ABSTRACT
The CRISPR-Cas system stands out as a promising genome editing tool due to its cost-effectiveness and time efficiency compared to other methods. This system has tremendous potential for treating various diseases, including genetic disorders and cancer, and promotes therapeutic research for a wide range of genetic diseases. Additionally, the CRISPR-Cas system simplifies the generation of animal models, offering a more accessible alternative to traditional methods. The CRISPR-Cas9 system can be used to cleave target DNA strands that need to be corrected, causing double-strand breaks (DSBs). DNA with DSBs can then be recovered by the DNA repair pathway that the CRISPR-Cas9 system uses to edit target gene sequences. High cleavage efficiency of the CRISPR-Cas9 system is thus imperative for effective gene editing. Herein, we explore several factors affecting the cleavage efficiency of the CRISPR-Cas9 system. These factors include the GC content of the protospacer-adjacent motif (PAM) proximal and distal regions, single-guide RNA (sgRNA) properties, and chromatin state. These considerations contribute to the efficiency of genome editing.
Introduction
Genome editing is a technology used to mutate a target gene for curing various diseases via deletion, insertion, and substitution of short nucleotides (Doudna Citation2020). Given the rapid developments in genome editing, there is growing demand for more efficient and accurate genome editing tools (Doudna Citation2020). The CRISPR-Cas system has been in the spotlight in this field because of its more efficient approach to genetic disorders and gene therapy compared to conventional genome editing tools, such as zinc-finger nuclease (ZFN) and transcription-activator-like effector nuclease (TALEN) (Chen and Gao Citation2014; Gilles and Averof Citation2014; Feng et al. Citation2015; Jo et al. Citation2015; Eid and Mahfouz Citation2016; Ho et al. Citation2018; Janik et al. Citation2020; Saifaldeen et al. Citation2020; Seah et al. Citation2022).
The CRISPR-Cas system is derived from the adaptive immune systems of bacteria and archaea to resist invading nucleic acids (Mali et al. Citation2013; Ran et al. Citation2013; Hryhorowicz et al. Citation2017; Janik et al. Citation2020; Zhang Citation2021). Upon invasion by foreign nucleic acids, bacteria use a Cas nuclease to cut the invading DNA into smaller fragments called spacers and incorporate these fragments into the clustered regularly interspaced short palindromic repeats (CRISPR) locus (Zhang et al. Citation2014). These fragments are then used as the transcription templates to produce CRISPR RNA (crRNA) in response to the foreign nucleic acids, and the crRNA interact with the complementary DNA sequences of the invading nucleic acids (Barrangou et al. Citation2007; Zhang et al. Citation2014). Cas nucleases cleave the DNA sequences interacting with the crRNA, causing immune effects through loss of function in the genes of the invading nucleic acids.
The CRISPR-Cas system is composed of guide RNA that specifically bind to the target DNA and the Cas nuclease (Kim et al. Citation2023) that can cleave the target (). The guide RNA uses crRNA and trans-activating crRNA (tracrRNA) (Wang et al. Citation2016), where the crRNA has a spacer part that is complementary to the target DNA sequence and has an important role in binding to the target DNA (Faure et al. Citation2019). The tracrRNA plays an important role in crRNA processing, Cas nuclease binding, and Cas-nuclease-mediated target cleavage (Wang et al. Citation2016). The repeat region of the crRNA hybridizes with the antirepeat region of the tracrRNA, which includes various bulges and a main bulge separating the upper stem from the lower stem adjacent to the distal part of the tracrRNA (Faure et al. Citation2019). The distal part of the tracrRNA folds into nexus stem loops and various stem loops that form the Rho-independent terminator (Faure et al. Citation2019). This sgRNA recognizes the target DNA and recruits the Cas9 protein to the protospacer-adjacent motif (PAM) sequences of the DNA (Brown et al. Citation2021).
Figure 1. CRISPR-Cas9 system generating DSBs to edit genes. The CRISPR-Cas9 system has sgRNA that recognizes the target and RuvC, HNH domains, which are the nuclease domains that cleave the DNA.
