. Induction of AMPs is primarily controlled by the Drosophila homolog of the Nuclear factor kappa B transcription factor. In flies, two different pathways respond to microbial infections: the Toll and IMD pathways. In general, the Toll pathway recognizes Gram-positive bacteria with lysine-type peptidoglycan in the cell wall, while the IMD pathway recognizes Gram-negative and Gram-positive bacteria with diaminopimelic acid (DAP)-type peptidoglycan (Leulier et al. Citation2003; Buchon et al. Citation2014). Over-activation of the IMD pathway during the juvenile growth period results in smaller final size, developmental delay, and disruption of metabolic homeostasis via decreased IIS/TOR signaling, but the mechanism underlying the influence of gut commensal bacteria on IMD pathway-mediated modulation of larval development remains unknown (Davoodi et al. Citation2019). By comparative transcriptomic analysis of GF flies and CR flies, it was reported that multiple peptidase genes are significantly elevated in the presence of gut commensal bacteria, and their expression was partly dependent on the IMD pathway (Erkosar et al. Citation2014). Based on this finding, the authors hypothesized that Lp, which has DAP-type peptidoglycan, regulates peptidase expression through the IMD pathway. Mono-association of Lp was sufficient to induce an overall increase in midgut-enriched peptidases, and ectopic overexpression of Jon66Cii peptidases in the ECs of GF larvae was sufficient to promote larval body growth (Erkosar et al. Citation2015). Consistent with previous observations, the loss of the IMD pathway partially abolished Lp-induced peptidase expression and growth promotion. Interestingly, D-alanylated teichoic acids (D-Ala-TAs) in the cell wall of Lp promote body growth and juvenile development via gut peptidase expression during chronic nutrition deficiency, and their effect is independent of the IMD pathway (Matos et al. Citation2017). Teichoic acids (TAs) are anionic polymers found within the cell walls of Gram-positive bacteria, contributing up to 50% of the cell envelope weight, and are present in two forms: wall TAs and lipoteichoic acids (LTAs). Encoded-Dlt genes from pbpX2-Dlt operon in other Lactiplantibacillus species are involved in bacterial cell wall biogenesis, and the inactivation of Dlt genes causes a significant reduction in the esterification of TAs by D-alanine. Deletion of the entire Dlt operon in the LpNC8 strain (Lactiplantibacillus plantarumNC8) leads to a reduction in the D-alanine esterification of TAs, which slightly increases gut peptidase expression and larval growth. Recently, only D-Ala-LTAs produced by Dlt genes have been shown to be direct triggers that promote intestinal peptidase expression and body growth (Nikolopoulos et al. Citation2023). This suggests that the detection of specific cell envelope structures of commensal bacteria is critical for Drosophila to benefit from their interactions with symbiotic microorganisms.
Reactive oxygen species (ROS) were initially thought to be harmful to cell integrity, being the underlying cause of numerous diseases and aging. However, ROS plays an important roles in maintaining proper physiology and homeostasis in insects and humans (Lopez-Otin et al. Citation2013; Liu et al. Citation2018; Sinenko et al. Citation2021). The gut microbiome affects many aspects of host physiology, such as intestinal cell proliferation and pathogen resistance, by producing ROS (Ballard and Towarnicki Citation2020). Specifically, Lp stimulates Drosophila NADPH oxidase-dependent ROS production in ECs and subsequently activates Nrf2-responsive cytoprotective genes (Jones et al. Citation2013, Citation2015). In addition to contributing to host nutrition, Lp-induced ROS effectively inhibits phenotypic variation in host developmental traits under low-nutrient stress (Ma et al. Citation2019). Consistently, Lp-associated larvae had fewer developmental trait abnormalities than GF larvae, including smaller larval body length and delayed developmental time. The mechanism underlying this buffering effect is still unclear; however, the antioxidant N-acetylcysteine compromises the buffering capacity of Lp (Ma et al. Citation2019).
Acetobacter species
Drosophila is often referred to as ‘vinegar flies,’ most likely because they are constantly present near vinegar manufacturers. Drosophila has been shown to play a key role in acetic acid synthesis through its function as a vector for microorganisms required for acetic acid production. Members of the genus Acetobacter, including Acetobacter pomorum, Acetobacter pasteurianus, Acetobacter tropicalis, and Acetobacter aceti are dominant in the gut microbiota of Drosophila and are Gram-negative acetic acid-producing bacteria (Ren et al. Citation2007; Ryu et al. Citation2008; Wong CNA et al. Citation2011). Shin et al. found A. pomorum (hereinafter referred to as Ap) modulates host homeostatic programs that control the developmental rate and final body size by regulating the IIS pathway. Genetic investigations have revealed that the pyrroquinoline quinone-dependent alcohol dehydrogenase (PQQ-ADH) enzyme in Ap, which catalyzes the oxidation of alcohols into acetic acid, plays a crucial role in the modulatory effect of Ap on the host IIS pathway (Yakushi and Matsushita Citation2010; Shin et al. Citation2011). However, extensive research has not yet been conducted on the underlying mechanisms through which AP-derived acetic acid regulates insulin signaling.