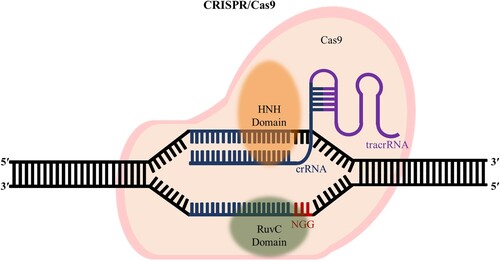
There are several variants of the Cas nuclease of the CRISPR-Cas system, among which Cas9 is the most widely used and studied type (Ma et al. Citation2014; Manghwar et al. Citation2019; Anzalone et al. Citation2020). The Cas9 nuclease has a catalytic domain that can cleave DNA, including the RuvC and HNH domains (Liu et al. Citation2021); these domains hydrolyze the double-stranded DNA and produce blunt ends or 1-bp staggered ends (Chang et al. Citation2017). DNA strands cut by the CRISPR-Cas9 system are repaired through a DNA repair pathway (Rath et al. Citation2015). The DNA repair mechanisms mainly result in nonhomologous end joining (NHEJ) or homology-directed repair (HDR) (Ran et al. Citation2013; Zhang Citation2021). NHEJ causes random insertion or deletion (indel) mutations, which cause frameshifts in the genes and disrupt gene functions (Wang et al. Citation2022). HDR occurs primarily in the cell-cycle S-G2 phase and is a pathway for DNA repair using endogenous or exogenous fragments of the homologous DNA as templates (Papadopoulos et al. Citation2016). NHEJ and HDR occur competitively with each other, and HDR occurs more infrequently than NHEJ (Karapurkar et al. Citation2021).
The CRISPR-Cas9 system is a versatile tool for genome editing. Unlike ZFN and TALEN, which are time consuming and expensive for engineering, the CRISPR-Cas9 system entails only changing the spacer part of the sgRNA, and this is an attractive advantage compared to conventional genome editing tools (Eid and Mahfouz Citation2016). Currently, the CRISPR-Cas9 system has greater advantages as a genome editing tool, but it also has limitations that must be overcome (Jo et al. Citation2015; Anzalone et al. Citation2020; Janik et al. Citation2020). The catalytic efficiency, limited PAM availability, low stability or durability of the Cas9-nuclease-RNA complex, large deletion, chromosome deletion, and high off-target effects are the main concerns and factors to consider for effective and accurate gene editing when using the CRISPR-Cas9 system (Chen and Gao Citation2014; Gilles and Averof Citation2014; Jo et al. Citation2015; Eid and Mahfouz Citation2016; Adikusuma et al. Citation2018; Anzalone et al. Citation2020; Janik et al. Citation2020; Tsuchida et al. Citation2023). These factors are obstacles to recognizing and cutting the desired DNA and may eventually reduce the editing efficiency of the CRISPR-Cas9 system. Many researchers have attempted to optimize this tool by engineering Cas9 and sgRNA to overcome their limitations while increasing the editing efficiency (Kleinstiver et al. Citation2015; Kleinstiver et al. Citation2016; Park et al. Citation2021; Dong et al. Citation2022; Riesenberg et al. Citation2022). Herein, we address considerations that affect the cleavage efficiency for optimizing the CRISPR-Cas9 system and provide directions for future improvements as well as applications to genetic diseases, therapeutics, and generation of animal models (Jin et al. Citation2021; Xie et al. Citation2022).
Cleavage mechanism
The CRISPR-Cas9 nuclease can cut the desired DNA and is mediated by the sgRNA. The complex of the Cas9 nuclease and sgRNA induces structural changes in the protein, during which the protein is converted to an active state (Feng et al. Citation2021). The Cas9-sgRNA complex has specificity for recognizing the target DNA through Cas9 interactions with PAM adjacent to the sgRNA target sites and binds to the target DNA through Watson–Crick base-pairing interactions between the target DNA and sgRNA spacer (Wang et al. Citation2016). The PAM recognition specificity of the Cas9 orthologs is determined by the base-dependent interactions (Wang et al. Citation2016). The Cas nuclease has different PAM sequences for each variant and can recognize a 5’-NGG-3’ (N = A or T or G or C) PAM sequence in the case of Streptococcus pyogenes Cas9 (SpCas9) (Wang et al. Citation2016; Anzalone et al. Citation2020).
When the Cas9-sgRNA complex finds a sequence recognized as PAM, the Cas nuclease separates the strands of the DNA double helix for bases upstream of the PAM site, and the nontarget strands with sequences consistent with PAM are combined with a PAM-interacting (PI) domain (Jinek et al. Citation2014). Thereafter, a RNA•DNA heteroduplex is formed between the sgRNA spacer and target DNA, which proceeds from the PAM-proximal to PAM-distal regions (Sternberg et al. Citation2014; Wang et al. Citation2016; Jiang and Doudna Citation2017; Anzalone et al. Citation2020). On the target DNA strand, an R-loop is formed by the sgRNA spacer, and the formation of the R-loop exposes the editing site, allowing other molecules to access this target site (Szczelkun et al. Citation2014; Jiang et al. Citation2016).
After formation of the R-loop, DNA–protein interactions are utilized to bring the nuclease domain close to the target DNA site in a form essential for DNA cleavage, and these structural changes allow the nuclease domain to be activated in the Cas9 nuclease (Anders et al. Citation2014; Jinek et al. Citation2014; Jiang et al. Citation2016). When the nuclease is activated, the RuvC and HNH nuclease domains hydrolyze the phosphodiester bonds of the DNA next to the 3–4 nucleotide base upstream of the PAM (Jiang et al. Citation2015). The nucleic sequences damaged by the nuclease for DNA cleavage are restored through the DNA repair mechanism (Moon et al. Citation2023). In the NHEJ pathway, the damaged DNA is immediately linked through a random indel (Molla et al. Citation2022); as a result, a different type of amino acid is produced than that existing one, changing or knocking out the gene function (Molla et al. Citation2022). When HDR occurs, the base is replicated using the donor DNA as a template (Xue and Greene Citation2021). In mammalian cells, the efficiency of HDR is highest when the template insertion site and double-strand break (DSB) are within 10 nucleotides of each other, with the efficiency of HDR decreasing as this distance increases (Elliott et al. Citation1998). Therefore, it is important to design the donor in consideration of these properties. HDR is capable of precise gene editing and is used to knock in the desired genes (Banan Citation2020; Xue and Greene Citation2021); these gene knock ins can be used to edit human DNA where mutations have occurred.
Factors affecting cleavage efficiency
The CRISPR-Cas9 system is easy to use as a genome editing tool, but has the limitations described previously. These limitations that affect editing efficiency are obstacles to research and development efforts concerned with genome editing, so these factors must be addressed for more effective gene therapy. In this work, we present the factors affecting cleavage efficiency among others related to the genome editing efficiency in SpCas9. Increasing the cleavage efficiency of the CRISPR-Cas9 system is a major concern when using this editing tool, and this efficiency is determined by a combination of factors.
GC contents of PAM proximal and distal regions
The GC contents of the PAM-proximal and PAM-distal regions is an indispensable factor correlated with cleavage efficiency. Here, the GC contents are the percentages of G and C in DNA or RNA molecules. The binding between G and C has three hydrogen bonds and is stronger (Yakovchuk et al. Citation2006) than that between A and T, which has two hydrogen bonds. Therefore, the GC bonds are more stable and have heat resistance than the AT bonds. The frequency of GC is preferable for RNA/DNA hybridization and is also important for initiation of the R-loop (Liu et al. Citation2016).
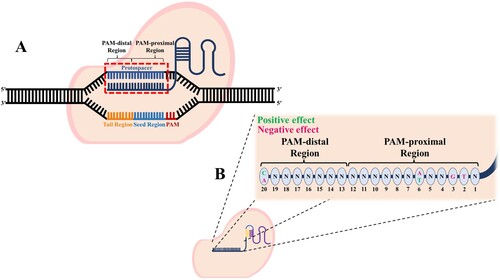
In the CRISPR-Cas9 system, the GC contents of the PAM-proximal and PAM-distal regions affect the cleavage efficiency ((a)). If these GC contents are exceedingly high or low, the Cas nuclease shows low cleavage effects, and most studies show higher efficiencies when the GC content is 40–60% (Liu et al. Citation2016; Malik et al. Citation2021). In addition, the presence of more than 56% GC content in the PAM-proximal seed region (1-12nt) further reduces the cleavage efficiency compared to the PAM-distal region (13-20nt) (Malik et al. Citation2021). Another study has shown that 51–100% GC content present in the PAM-distal region has 1.8-fold higher editing efficiency of sgRNA than 26–50% GC content (Labuhn et al. Citation2018). These studies suggest that the presence of 40–60% GC content allows higher cleavage efficiency in the CRISPR-Cas9 system. In addition, it is recommended that the GC content not exceed 56% in the PAM-proximal region and be more than 50% in the PAM-distal region. These results are important considerations for achieving higher SpCas9 cleavage efficiency.
Figure 2. The CRISPR-Cas9 system identifies the target by recognizing the PAM site present in the DNA. The 1st to 12th nucleotides of the sgRNA spacer belong to the PAM-proximal region, and the 13th to 20th nucleotides belong to the PAM-distal region (A). Depending on the type and location of the nucleotide present in the spacer of the sgRNA, the cleavage efficiency is affected (B).