Acetate, a primary short-chain fatty acid, is secreted by gut bacteria, including Acetobacter species. Acetate deficiency in the gut interferes with the maintenance of several physiological functions, including body weight control (Hernandez et al. Citation2019; Fan and Pedersen Citation2021). Watnick et al. proposed a mechanism by which acetate produced by bacteria, including Ap, regulates host insulin signaling. Initially, they found that Vibrio cholerae, a pathogen that causes the diarrheal illness cholera in humans, affects host insulin signaling and lipid metabolism via acetate depletion through the acetate-switch process in the Drosophila gut (Hang et al. Citation2014). Subsequent studies have shown that microbial-derived acetate activates host insulin signaling by modulating the secretion of tachykinin through the IMD pathway in EEs in the gut (Kamareddine et al. Citation2018). Tachykinin is a gut endocrine peptide hormone produced by EEs that regulates lipogenesis in ECs via the PKA/SREBP signaling pathway (Song et al. Citation2014). Mono-association of Ap and supplementation with dietary acetate activates the IMD pathway and restores body growth and puparium morphogenesis in GF larvae by elevating IIS signaling, but these effects were abolished in the EE-specific IMD pathway knockdown mutant. Acetate generated by gut bacteria enters EEs through the monocarboxylic acid transporter Tarag, and is converted into acetyl-CoA by acetyl-CoA synthase. Tip60 histone acetyltransferase activates EcR signaling by acetylating H2Av using intracellular Acetyl-CoA as a substrate, thereby activating the IMD pathway (Jugder et al. Citation2021).
In line with this, our group recently reported that Ap can modulate larval development independently of acetate derived from microbes. Mono-association of Ap increases systemic IIS activity by suppressing Imp-L2 production in ECs in the larval gut (Lee J et al. Citation2022). Increased production of Imp-L2 in ECs was followed by a decrease in the activity of the PI3K–Akt axis in both the larval gut and fat bodies of GF larvae; however, these effects were restored by mono-association with Ap in GF larvae. Neither the mono-association of the PQQ-ADH mutant nor acetate treatment altered Imp-L2 production. Ap modulates ecdysone signaling-dependent Imp-L2 production from ECs, as indicated by the observation that inactivation of EcR prevents the decrease in Imp-L2 production noted in mono-association of Ap. Additionally, our group reported that Ap can regulate the rate of fly larval development by modulating the expression of Larval serum proteins (Lsps) (Lee J et al. Citation2023). During the larval stage, Lsps provide a source of amino acids, allowing insects to efficiently store excess nutrients in the hemolymph (Roberts et al. Citation1977; Powell et al. Citation1984; Telfer and Kunkel Citation1991; Burmester Citation1999; Short et al. Citation2020). We found that mono-association of Ap increased the expression of Lsps in Drosophila larval gut, which is suppressed by IMD immune pathway. Knockdown of Lsps in gut enterocytes delayed larval developmental rate, which became severe when Ap was mono-associated. Thus, multiple pathways, including IMD, appear to regulate Ap's impact on larval developmental rate in opposing directions (Lee J et al. Citation2023).
Vitamins are essential micronutrients that function as precursors to various coenzymes required for important biochemical reactions in all living cells. It has been hypothesized that the gut microbiota provides the host with various types of vitamin B, such as thiamine, riboflavin, and folate (Hill Citation1997; LeBlanc et al. Citation2013). Thiamine is an essential cofactor for several metabolic pathways. Specifically, thiamine pyrophosphate is a coenzyme involved in various enzymatic activities associated with cell regulatory processes and biosynthesis (Rindi and Laforenza Citation2000). A recent study reported that the gut microbiota of Drosophila, particularly Ap, produces thiamine, which is involved in juvenile development of the host (Sannino et al. Citation2018). The authors found that although GF larvae rarely develop on a thiamine-free diet, dietary thiamine promotes the larval development of Drosophila in a dose-dependent manner. Further, they found that Ap could restore the developmental failure of GF larvae by producing thiamine. Although it is unknown how thiamine affects developmental processes in Drosophila during the juvenile stage, a few studies have suggested that thiamine regulates the IIS pathway. Thiamine deficiency significantly inhibits insulin production and secretion in vertebrates. High levels of thiamine are stored in human pancreatic beta cells and islets, and its absorption is carrier-mediated and adaptively controlled by the prevailing vitamin levels via transcriptional processes (Rathanaswami et al. Citation1991; Mee et al. Citation2009). Consistently, rats fed a thiamine-deficient diet had lower blood insulin levels and transmembrane glucose transport (Debski et al. Citation2011). High-dose thiamine supplementation may relieve clinical signs of the condition, and may reduce or ablate the requirement for exogenous insulin (Olsen et al. Citation2007). These findings indicate that metabolites, such as acetate and thiamine, secreted by Ap regulate body growth and developmental processes of the host by activating various signaling pathways.
Enterococcus species
Enterococci are lactic acid-producing Gram-positive bacteria (Hanchi et al. Citation2018) that reside in the gastrointestinal tract of a wide range of mammals (Mundt Citation1963) and insects (Steinhaus Citation1941; Martin and Mundt Citation1972). A small proportion of Enterococcus species inhabit the Drosophila gut (Han et al. Citation2017). The most prevalent of these are Enterococcus faecalis, followed by Enterococcus durans, Enterococcus faecium, and Enterococcus gallinarum, which together account for less than 10% of the overall enterococcal population (Cox and Gilmore Citation2007). Although Enterococcus are known as pathogenic bacteria that cause hospital-acquired infections, and infection with E. faecalis reduces adult Drosophila survival (Huycke et al. Citation1998; Cabrera et al. Citation2023), a few species have also been utilized as probiotics (Huycke et al. Citation1998; Edwards Citation2000; Hanchi et al. Citation2018). Little is known about the effects of colonized Enterococci in the gut on juvenile development of other insects. Inoculation of E. mundtii and E. gallinarum in fall armyworm promoted body growth compared with GF larvae. Colonization of E. faecium in the gut of honey bees (Apis mellifera) increases midgut and hindgut weight and transcriptome analysis and microRNA profiling showed E.faecium in the gut influences host developmental genes including TOR pathway (Du et al. Citation2021). However, E. faecalis did not directly affect the body growth and pupation time of Bombyx mori (Chen et al. Citation2022; Zhang et al. Citation2022). These findings suggest that most enterococcus species have distinct functions, and the effect changes depending on the type of host. Thus, it is necessary to further investigate the characteristics of each Enterococcus species as well as their effects on host physiology, including body growth.
Effects of microbial metabolites produced from gut microbiome on host growth and gut microbial community
The gut microbiome connects the hosts to themselves through a network of nutrition and metabolism, allowing these interactions to directly affect host nutrition and metabolism. It is believed that an imbalance in gut microbiome composition is involved in the pathogenesis of a variety of diseases, such as cancers, metabolic disorders, and developmental disabilities, and that the gut microbiome composition that is favorable to the host or functions as a pathogen will exist separately (Gagniere et al. Citation2016; Gomaa Citation2020; Fan and Pedersen Citation2021). Microbial metabolites are considered major environmental factors that determine the genetically encoded growth potential of organisms and enable them to sustain distinct gut microbiota communities (Sommer and Backhed Citation2013; Moya and Ferrer Citation2016; Krautkramer et al. Citation2021). Malnourished children have an immature gut microbiome, and the gut microbiome compositions of healthy and growth-stunted twins are clearly different (Smith et al. Citation2013). When the fecal microbiome of the healthy or stunted twin is transplanted into genetically identical GF mice fed a poor diet, recipients of the microbiome from stunted twins exhibit lower growth and body weight recovery. Even the immature gut microbiome in severely malnourished children interferes with the restoration of healthy growth through classical re-nurturing treatments (Subramanian et al. Citation2014). Previous studies have proposed the following: (1) there may be an optimal gut microbiome composition that supports body growth; (2) microbial metabolites produced in the gut microbiome are used as nutrients for body growth and may be difficult to replace; (3) microbial metabolites play key roles in maintaining or modifying the composition of the gut microbiome. Therefore, understanding the types and mechanisms of microbial metabolites produced in the gut microbiome will broaden insights into host–gut microbiota interactions.
The Drosophila model system can be used to analyze the contribution of gut commensal bacteria to organismal body development and their composition in the gut under strictly controlled nutritional conditions using a holidic diet (HD) with a chemically specified fly medium. It was reported that GF larvae are auxotrophic for ten essential amino acids, one non-essential amino acid (Asn), six types of vitamin B (1, 2, 5, 6, 7, and 9), vitamin E, cholesterol, choline, Mg ions, and Zn ions during the juvenile phase, and mono-association with Lp or Ap differentially allows GF larvae to develop normally (Consuegra, Grenier, Baa-Puyoulet, et al. Citation2020). Except for in liquid HD without a few traces, Ap grew well in other conditions; however, LpNC8 did not grow in an environment lacking the most necessary amino acids (Arg, Ile, Leu, Met, Phe, Thr, and Val), several non-essential amino acids (Ala, Asp, and Cys), biotin, and pantothenate. Given that Ap and Lp are prevalent in the fly gut microbiome independent of changes in the majority of the internal/external environment, it is speculated that Ap and Lp aid each other's growth by supplying essential nutrients, and that their metabolic cooperation enhances the development of the host. A subsequent study found that the bi-association of Ap and LpNC8 not only increased body growth during larval development but also enhanced bacterial growth in the gut through exchange of microbial metabolites (Consuegra, Grenier, Akherraz, et al. Citation2020). During nutrient scarcity, Ap helps the growth of LpNC8 by supplying essential amino acids and vitamins. Conversely, LpNC8 secretes lactate, which promotes the production of amino acids by Ap. This metabolic cooperation enhances the promotive effects of body growth; however, the amino acids generated by Ap using lactate had no direct effect on larval body growth. Together, these findings suggest that microbial metabolites from gut commensal bacteria act as essential nutrients for the developmental processes of the host and for bacterial growth in the gut.