Types of nucleotides according to sgRNA spacer sequence position
The type of nucleotide according to the sgRNA spacer sequence position affects the cleavage efficiency, and this factor must be adequately considered to avoid low gene-editing efficiency. sgRNA activates the endonuclease activity of the Cas nuclease and can be programmed to guide Cas nucleases to the desired targets (Liu et al. Citation2016). For such programming, the sgRNA has a part called the spacer, which allows targeting and binding to specific sequences of genes close to the PAM (Briner et al. Citation2014). To perform this role, the spacer comprises a base sequence complementary to the target DNA sequence, and the spacer length of SpCas9 is optimized to 20nt (Zhang et al. Citation2016). Nucleotides positioned at 8-12nt in the PAM-proximal region form the seed sequence for determining target specificity of the SpCas9 nuclease (Wu et al. Citation2014).
Liu et al. performed on-target efficiency tests according to the nucleotide type of the PAM-proximal region. If T or G is positioned in the 2-3nt PAM-proximal region, it tends to have a negative effect on cleavage (Liu et al. Citation2016). In addition, if the nucleotide at position 6 is A, it negatively affects cleavage efficiency. However, if the nucleotide at position 6 is T, it has a positive effect on cleavage efficiency. The PAM-distal region of sgRNA is believed to be relatively less important but has been found to influence cleavage efficiency. The nucleotide positioned at 20 in the spacer plays a role in inducing denaturation of the DNA double strand initiating R-loop formation; if A is at this position, then the cleavage efficiency is decreased by almost 50%. However, when nucleotide C is in this position, the cleavage efficiency is improved. (b) summarizes the key nucleotide positions in the spacer and their recommended bases.
Scaffold of sgRNA
The scaffold forms sgRNA with spacers and interacts with the SpCas9 nuclease. The scaffold consists of five parts: lower stem, upper stem, bulk, nexus, and hairpin ((a)). Scaffolds can also be engineered to increase the efficiency of the CRISPR-Cas system. The bulk and nexus parts are essential for sgRNA activity, so the lower stem, upper stem, and hairpin parts can be engineered (Dong et al. Citation2022).
Figure 3. Structure and components of the sgRNA (A). Secondary structures of the sgRNA that affect cleavage efficiency (B).
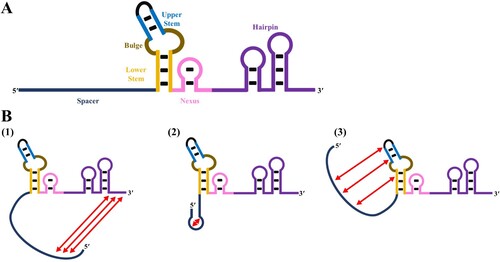
The continuity of thymidine in a scaffold affects the cleavage efficiency. The scaffold of sgRNA with thymine trinucleotide (TTT) or poly T sequences reduces the activity of sgRNA and cleavage efficiency. This is assumed to be because RNA pol III transcription termination occurs when there are three or more T sequences, and the poly T termination signal does not cause termination by itself but may cause backtracking and catalytic inactivation of RNA pol III (Nielsen et al. Citation2013; Xu et al. Citation2015; Zarate et al. Citation2022). Additionally, RNA aptamers with hairpin structures can be added to engineer the upper stem or hairpin of the scaffold to increase sgRNA activity (Dong et al. Citation2022). In this case, divalent modification of the aptamer in the scaffold increases sgRNA activity but trivalent modification would reduce such activity (Dong et al. Citation2022). Therefore, excessive hairpin formation may adversely affect the cleavage efficiency of CRISPR-Cas9 (Dong et al. Citation2022).
Stabilization of the hairpin structure in the sgRNA affects the ability to cleave SpCas9. In a previous study, the 34th U of tracrRNA was changed to A or the U-T bond to the stem loop was replaced with G-C to achieve a more stable structure, increasing the activity of SpCas9 by 40% (Scott et al. Citation2019). There is also another study on elongating the first hairpin 3’ of the nexus part to form a locked tracrRNA (t-lock) with an super stable loop, allowing a folding nucleation site of sgRNA and increasing the cleavage efficiency by preventing sgRNA misfolding; the efficiency was reported to have increased 7.4-fold (Riesenberg et al. Citation2022). This engineering is also closely related to the secondary structure of the sgRNA.
Secondary structure of sgRNA
The secondary structure of the sgRNA significantly impacts the cleavage efficiency of the CRISPR system and is determined by the scaffold design as well as spacer sequence. The secondary structure of RNA can be predicted by calculating various interactions. Because RNA has a single chain, it folds on itself and has the property of binding complementary base pairs with each other.