Concluding remarks
The juvenile stage is a key phase in most metazoans that determines the final adult size, as well as defines the quality of life and ability to survive in the wild. Many environmental factors, including nutritional conditions, affect body development during the juvenile stage. The IIS/TOR pathway is a core effector of body growth and developmental maturation, and is regulated by nutritional availability. Over the past decade, it has been highlighted that the relationship between host nutrition and gut microbiome is crucial for maintaining host physiology. The physiological systems of animals, including tissue structure/function and developmental processes, are well conserved in the Drosophila model system. Furthermore, Drosophila is recognized as an excellent model for studying the interactions between gut commensal bacteria and the host because it has a relatively simple gut microbiota complex and can be readily sterilized and colonized by specific bacterial species. Lactiplantibacillus and Acetobacter species are major constituents of the fly gut microbiome, and both Lp and Ap contribute to body growth and developmental maturation of the host by primarily regulating the IIS/TOR pathway under nutrient-deficient conditions. In short, gut commensal bacteria affect host physiology in three main ways: (1) through recognition of the bacterial cell wall structure via immune pathways; (2) production of bacteria-derived metabolites that confer beneficial effects to the host and/or other gut commensal bacteria; and (3) modulation of the nutritional availability and other physiological pathways in the gut. In particular, recent studies have extensively explored the types and formation principles of gut microbiome-derived or gut microbiome-modified metabolites, and have challenged us to expand our understanding of the functions and mechanisms underlying metabolic activities in host–gut microbiome interactions (Agus et al. Citation2021; Krautkramer et al. Citation2021). Future elucidation of the mechanisms underlying the regulation of body growth and microbial metabolites during the juvenile phase in Drosophila could provide new insights into the pathogenesis of various growth disorders, hormonal imbalances, and metabolic dysfunctions.
Acknowledgments
This research was supported by the Chung-Ang University Graduate Research Scholarship in 2016. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (Grant Number: 2023R1A2C1005908).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Agus A, Clement K, Sokol H. 2021. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. 70(6):1174–1182. doi:10.1136/gutjnl-2020-323071.
- Alic N, Hoddinott MP, Vinti G, Partridge L. 2011. Lifespan extension by increased expression of the the IGFBP7 tumour suppressor. Aging Cell. 10(1):137–147. doi:10.1111/j.1474-9726.2010.00653.x.
- Apidianakis Y, Rahme LG. 2011. Drosophila melanogaster as a model for human intestinal infection and pathology. Dis Model Mech. 4(1):21–30. doi:10.1242/dmm.003970.
- Arden KC. 2008. FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene. 27(16):2345–2350. doi:10.1038/onc.2008.27.
- Auger KR, Serunian LA, Soltoff SP, Libby P, Cantley LC. 1989. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 57(1):167–175. doi:10.1016/0092-8674(89)90182-7.
- Ballard JWO, Towarnicki SG. 2020. Mitochondria, the gut microbiome and ROS. Cell Signal. 75:109737. doi:10.1016/j.cellsig.2020.109737.
- Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. 2002. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2(2):239–249. doi:10.1016/S1534-5807(02)00117-X.
- Buchon N, Silverman N, Cherry S. 2014. Immunity in Drosophila melanogaster – from microbial recognition to whole-organism physiology. Nat Rev Immunol. 14(12):796–810. doi:10.1038/nri3763.
- Burmester T. 1999. Evolution and function of the insect hexamerins. Eur J Entomol. 96(3):213–225.
- Cabrera K, Hoard DS, Gibson O, Martinez DI, Wunderlich Z. 2023. Drosophila immune priming to Enterococcus faecalis relies on immune tolerance rather than resistance. PLoS Pathog. 19(8):e1011567. doi:10.1371/journal.ppat.1011567.
- Calcaterra V, Verduci E, Magenes VC, Pascuzzi MC, Rossi V, Sangiorgio A, Bosetti A, Zuccotti G, Mameli C. 2021. The role of pediatric nutrition as a modifiable risk factor for precocious puberty. Life (Basel). 11(12).
- Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. 2011. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet. 7(9):e1002272. doi:10.1371/journal.pgen.1002272.
- Chaston JM, Newell PD, Douglas AE. 2014. Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. mBio. 5(5):e01631–e01614. doi:10.1128/mBio.01631-14.
- Chen B, Mason CJ, Peiffer M, Zhang D, Shao Y, Felton GW. 2022. Enterococcal symbionts of caterpillars facilitate the utilization of a suboptimal diet. J Insect Physiol. 138:104369. doi:10.1016/j.jinsphys.2022.104369.