When sequences are folded in the seed region, their target cleavage efficiencies are reduced (Liu et al. Citation2016). Among them, misfolding of sgRNA with extremely low cleavage efficiencies are as follows. (1) The sgRNA spacer is complementary to the 3’-end of tracrRNA. (2) The sgRNA spacer forms its own hairpin structure. (3) The sgRNA spacer is complementary to the 3’-end of crRNA (Riesenberg et al. Citation2022) ((b)). Thus, sgRNA can be inactivated by the hairpin structure in the sgRNA spacer or interactions between the spacer and sgRNA backbone. In the sgRNA structure, misfolded sgRNA competes with active sgRNA to bind with the Cas nuclease, which is another factor that reduces cleavage efficiency.
As described above, the reduction in cleavage efficiency due to undesired interactions between the secondary structures of the sgRNA can be prevented by replacing the nucleotide sequence. When engineering the secondary structure of sgRNA to increase cleavage efficiency, a highly stable hairpin can be created in the secondary structure using genome-editing optimizes locked design (GOLD), such as t-lock, described in the scaffold of the sgRNA part or chemical modifications can be introduced with phosphorothioate bonds for the terminal nucleotides in the sgRNA. The chemical modifications using phosphorothioate bonds and 2’OMe modifications (t-2′OMe*) allow the nexus portion of tracrRNA to be preserved, optimizing the structure of tracrRNA for high cleavage efficiency (Riesenberg et al. Citation2022).
Thus, formation of highly stable hairpins provides efficient nucleation sites for RNA folding, which can prevent misfolding of sgRNA regardless of the spacer sequence. In addition, chemical modification of tracrRNA can increase the cleavage efficiency of the CRISPR-Cas9 system. These tracrRNA structural modifications can induce a secondary structure of sgRNA with higher cleavage efficiency for better editing efficiency of the CRISPR-Cas9 system.
Chromatin state
The chromatin state is a factor that affects the binding of the SpCas9 nuclease. Chromatin is present in eukaryotes and is a complex composed of DNA and histone proteins. Additionally, chromatin regulates gene expression and DNA replication. Histone proteins pack large amounts of DNA into dense forms that can fit inside a cell nucleus and keep them in the long form to prevent the strands from getting tangled. The lightly condensed part of the chromatin is called euchromatin, and the highly condensed part is called heterochromatin. Owing to these characteristics, euchromatin is an unpackaged, accessible, and transcriptionally facilitated chromatin segment but heterochromatin has packaged nucleosomes and inhibited transcription.
Among these two chromatin states, heterochromatin lowers the editing efficiency of the CRISPR-Cas9 system. The heterochromatin state is a nucleosome form enclosed in other proteins, which prevents the Cas nuclease from accessing and binding to the target DNA. The decrease in editing efficiency by the nucleosome is noticeable in vitro. In fact, the decreased editing efficiency by the nucleosome is lesser in vivo because cell nucleosomes are dynamic structures (Verkuijl and Rots Citation2019). Nucleosomal DNA shows ‘nucleosome breathing,’ in which temporary unwrapping occurs in vivo. At this time, both ends of the DNA that are wrapped by nucleosomes are exposed, and the SpCas9 nuclease can interact with the target. This nucleosome breathing is assumed to occur frequently. In actual experiments, there is almost no editing efficiency in the unexposed center of the DNA, but cleavages occur at the edges of the DNA or in areas with low nucleosome occupancy. In fact, there is research showing increased editing efficiency when using SpCas9 nuclease and chromatin-modulating peptides (CMPs), which can open the chromatin structure in consideration of these in vivo characteristics (Park et al. Citation2021). Thus, the editing efficiency of CRISPR-Cas9 varies depending on the chromatin state and occurrence position. In the euchromatin state, the cleavage effect is higher than that in the heterochromatin state. These results show that binding of the Cas nuclease is affected by the nucleosomes. Additionally, it suggests that there is a need to consider the chromatin state in vivo.
Others
Position of the target DNA site: The cleavage efficiency varies depending on the DNA sequence location to be corrected in the gene. If the target DNA sequence is located at the transcription start site of the promoter, the efficiency is higher than that of a sequence located in the intergenic region (Liu et al. Citation2016). This is caused by differences in the chromatin accessibility of the genomes (Liu et al. Citation2016).
Transcriptional state of the gene: The transcriptional state of a gene is correlated with the cleavage efficiency of the CRISPR-Cas system. Studies have shown that the Cas9:sgRNA complex, which forms a base pair with the DNA strand that acts as a template for RNA polymerase, increases the incidence of mutagenesis (Clarke et al. Citation2018). This indicates that RNA polymerase, the main transcription enzyme, replaces Cas nuclease and cuts the other DNA molecules (Clarke et al. Citation2018).
Cas nuclease domain: The Cas nuclease domain is related to the cleavage efficiency, and the cleavage efficiency of the CRISPR-Cas9 system can be increased by replacing the amino acids to cause mutations (Kleinstiver et al. Citation2015). However, it is important to optimize the combination because engineering all multiple domains can reduce the efficiency.
Conclusion
CRISPR-Cas9 is a widely used tool in gene editing, but there are still many issues that must be solved regarding cleavage efficiency. Therefore, it is important to understand and engineer the CRISPR-Cas9 system accordingly. Furthermore, precise gene editing technologies like base editor and prime editor, which are based on the CRISPR-Cas9 system, are being utilized in biological and medical research. Therefore, it is essential to understand the factors affecting the cleavage efficiency of the CRISPR-Cas9 system as the ability to cut the target DNA is directly linked to the efficacy of this tool.
GC contents in the PAM-proximal and PAM-distal regions is another factor affecting the cleavage efficiency of SpCas9. The cleavage efficiency decreases for GC content lower than 56% in the PAM-proximal region and increases when 51–100% GC content is present in the PAM-distal region.
The sgRNA is composed of a spacer and a scaffold, and several factors affect the cleavage efficiency of SpCas9. The type of nucleotide according to the sgRNA spacer sequence position affects the cleavage efficiency. If nucleotides T, G, and A are present at positions 2, 3, and 6, which are PAM-proximal regions, the cleavage efficiency is lower. In addition, when nucleotide A is in position 20, which is the PAM-distal region, the cleavage efficiency decreases. If the scaffold has a thymine trinucleotide or poly T sequence, the activity of sgRNA is reduced to negatively affect the cleavage efficiency. In addition, if the spacer sequence forms a hairpin structure with itself or interacts with the sgRNA backbone in the secondary structure of the sgRNA, it negatively affects the cleavage efficiency. Therefore, many things should be considered when designing sgRNA, and it is important to modify areas that can be improved.
If the location of the target DNA sequence is in the heterochromatin state, it negatively affects the cleavage efficiency. This effect by the nucleosome occurs more strongly in vitro than in vivo. When the target DNA is present at the transcription start site, it shows positive cleavage efficiency more than at the intergenic region; further, the transcription state of the gene affects the cleavage efficiency. Therefore, the CRISPR-Cas system can be affected by the location and condition of the target gene, so these factors should be considered.
Currently, the CRISPR-Cas system is developing rapidly, but it still faces obstacles such as catalytic efficiency, sequence specificity, delivery efficiency, PAM availability, stability, durability of the RNA-Cas9 nuclease complex, and off-target effects. After carefully considering all relevant factors as discussed above, we would like to utilize the optimized CRISPR-Cas9 system to obtain clear research results. Therefore, when conducting experiments, it is necessary to have a clear understanding of the physiological rationale for the target location of the gene and design of the sgRNA. This will significantly aid in researching unknown pathogenesis, discovering gene roles, and developing disease therapies.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, Beard MR, Hughes J, Pomerantz RT, Thomas PQ. 2018. Large deletions induced by Cas9 cleavage. Nature. 560(7717):E8–E9.
- Anders C, Niewoehner O, Duerst A, Jinek M. 2014. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 513(7519):569–573.
- Anzalone AV, Koblan LW, Liu DR. 2020. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 38(7):824–844.
- Banan M. 2020. Recent advances in CRISPR/Cas9-mediated knock-ins in mammalian cells. J Biotechnol. 308:1–9.
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 315(5819):1709–1712.
- Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, Haurwitz RE, Beisel CL, May AP, Barrangou R. 2014. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 56(2):333–339.
- Brown W, Zhou W, Deiters A. 2021. Regulating CRISPR/Cas9 function through conditional guide RNA control. Chembiochem. 22(1):63–72.
- Chang HHY, Pannunzio NR, Adachi N, Lieber MR. 2017. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 18(8):495–506.
- Chen K, Gao C. 2014. Targeted genome modification technologies and their applications in crop improvements. Plant Cell Rep. 33(4):575–583.
- Clarke R, Heler R, MacDougall MS, Yeo NC, Chavez A, Regan M, Hanakahi L, Church GM, Marraffini LA, Merrill BJ. 2018. Enhanced bacterial immunity and mammalian genome editing via RNA-polymerase-mediated dislodging of Cas9 from double-strand DNA breaks. Mol Cell. 71(1):42–55e48.