- Chiang MH, Ho SM, Wu HY, Lin YC, Tsai WH, Wu T, Lai CH, Wu CL. 2022. Drosophila model for studying Gut microbiota in behaviors and neurodegenerative diseases. Biomedicines. 10(3):596. doi:10.3390/biomedicines10030596.
- Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, Carre C, Noselli S, Leopold P. 2005. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 310(5748):667–670. doi:10.1126/science.1119432.
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. 2003. A nutrient sensor mechanism controls Drosophila growth. Cell. 114(6):739–749. doi:10.1016/S0092-8674(03)00713-X.
- Consuegra J, Grenier T, Akherraz H, Rahioui I, Gervais H, da Silva P, Leulier F. 2020. Metabolic cooperation among commensal bacteria supports Drosophila juvenile growth under nutritional stress. Iscience. 23(6):101232. doi:10.1016/j.isci.2020.101232.
- Consuegra J, Grenier T, Baa-Puyoulet P, Rahioui I, Akherraz H, Gervais H, Parisot N, da Silva P, Charles H, Calevro F, et al. 2020. Drosophila-associated bacteria differentially shape the nutritional requirements of their host during juvenile growth. PLoS Biol. 18(3):e3000681. doi:10.1371/journal.pbio.3000681.
- Cox CR, Gilmore MS. 2007. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun. 75(4):1565–1576. doi:10.1128/IAI.01496-06.
- Davoodi S, Galenza A, Panteluk A, Deshpande R, Ferguson M, Grewa S, Foley E. 2019. The immune deficiency pathway regulates metabolic homeostasis in Drosophila. J Immunol. 202(9):2747–2759. doi:10.4049/jimmunol.1801632.
- Debski B, Kuryl T, Gralak MA, Pierzynowska J, Drywien M. 2011. Effect of inulin and oligofructose enrichment of the diet on rats suffering thiamine deficiency. J Anim Physiol Anim Nutr. 95(3):335–342. doi:10.1111/j.1439-0396.2010.01059.x.
- Donaldson GP, Lee SM, Mazmanian SK. 2016. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 14(1):20–32. doi:10.1038/nrmicro3552.
- Du Y, Luo S, Zhou X. 2021. Enterococcus faecium regulates honey Bee developmental genes. Int J Mol Sci. 22(22).
- Edwards DD. 2000. Enterococci attract attention of concerned microbiologists (vol 66, pg 540, 2000). Asm News. 66(11):653–653.
- Erkosar B, Defaye A, Bozonnet N, Puthier D, Royet J, Leulier F. 2014. Drosophila microbiota modulates host metabolic gene expression via IMD/NF-κB signaling. PLoS One. 9(4):e94729. doi:10.1371/journal.pone.0094729.
- Erkosar B, Storelli G, Mitchell M, Bozonnet L, Bozonnet N, Leulier F. 2015. Pathogen virulence impedes mutualist-mediated enhancement of host juvenile growth via inhibition of protein digestion. Cell Host Microbe. 18(4):445–455. doi:10.1016/j.chom.2015.09.001.
- Fan Y, Pedersen O. 2021. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 19(1):55–71. doi:10.1038/s41579-020-0433-9.
- Fink C, Staubach F, Kuenzel S, Baines JF, Roeder T. 2013. Noninvasive analysis of microbiome dynamics in the fruit fly Drosophila melanogaster. Appl Environ Microbiol. 79(22):6984–6988. doi:10.1128/AEM.01903-13.
- Gagniere J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, Bringer MA, Pezet D, Bonnet M. 2016. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 22(2):501–518. doi:10.3748/wjg.v22.i2.501.
- Gomaa EZ. 2020. Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek. 113(12):2019–2040. doi:10.1007/s10482-020-01474-7.
- Han G, Lee HJ, Jeong SE, Jeon CO, Hyun S. 2017. Comparative analysis of Drosophila melanogaster Gut microbiota with respect to host strain, sex, and age. Microb Ecol. 74(1):207–216. doi:10.1007/s00248-016-0925-3.
- Hanchi H, Mottawea W, Sebei K, Hammami R. 2018. The genus enterococcus: between probiotic potential and safety concerns-An update. Front Microbiol. 9:1791. doi:10.3389/fmicb.2018.01791.
- Hang S, Purdy AE, Robins WP, Wang Z, Mandal M, Chang S, Mekalanos JJ, Watnick PI. 2014. The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe. 16(5):592–604. doi:10.1016/j.chom.2014.10.006.
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 110(2):177–189. doi:10.1016/S0092-8674(02)00833-4.
- Hernandez MAG, Canfora EE, Jocken JWE, Blaak EE. 2019. The short-chain fatty acid acetate in body weight control and insulin sensitivity. Nutrients. 11(8).
- Hietakangas V, Cohen SM. 2007. Re-evaluating AKT regulation: role of TOR complex 2 in tissue growth. Genes Dev. 21(6):632–637. doi:10.1101/gad.416307.