- Dong C, Gou Y, Lian J. 2022. SgRNA engineering for improved genome editing and expanded functional assays. Curr Opin Biotechnol. 75:102697.
- Doudna JA. 2020. The promise and challenge of therapeutic genome editing. Nature. 578(7794):229–236.
- Eid A, Mahfouz MM. 2016. Genome editing: the road of CRISPR/Cas9 from bench to clinic. Exp Mol Med. 48(10):e265.
- Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. 1998. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 18(1):93–101.
- Faure G, Shmakov SA, Makarova KS, Wolf YI, Crawley AB, Barrangou R, Koonin EV. 2019. Comparative genomics and evolution of trans-activating RNAs in class 2 CRISPR-Cas systems. RNA Biol. 16(4):435–448.
- Feng W, Dai Y, Mou L, Cooper DK, Shi D, Cai Z. 2015. The potential of the combination of CRISPR/Cas9 and pluripotent stem cells to provide human organs from chimaeric pigs. Int J Mol Sci. 16(3):6545–6556.
- Feng Y, Liu S, Chen R, Xie A. 2021. Target binding and residence: a new determinant of DNA double-strand break repair pathway choice in CRISPR/Cas9 genome editing. J Zhejiang Univ Sci B. 22(1):73–86.
- Gilles AF, Averof M. 2014. Functional genetics for all: engineered nucleases, CRISPR and the gene editing revolution. Evodevo. 5:43.
- Ho BX, Loh SJH, Chan WK, Soh BS. 2018. In vivo genome editing as a therapeutic approach. Int J Mol Sci. 19(9.
- Hryhorowicz M, Lipinski D, Zeyland J, Slomski R. 2017. CRISPR/cas9 immune system as a tool for genome engineering. Arch Immunol Ther Exp (Warsz. 65(3):233–240.
- Janik E, Niemcewicz M, Ceremuga M, Krzowski L, Saluk-Bijak J, Bijak M. 2020. Various aspects of a gene editing system-CRISPR-Cas9. Int J Mol Sci. 21(24).
- Jiang F, Doudna JA. 2017. CRISPR-Cas9 Structures and mechanisms. Annu Rev Biophys. 46:505–529.
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. 2016. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 351(6275):867–871.
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. 2015. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 348(6242):1477–1481.
- Jin S, Na H, Jeon H, Park J, Choe CP. 2021. Egfl6 expression in the pharyngeal pouch is dispensable for craniofacial development. Animal Cells Syst (Seoul). 25(5):255–263.
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. 2014. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 343(6176):1247997.
- Jo YI, Suresh B, Kim H, Ramakrishna S. 2015. CRISPR/cas9 system as an innovative genetic engineering tool: enhancements in sequence specificity and delivery methods. Biochim Biophys Acta. 1856(2):234–243.
- Karapurkar JK, Antao AM, Kim K-S, Ramakrishna S. 2021. Chapter nine - CRISPR-Cas9 based genome editing for defective gene correction in humans and other mammals. In: Singh V, editor. Progress in molecular biology and translational science. Academic Press; p. 185–229.
- Kim S-H, Yang S, Yang E, Kang M, Joo J-Y. 2023. Potent of strategic approaches for tauopathies ranging from single-cell transcriptome to microbiome. Animal Cells Syst (Seoul). 27(1):378–393.
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. 2016. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 529(7587):490–495.
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, et al. 2015. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 523(7561):481–485.
- Labuhn M, Adams FF, Ng M, Knoess S, Schambach A, Charpentier EM, Schwarzer A, Mateo JL, Klusmann JH, Heckl D. 2018. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR-Cas9 applications. Nucleic Acids Res. 46(3):1375–1385.
- Liu R, Liang L, Freed EF, Gill RT. 2021. Directed evolution of CRISPR/Cas systems for precise gene editing. Trends Biotechnol. 39(3):262–273.
- Liu X, Homma A, Sayadi J, Yang S, Ohashi J, Takumi T. 2016. Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci Rep. 6:19675.
- Ma Y, Zhang L, Huang X. 2014. Genome modification by CRISPR/Cas9. FEBS J. 281(23):5186–5193.
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science. 339(6121):823–826.
- Malik A, Gul A, Munir F, Amir R, Alipour H, Babar MM, Bakhtiar SM, Paracha RZ, Khalid Z, Hayat MQ. 2021. Evaluating the cleavage efficacy of CRISPR-Cas9 sgRNAs targeting ineffective regions of arabidopsis thaliana genome. PeerJ. 9:e11409.
- Manghwar H, Lindsey K, Zhang X, Jin S. 2019. CRISPR/cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 24(12):1102–1125.