- Hill MJ. 1997. Intestinal flora and endogenous vitamin synthesis. Eur J Cancer Prev. 6:S43–S45. doi:10.1097/00008469-199703001-00009.
- Honegger B, Galic M, Kohler K, Wittwer F, Brogiolo W, Hafen E, Stocker H. 2008. Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J Biol. 7(3):10. doi:10.1186/jbiol72.
- Huang JH, Douglas AE. 2015. Consumption of dietary sugar by gut bacteria determines Drosophila lipid content. Biol Lett. 11(9):20150469. doi:10.1098/rsbl.2015.0469.
- Huycke MM, Sahm DF, Gilmore MS. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 4(2):239–249. doi:10.3201/eid0402.980211.
- Hyun S. 2013. Body size regulation and insulin-like growth factor signaling. Cell Mol Life Sci. 70(13):2351–2365. doi:10.1007/s00018-013-1313-5.
- Hyun S. 2018. Body size regulation by maturation steroid hormones: a Drosophila perspective. Front Zool. 15. doi:10.1186/s12983-018-0290-9.
- Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. 2009. Conserved MicroRNA miR-8/miR-200 and Its target USH/FOG2 control growth by regulating PI3K. Cell. 139(6):1096–1108. doi:10.1016/j.cell.2009.11.020.
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 6(11):1122–1128. doi:10.1038/ncb1183.
- Jevtov I, Zacharogianni M, van Oorschot MM, van Zadelhoff G, Aguilera-Gomez A, Vuillez I, Braakman I, Hafen E, Stocker H, Rabouille C. 2015. TORC2 mediates the heat stress response in Drosophila by promoting the formation of stress granules. J Cell Sci. 128(14):2497–2508.
- Jin H, Kim VN, Hyun S. 2012. Conserved microRNA miR-8 controls body size in response to steroid signaling in Drosophila. Genes Dev. 26(13):1427–1432. doi:10.1101/gad.192872.112.
- Jones RM, Desai C, Darby TM, Luo L, Wolfarth AA, Scharer CD, Ardita CS, Reedy AR, Keebaugh ES, Neish AS. 2015. Lactobacilli modulate epithelial cytoprotection through the Nrf2 pathway. Cell Rep. 12(8):1217–1225. doi:10.1016/j.celrep.2015.07.042.
- Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA, et al. 2013. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 32(23):3017–3028. doi:10.1038/emboj.2013.224.
- Jugder BE, Kamareddine L, Watnick PI. 2021. Microbiota-derived acetate activates intestinal innate immunity via the Tip60 histone acetyltransferase complex. Immunity. 54(8):1683–1697.e3 e1683. doi:10.1016/j.immuni.2021.05.017.
- Kamareddine L, Robins WP, Berkey CD, Mekalanos JJ, Watnick PI. 2018. The Drosophila immune deficiency pathway modulates enteroendocrine function and host metabolism. Cell Metab. 28(3):449–462.e5 e445. doi:10.1016/j.cmet.2018.05.026.
- Kramer JM, Davidge JT, Lockyer JM, Staveley BE. 2003. Expressionof Drosophila FOXO regulates growth and can phenocopy starvation. BMC Dev Biol. 3:5. doi:10.1186/1471-213X-3-5.
- Krautkramer KA, Fan J, Backhed F. 2021. Gut microbial metabolites as multi-kingdom intermediates. Nat Rev Microbiol. 19(2):77–94. doi:10.1038/s41579-020-0438-4.
- Layalle S, Arquier N, Leopold P. 2008. The TOR pathway couples nutrition and developmental timing in Drosophila. Dev Cell. 15(4):568–577. doi:10.1016/j.devcel.2008.08.003.
- LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. 2013. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 24(2):160–168. doi:10.1016/j.copbio.2012.08.005.
- Lee GJ, Han G, Yun HM, Lim JJ, Noh S, Lee J, Hyun S. 2018. Steroid signaling mediates nutritional regulation of juvenile body growth via IGF-binding protein in Drosophila. Proc Natl Acad Sci USA. 115(23):5992–5997. doi:10.1073/pnas.1718834115.
- Lee J, Han G, Kim JW, Jeon CO, Hyun S. 2020. Taxon-specific effects of lactobacillus on Drosophila host development. Microb Ecol. 79(1):241–251. doi:10.1007/s00248-019-01404-9.
- Lee J, Song X, Hyun B, Jeon CO, Hyun S. 2023. Drosophila gut immune pathway suppresses host development-promoting effects of acetic acid bacteria. Mol Cells. 46(10):637–653. doi:10.14348/molcells.2023.0141.
- Lee J, Yun HM, Han G, Lee GJ, Jeon CO, Hyun S. 2022. A bacteria-regulated gut peptide determines host dependence on specific bacteria to support host juvenile development and survival. BMC Biol. 20(1):258. doi:10.1186/s12915-022-01458-1.
- Lee JE, Lim YH, Kim JH. 2021. UCH-L1 and UCH-L3 regulate the cancer stem cell-like properties through PI3 K/Akt signaling pathway in prostate cancer cells. Animal Cells Syst (Seoul). 25(5):312–322. doi:10.1080/19768354.2021.1987320.
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. 1999. Crystal structure of the PTEN tumor suppressor. Cell. 99(3):323–334. doi:10.1016/S0092-8674(00)81663-3.
- Lee SH, Kim EY. 2021. Short-term maintenance on a high-sucrose diet alleviates aging-induced sleep fragmentation in Drosophila. Animal Cells Syst (Seoul). 25(6):377–386. doi:10.1080/19768354.2021.1997801.
- Leopold P, Perrimon N. 2007. Drosophila and the genetics of the internal milieu. Nature. 450(7167):186–188. doi:10.1038/nature06286.
- Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, Mengin-Lecreulx D, Lemaitre B. 2003. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 4(5):478–484. doi:10.1038/ni922.
- Liu Z, Ren Z, Zhang J, Chuang CC, Kandaswamy E, Zhou T, Zuo L. 2018. Role of ROS and nutritional antioxidants in human diseases. Front Physiol. 9:477. doi:10.3389/fphys.2018.00477.
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell. 153(6):1194–1217. doi:10.1016/j.cell.2013.05.039.
- Ma D, Bou-Sleiman M, Joncour P, Indelicato CE, Frochaux M, Braman V, Litovchenko M, Storelli G, Deplancke B, Leulier F. 2019. Commensal gut bacteria buffer the impact of host genetic variants on Drosophila developmental traits under nutritional stress. iScience. 19:436–447. doi:10.1016/j.isci.2019.07.048.
- Martin JD, Mundt JO. 1972. Enterococci in insects. Appl Microbiol. 24(4):575–580. doi:10.1128/am.24.4.575-580.1972.
- Matos RC, Schwarzer M, Gervais H, Courtin P, Joncour P, Gillet B, Ma D, Bulteau AL, Martino ME, Hughes S, et al. 2017. D-Alanylation of teichoic acids contributes to Lactobacillus plantarum-mediated Drosophila growth during chronic undernutrition. Nat Microbiol. 2(12):1635–1647. doi:10.1038/s41564-017-0038-x.
- Mee L, Nabokina SM, Sekar VT, Subramanian VS, Maedler K, Said HM. 2009. Pancreatic beta cells and islets take up thiamin by a regulated carrier-mediated process: studies using mice and human pancreatic preparations. Am J Physiol-Gastr L. 297(1):G197–G206.
- Miguel-Aliaga I, Jasper H, Lemaitre B. 2018. Anatomy and physiology of the digestive tract of Drosophila melanogaster. Genetics. 210(2):357–396. doi:10.1534/genetics.118.300224.
- Mora A, Komander D, van Aalten DM, Alessi DR. 2004. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol. 15(2):161–170. doi:10.1016/j.semcdb.2003.12.022.
- Moya A, Ferrer M. 2016. Functional redundancy-induced stability of Gut microbiota subjected to disturbance. Trends Microbiol. 24(5):402–413. doi:10.1016/j.tim.2016.02.002.
- Mundt JO. 1963. Occurrence of enterococci in animals in a wild environment. Appl Microbiol. 11(2):136–140. doi:10.1128/am.11.2.136-140.1963.
- Nikolopoulos N, Matos RC, Ravaud S, Courtin P, Akherraz H, Palussiere S, Gueguen-Chaignon V, Salomon-Mallet M, Guillot A, Guerardel Y, et al. 2023. Structure-function analysis of Lactiplantibacillus plantarum DltE reveals D-alanylated lipoteichoic acids as direct cues supporting Drosophila juvenile growth. Elife. 12. doi:10.7554/eLife.84669.
- Oelschlaeger TA. 2010. Mechanisms of probiotic actions – a review. Int J Med Microbiol. 300(1):57–62. doi:10.1016/j.ijmm.2009.08.005.
- Oldham S, Hafen E. 2003. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 13(2):79–85. doi:10.1016/S0962-8924(02)00042-9.
- Olsen BS, Hahnemann JMD, Schwartz M, Ostergaard E. 2007. Thiamine-responsive megaloblastic anaemia: a cause of syndromic diabetes in childhood. Pediatr Diabetes. 8(4):239–241. doi:10.1111/j.1399-5448.2007.00251.x.
- Owusu-Ansah E, Perrimon N. 2014. Modeling metabolic homeostasis and nutrient sensing in Drosophila: implications for aging and metabolic diseases. Dis Model Mech. 7(3):343–350. doi:10.1242/dmm.012989.
- Pereira FC, Berry D. 2017. Microbial nutrient niches in the gut. Environ Microbiol. 19(4):1366–1378. doi:10.1111/1462-2920.13659.
- Powell D, Sato JD, Brock HW, Roberts DB. 1984. Regulation of synthesis of the larval serum proteins of Drosophila melanogaster. Dev Biol. 102(1):206–215. doi:10.1016/0012-1606(84)90185-4.