- Molla KA, Shih J, Wheatley MS, Yang Y. 2022. Predictable NHEJ insertion and assessment of HDR editing strategies in plants. Front Genome Ed. 4:825236.
- Moon J, Kitty I, Renata K, Qin S, Zhao F, Kim W. 2023. DNA damage and its role in cancer therapeutics. Int J Mol Sci. 24(5).
- Nielsen S, Yuzenkova Y, Zenkin N. 2013. Mechanism of eukaryotic RNA polymerase III transcription termination. Science. 340(6140):1577–1580.
- Papadopoulos KI, Wattanaarsakit P, Prasongchean W, Narain R. 2016. 10 - Gene therapies in clinical trials. In: Narain R, editor. Polymers and nanomaterials for gene therapy. Woodhead Publishing; p. 231–256.
- Park SJ, Jeong TY, Shin SK, Yoon DE, Lim SY, Kim SP, Choi J, Lee H, Hong JI, Ahn J, et al. 2021. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor. Genome Biol. 22(1):170.
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 8(11):2281–2308.
- Rath D, Amlinger L, Rath A, Lundgren M. 2015. The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie. 117:119–128.
- Riesenberg S, Helmbrecht N, Kanis P, Maricic T, Paabo S. 2022. Improved gRNA secondary structures allow editing of target sites resistant to CRISPR-Cas9 cleavage. Nat Commun. 13(1):489.
- Saifaldeen M, Al-Ansari DE, Ramotar D, Aouida M. 2020. CRISPR foki dead Cas9 system: principles and applications in genome engineering. Cells. 9(11).
- Scott T, Urak R, Soemardy C, Morris KV. 2019. Improved Cas9 activity by specific modifications of the tracrRNA. Sci Rep. 9(1):16104.
- Seah I, Goh D, Chan HW, Su X. 2022. Developing Non-human primate models of inherited retinal diseases. Genes (Basel). 13(2.
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. 2014. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 507(7490):62–67.
- Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. 2014. Direct observation of R-loop formation by single RNA-guided Cas9 and cascade effector complexes. Proc Natl Acad Sci U S A. 111(27):9798–9803.
- Tsuchida CA, Brandes N, Bueno R, Trinidad M, Mazumder T, Yu B, Hwang B, Chang C, Liu J, Sun Y, et al. 2023. Mitigation of chromosome loss in clinical CRISPR-Cas9-engineered T cells. Cell. 186(21):4567–4582e4520.
- Verkuijl SA, Rots MG. 2019. The influence of eukaryotic chromatin state on CRISPR-Cas9 editing efficiencies. Curr Opin Biotechnol. 55:68–73.
- Wang H, La Russa M, Qi LS. 2016. CRISPR/cas9 in genome editing and beyond. Annu Rev Biochem. 85:227–264.
- Wang SW, Gao C, Zheng YM, Yi L, Lu JC, Huang XY, Cai JB, Zhang PF, Cui YH, Ke AW. 2022. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol Cancer. 21(1):57.
- Wu X, Kriz AJ, Sharp PA. 2014. Target specificity of the CRISPR-Cas9 system. Quant Biol. 2(2):59–70.
- Xie Y, Hu B, Gao Y, Tang Y, Chen G, Shen J, Jiang Z, Jiang H, Han J, Yan J, et al. 2022. Yap signalling regulates ductular reactions in mice with CRISPR/Cas9-induced glycogen storage disease type Ia. Anim Cells Syst (Seoul. 26(6):300–309.
- Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, Wu D, Cong L, Zhang F, Liu JS, et al. 2015. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25(8):1147–1157.
- Xue C, Greene EC. 2021. DNA repair pathway choices in CRISPR-Cas9-mediated genome editing. Trends Genet. 37(7):639–656.
- Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. 2006. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 34(2):564–574.
- Zarate OA, Yang Y, Wang X, Wang JP. 2022. BoostMEC: predicting CRISPR-Cas9 cleavage efficiency through boosting models. BMC Bioinformatics. 23(1):446.
- Zhang B. 2021. CRISPR/cas gene therapy. J Cell Physiol. 236(4):2459–2481.
- Zhang F, Wen Y, Guo X. 2014. CRISPR/cas9 for genome editing: progress, implications and challenges. Hum Mol Genet. 23(R1):R40–R46.
- Zhang JP, Li XL, Neises A, Chen W, Hu LP, Ji GZ, Yu JY, Xu J, Yuan WP, Cheng T, et al. 2016. Different effects of sgRNA length on CRISPR-mediated gene knockout efficiency. Sci Rep. 6:28566.