- Puig O, Marr MT, Ruhf ML, Tjian R. 2003. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17(16):2006–2020. doi:10.1101/gad.1098703.
- Rathanaswami P, Pourany A, Sundaresan R. 1991. Effects of thiamine-deficiency on the secretion of insulin and the metabolism of glucose in isolated Rat pancreatic-islets. Biochem Int. 25(3):577–583.
- Ren C, Webster P, Finkel SE, Tower J. 2007. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 6(2):144–152. doi:10.1016/j.cmet.2007.06.006.
- Rindi G, Laforenza U. 2000. Thiamine intestinal transport and related issues: recent aspects. Proc Soc Exp Biol Med. 224(4):246–255. doi:10.1111/j.1525-1373.2000.22428.x.
- Roberts DB, Wolfe J, Akam ME. 1977. The developmental profiles of two major haemolymph proteins from Drosophila melanogaster. J Insect Physiol. 23(7):871–878. doi:10.1016/0022-1910(77)90013-0.
- Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. 2008. Innate immune homeostasis by the homeobox gene Caudal and commensal-gut mutualism in Drosophila. Science. 319(5864):777–782. doi:10.1126/science.1149357.
- Sannino DR, Dobson AJ, Edwards K, Angert ER, Buchon N. 2018. The Drosophila melanogaster gut microbiota provisions thiamine to its host. mBio. 9(2). doi:10.1128/mBio.00155-18.
- Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. 2011. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 334(6056):670–674. doi:10.1126/science.1212782.
- Short CA, Hatle JD, Hahn DA. 2020. Protein stores regulate when reproductive displays begin in the male Caribbean fruit Fly. Front Physiol. 11. doi:10.3389/fphys.2020.00991.
- Sinenko SA, Starkova TY, Kuzmin AA, Tomilin AN. 2021. Physiological signaling functions of reactive oxygen species in stem cells: from flies to Man. Front Cell Dev Biol. 9:714370. doi:10.3389/fcell.2021.714370.
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. 2013. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 339(6119):548–554. doi:10.1126/science.1229000.
- Sommer F, Backhed F. 2013. The gut microbiota – masters of host development and physiology. Nat Rev Microbiol. 11(4):227–238. doi:10.1038/nrmicro2974.
- Song W, Veenstra JA, Perrimon N. 2014. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 9(1):40–47. doi:10.1016/j.celrep.2014.08.060.
- Staubach F, Baines JF, Kunzel S, Bik EM, Petrov DA. 2013. Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural environment. PLoS One. 8(8):e70749. doi:10.1371/journal.pone.0070749.
- Steinhaus EA. 1941. A study of the bacteria associated with thirty species of insects. J Bacteriol. 42(6):757–790. doi:10.1128/jb.42.6.757-790.1941.
- Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. 2011. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14(3):403–414. doi:10.1016/j.cmet.2011.07.012.
- Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, et al. 2014. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 510(7505):417–421. doi:10.1038/nature13421.
- Tang J, Xue P, Huang X, Lin C, Liu S. 2022. Diet and nutrients intakes during infancy and childhood in relation to early puberty: a systematic review and meta-analysis. Nutrients. 14(23):5004. doi:10.3390/nu14235004.
- Tanner JM, Davies PS. 1985. Clinical longitudinal standards for height and height velocity for North American children. J Pediatr. 107(3):317–329. doi:10.1016/S0022-3476(85)80501-1.
- Telfer WH, Kunkel JG. 1991. The function and evolution of insect storage hexamers. Annu Rev Entomol. 36:205–228. doi:10.1146/annurev.en.36.010191.001225.
- Wang P, Hou SX. 2010. Regulation of intestinal stem cells in mammals and Drosophila. J Cell Physiol. 222(1):33–37. doi:10.1002/jcp.21928.
- Wong AC, Chaston JM, Douglas AE. 2013. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 7(10):1922–1932. doi:10.1038/ismej.2013.86.
- Wong CNA, Ng P, Douglas AE. 2011. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ Microbiol. 13(7):1889–1900. doi:10.1111/j.1462-2920.2011.02511.x.
- Wullschleger S, Loewith R, Hall MN. 2006. TOR signaling in growth and metabolism. Cell. 124(3):471–484. doi:10.1016/j.cell.2006.01.016.
- Yakushi T, Matsushita K. 2010. Alcohol dehydrogenase of acetic acid bacteria: structure, mode of action, and applications in biotechnology. Appl Microbiol Biotechnol. 86(5):1257–1265. doi:10.1007/s00253-010-2529-z.
- Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. 2013. mTOR kinase structure, mechanism and regulation. Nature. 497(7448):217–223. doi:10.1038/nature12122.
- Zhang X, Feng H, He J, Muhammad A, Zhang F, Lu X. 2022. Features and colonization strategies of Enterococcus faecalis in the Gut of Bombyx mori. Front Microbiol. 13:921330. doi:10.3389/fmicb.2022.921330.
